ABSTRACT
Plants have evolved a network of complex signaling pathways that allow them to cope with the fluctuations of internal and external environmental cues. GIGANTEA (GI), a well-known, highly conserved plant nuclear protein, has been shown to regulate multiple biological functions in plants such as circadian rhythm, light signaling, cold tolerance, hormone signaling, and photoperiodic flowering. Recently, the role of GI in disease tolerance against different pathogens has come to light; however, a detailed mechanism to understand the role of GI in pathogen defense remains largely unexplained. Here, we report that GIGANTEA is upregulated upon infection with a virulent oomycete pathogen, Hyaloperonospora arabidopsidis (Hpa), in Arabidopsis thaliana accession Col-0. To investigate the role of GI in Arabidopsis defense, we examined the pathogen infection phenotype of gi mutant plants and found that gi-100 mutant was highly susceptible to Hpa Noco2 infection. Notably, the quantitative real-time PCR showed that PHYTOALEXIN DEFICIENT4 (PAD4) and several PAD4-regulated downstream genes were downregulated upon Noco2 infection in gi-100 mutant as compared to Col-0 plants. Furthermore, the chromatin immunoprecipitation results show that GI can directly bind to the intronic region of the PAD4 gene, which might explain the mechanism of GI function in regulating disease resistance in plants. Taken together, our results suggest that GI expression is induced upon Hpa pathogen infection and GI can regulate the expression of PAD4 to promote resistance against the oomycete pathogen Hyaloperonospora arabidopsidis in Arabidopsis thaliana.
1. Introduction
Plants are under constant attack from various pathogens. To maintain their propagation, alterations in flowering time have been observed during plant-pathogen interactions. For instance, bacterial pathogens like Pseudomonas syringae and Xanthomonas campestris and an oomycete, Peronospora parasitica infection, cause earlier flowering than uninfected plants in susceptible Arabidopsis plants.Citation1 In contrast, in Brassica rapa, herbivory by the invasive Spodoptera littoralis enhances glucobrassicanapin, leading to delayed flowering.Citation2 To ensure their survival and reproduction upon pathogen infection, plants induce defense pathways mainly by salicylic acid (SA) and jasmonic acid (JA) signaling pathways. Salicylic acid (SA) signaling-associated mutants such as SA INDUCTION-DEFICIENT2 (sid2), eds5, and nahG suppress flowering by elevating the expression of floral repressor FLOWERING LOCUS C (FLC) gene.Citation3 On the other hand, SA regulatory genes like HOPW1-1-INTERACTING3 (WIN3) and NONEXPRESSOR OF PR GENES1 (NPR1) synergistically affect flowering time by altering the expression of flowering regulatory genes, FLC and FLOWERING LOCUS T (FT).Citation4 SUMO E3 ligase SIZ1, PLANT U-BOX 13 (PUB13), and MYB30, regulators of SA-mediated defense, have also been documented to regulate flowering time under biotic stress.Citation5–7 Like the SA pathway, another defense signaling pathway, jasmonate (JA) signaling, also regulates both negative and positive Fusarium oxysporum (F. oxysporum) resistance in Arabidopsis thaliana (At). Upon infection, bHLH transcription factors that suppress JA-mediated defense response promote flowering, while the JA receptor mutant coi1 shows extreme resistance to F. oxysporum and causes early flowering.Citation8 Additionally, ethylene (ET)-insensitive mutants cause a delay in flowering time,Citation9 whereas HDA6 and HDA19, the histone deacetylases that regulate JA and ET-mediated defense responses, have been shown to enhance the transition to flowering.Citation10–12
In plants, the successful transition from vegetative to reproductive growth is a multifaceted trait regulated by a complex network of different genetic pathways, including the vernalization, photoperiod, autonomous, and gibberellin (GA) pathways.Citation13 Additionally, several experiments have demonstrated the crucial role of flowering-associated genes in the defense signaling pathway. Regulators of the autonomous pathway, including FPA (an RNA binding protein) and FLOWERING LOCUS D (FLD), promote susceptibility to the bacterial pathogen Pseudomonas syringaeCitation14–16 while LEAFY, the floral meristem identity gene, represses key regulators of basal immunity.Citation17 The phytohormone GA, which promotes the flowering in Arabidopsis,Citation18 has also been shown to increase resistance to the bacterial pathogen Pseudomonas syringae and confers susceptibility to the necrotrophic fungus Alternaria brassica.Citation19
GIGANTEA (GI) is one such photoperiodic pathway regulator, and previous studies have shown that GI promotes susceptibility to F. oxysporum.Citation20 In Arabidopsis, mutations in GIGANTEA (gi-1 and gi-2) lead to increased resistance to the F. oxysporum infection compared to wild-type plants, but the detailed mechanism is unknown.Citation20 Throughout numerous stages of plant development, GI plays a role in diverse physiological processes such as flowering time regulation, circadian rhythm, light signaling, starch accumulation, miRNA processing, chlorophyll accumulation, and transpiration.Citation21–24 GI has also been shown to regulate abiotic stresses like cold, salt, drought, and oxidative stresses in plants, but its role in pathogen infection remains to be elucidated.Citation24–29
Pathogen attack in plants is recognized by innate immune receptors located at the host cell surface or in the cytoplasm. These receptors bind to the conserved microbial molecules (pathogen‐associated molecular patterns, PAMPs) and induce PAMP‐triggered immunity (PTI), which provides early protection from pathogens.Citation30 In the course of host-pathogen co-evolution, PTI becomes suppressed by pathogen-derived virulence factors known as effectors to promote infection in the host cell.Citation31 These pathogen effectors are primarily sensed by intracellular nucleotide‐binding/leucine‐rich‐repeat (NLR) receptors, which trigger effector‐triggered immunity (ETI). PTI and ETI signaling are associated and increase defense pathways, including mobilization of Ca2+‐dependent protein kinase, production of reactive oxygen species (ROS), activation of mitogen‐activated protein kinase (MAPK) signaling cascades, transcriptional reprogramming, and generation of salicylic acid (SA).Citation32,Citation33 SA contributes to PTI and ETI, and its biosynthesis upon pathogen recognition is mainly regulated by the SA biosynthetic enzyme gene, ICS1.Citation34–37 In basal immunity, ENHANCED DISEASE SUSCEPTIBILITY 1 (EDS1) directly binds to PHYTOALEXIN DEFICIENT4 (PAD4) and upregulates ISOCHORISMATE SYNTHASE 1 (ICS1) expression leading to SA accumulation. The downstream events of SA-mediated signaling are executed by the nucleocytoplasmic regulator NONEXPRESSOR OF PR GENES1 (NPR1), a transcriptional co‐activator of SA‐dependent immunity pathways.Citation38,Citation39
Hyaloperonospora arabidopsidis (Hpa) is an obligate biotrophic pathogen of the model plant Arabidopsis thaliana and has been extensively used to study host/pathogen co-evolution. Upon infection, it causes downy mildew disease in Arabidopsis.Citation40 EDS1-PAD4 module activates two branches of immune responses, namely, (i) SA-dependent signaling in which pathogen-induced SA accumulation via ISOCHORISMATE SYNTHASE 1 (ICS1) provides resistance and (ii) SA-independent signaling, which provides resistance via FLAVIN-DEPENDENT MONOOXYGENASE 1 (FMO1).Citation41–43 Constitutively overexpressing FMO1 in Arabidopsis revealed enhanced resistance to Hpa pathogens, whereas loss-of-function fmo1 mutants were compromised in resistance to Hpa.Citation41,Citation43,Citation44 FMO is a pipecolate N-hydroxylase and catalyzes the biochemical conversion of pipecolic acid to N-hydroxypipecolic acid (NHP). NHP systemically accumulates in the plant foliage and induces systemic acquired resistance to pathogen infection.Citation45,Citation46
Here, we have explored the role of GIGANTEA (GI) in resistance to biotic stress. Our results indicate that the infection with virulent Noco2 strain of Hpa in Col-0 results in increased expression of GI. Further, Noco2 infections also lead to increased expression of PAD4 and its downstream defense signaling genes like ICS1, PR1, FMO1, and PBS3. To confirm our hypothesis that GI mediates the activation of defense signaling pathways, we tested the resistance response of gi-100 mutant lines against Hpa Noco2 infection. In the absence of GI, the pathogen-induced SA-dependent and SA-independent signaling pathways were suppressed leading to increased disease susceptibility. Further, chromatin immunoprecipitation of GI followed by quantitative real-time PCR of PAD4 shows that GI binds to the intronic region of the PAD4 gene. On the other hand, expression analysis of PAD4 in gi-100 mutant indicates that GIGANTEA positively regulates PAD4 expression after Noco2 infection. Therefore, our study provides a novel insight into the role of GIGANTEA that likely involves PAD4-mediated plant defense responses upon pathogen perception in Arabidopsis.
2. Material and methods
2.1. Plant lines and growth conditions
Arabidopsis thaliana lines used in this study were Columbia (Col-0), eds1-2, Ws-2, gi-2 and gi-100. Plants were grown on potting soil in a growth chamber at 22°C with 8 h of light (100 μE/m2/s) and a relative humidity of 75%.
2.2. Growth and infection of downy mildew
Arabidopsis plants were grown at 22°C with ∼75% relative humidity (RH) and an 8 h light period. Virulent Noco2 strain of Hpa was used for infections. For infection, conidiospore suspensions (5 × 104 conidiospores ml−1) were sprayed on 2-week-old Arabidopsis seedlings grown on potting soil. Plants were allowed to dry for 1 h and kept at 100% RH for 24 h in a growth chamber with 8 h light at 22°C. Plants were then moved to ∼75% RH for infection to progress, where Hpa growth on Arabidopsis leaves was scored 6 d post-inoculation by counting spore numbers using a hemocytometer.Citation47,Citation48
2.3. Microscopy
The infection of Noco2 pathogen in the leaves was visualized by trypan blue staining. Infected leaves were collected in a 1.5-ml centrifuge tube. A 1:1:1:1 volume of lactic acid/glycerol/phenol/H2O with trypan blue (1 mg/ml) was added. The tubes were placed in a boiling water bath for 1 min. This was followed by the destaining of leaves in chloral hydrate. The tubes were placed in a speed-vacuum infiltrator for 1 min to remove air bubbles from the leaves.Citation49 Hpa growth was detected by differential interference contrast microscopy.
2.4. RNA extraction and real-time PCR
For RNA isolation, ~50 mg of leaf tissue was harvested in liquid nitrogen and immediately frozen at −80°C. Leaf samples were processed using a Qiagen Plant RNeasy Plant Mini Kit (Cat #74104) according to the manufacturer’s protocol. RNA was quantified using a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific, USA), and up to 1 μg of RNA was treated with DNase I (TURBO DNA-free kit, Invitrogen) to remove genomic DNA. The cDNA was prepared from 1 µg of RNA of each sample using iScriptTM Reverse Transcription Super-mix (Cat #1708840, Bio-rad, USA). The quantitative RT-PCR (qRT-PCR) was performed using the CFX384TouchTM Real-time detection system (Bio-Rad, USA). Primers used in qRT-PCR were designed using the Primer Quest tool. All reactions were carried out in Hard-shell 384-well PCR plates (supplied by Bio-Rad, Cat #HSP3805), with a reaction volume of 10 µl per well. The PCR mix and thermocycler program for the qRT-PCR were similar to those in Ó’Maoiléidigh et al. 2021.Citation50 Transcript levels were normalized with the housekeeping gene, ACTIN1. Each qRT-PCR reaction was performed on three biological replicates, and all data were presented as mean ± SEM. Primers used in the qRT-PCR are listed in Supplementary Table 1.
2.5. Chromatin immunoprecipitation (ChIP) followed by qPCR
ChIP experiment was performed as published previouslyCitation51 with minor modifications. Three independent biological replicates for Col-0 and 35S::HA:GI were generated. For each sample, 1 g of 10-d-old seedlings were harvested and cross-linked twice by infiltration with 1% formaldehyde under a vacuum for 10 minutes. The material was collected from plants grown in LDs at 22°C for 10 days (16 hlight and 8 h dark). Nuclei were disrupted by sonication four times for 5 min in cycles of 15 sec “on”/15 sec “off,” with a 1 min incubation between each sonication treatment using a water bath Bioruptor (Diagenode). Chromatin immunoprecipitation was performed by using HA antibodies (Abcam, ab9110) and Protein A agarose beads (11719408001, Roche). This was followed by DNA precipitation using 3 mM sodium acetate and absolute ethanol. DNA pellets were washed with 70% ethanol, air-dried, and resuspended in water before qRT-PCR. A small aliquot of untreated sonicated chromatin was used as the total input DNA. ChIP-qPCR was analyzed, and the relative enrichment of the IP/Input was normalized to ACTIN8. Primers were designed from the exon 2 region, intron 1 region and transcription start site (TSS) of the PAD4 gene, and qRT-PCR was performed in three biological replicates. Primers used in the ChIP-qPCR are listed in Supplementary Table 2.
3. Results
3.1. Loss-of-function mutant, gi-100 shows an enhanced susceptibility to Hpa Noco2 infection
To explore the function of GI in activating defense during plant-biotrophic pathogen interaction, we first evaluated the enrichment of GI expression upon pathogen infection using downy mildew pathogen Hyaloperonospora arabidopsidis (Hpa). The most common symptom for this obligate biotrophic pathogen is the aerial conidiophores. Here, we have used virulent Noco2 strain of Hpa for infection. To investigate the possible alteration in GI expression, transcript abundance was analyzed by qRT-PCR in the Col-0 leaf sample after 48 h of infection with Noco2 strain. An approximately >1.5-fold increase in the level of GI transcripts was observed compared to its uninfected control (). This result revealed that GI is a pathogen-inducible gene (Hpa Noco2).
Figure 1. Involvement of GIGANTEA in regulating Noco2 infection in Arabidopsis. (a) Relative abundance of GI transcripts in uninfected (0 hpi) and infected (48 hpi) Col-0 plants using qRT-PCR. Transcripts levels were normalized with the internal control gene, ACTIN1. Three biological replicates were used for the experiments. Data represented as Mean ± SE. Student’s paired T-test was used to evaluate the significant differences as *p < .05. ** p < .01, *** p < .001 compared between uninfected and infected Col-0 plants. (b) Graph showing conidiospores count (*104/g) after 6 d of Noco2 infection in Columbia (Col-0), eds1-2, Ws-2 and gi-100. Plants were grown on potting soil in a growth chamber at 22°C with 8 h of light and relative humidity of 75%. For Noco2 infections, 2-weeks-old Arabidopsis seedlings of the indicated genotype were infected with conidiospores suspension (5 × 104 conidiospores/ml) and oomycete sporulation was measured using hemocytometer at 6 dpi. Three biological replicates were used for the experiments. To test for significance among the dataset, a one-way analysis of variance (ANOVA) followed by Sidak’s multiple comparisons test was performed using GraphPad prism software at *p < .05. ** p < .01, *** p < .001.
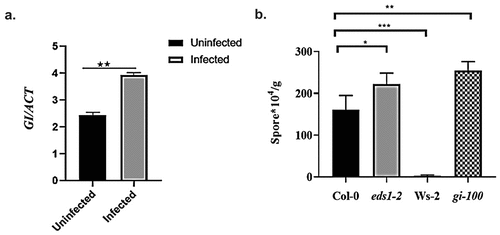
To further confirm the role of GI as a resistance factor during plant-biotrophic pathogen interactions, the well-characterized GI T-DNA insertional null mutant gi-100Citation52 and the wild-type Col-0 plants were challenged with Hpa Noco2. It is known that Ws-2 plants show resistance to Noco2 strain due to the presence of functional RPP1 receptor proteins and hence were used as a negative control for this experiment.Citation53 EDS1 acts as a positive regulator of plant defense signaling, while its absence makes the plant more susceptible to pathogens, and hence, eds1-2 mutant plants were used as a positive control.Citation42 The growth of Hpa is estimated in two ways: counting conidiosporesCitation54 or counting sporangiophores after trypan blue staining.Citation55 Plants were infected with the Hpa, Noco2 strain (104 conidiospores per milliliter), which is virulent on Col-0, and the appearance of conidiospores was scored 6 d later using a hemocytometer. We observed that gi-100 mutant plants were highly susceptible to Noco2 infection than the wild-type Col-0 control (). The infection was further evaluated by trypan blue staining of the conidiospores, which confirmed the higher colonization of conidiospores in gi-100 mutant compared to Col-0 (Supplementary Figure S1). Hence, the gi-100 mutant demonstrated enhanced susceptibility to the Hpa Noco2 infection in comparison to Col-0 plants. Thus, our results confirmed the positive role of GI in conferring pathogen resistance during plant-pathogen interaction. We have also checked the Noco2 fungal infection severity using another well-characterized gi mutant, gi-2.Citation56 Similar to gi-100, gi-2 also cause increased susceptibility to Hpa infection; however, gi-2 mutant has a weaker infection phenotype than gi-100. The trend of spore count after Noco2 infection in gi-2 was in the same direction as gi-100 when compared to Col-0. But, unlike gi-100 mutant plants, gi-2 mutant could not surpass the eds1-2 mutant spore count number after infection (Supplementary Figure S3). This difference in the severity of phenotypes could be because both the gi mutants, gi-2 and gi-100, have been shown to form different sizes of transcripts after T-DNA insertion.Citation24
3.2. GI binds to the intronic region of PAD4
We further explored the detailed mechanism by which GI confers resistance against Hpa pathogen infection. In a study by Nohales et al. 2019, putative GI binding target sites were identified by chromatin immunoprecipitation sequencing (ChIP-seq) using a pull-down of GI protein from the Arabidopsis chromatin preparation. To know the potential immunity pathway targets of GI, we explored the ChIP-seq data available onlineCitation57 and identified PHYTOALEXIN DEFICIENT4 (PAD4) as one of the putative targets of GI (). Several studies show that PAD4 along with the EDS1 regulates plant basal immunity against virulent biotrophic pathogens.Citation42 It is documented that the regulatory sequences of the PAD4 gene are present in the promoter as well as the intronic region of the gene and these regulatory sequences of PAD4 contain a G‐box element.Citation58 G-box-like elements are considered as potential binding sites of GI.Citation57 Based on the above pieces of evidence, we hypothesized that GI functions upstream of PAD4 and might regulate PAD4 by binding to the G-box element present in the intronic regulatory sequences.
Figure 2. Chromatin immunoprecipitation assay to show binding of GI to PAD4. (a) Visualization of GI ChIP-seq data in the genomic region encompassing the PAD4 locus. Peaks represent the sequence enrichment of PAD4 in Col-0 (red) and GI-overexpressed (blue) lines. (b) Schematic picture showing the regions of the PAD4 gene from which the primers were designed. (c) Graph showing enrichment of PAD4 gene after qRT-PCR using chromatin isolated from plants expressing HA-tagged GI (35S::HA:GI) and wild type (Col-0). Immunoprecipitation was done using HA-beads followed by qPCR. Gene expression was normalized to the internal housekeeping ACTIN8 gene. In this experiment, Heat shock protein (HSF) gene was used as a negative experimental control to show that binding of GI to PAD4 is specific and it is not binding to any random gene sequences. (d) qRT-PCR analysis of different regions of PAD4 gene using HA pull-down chromatin samples from Col-0 and 35S::HA:GI. Primers were designed from the exon 2 region, intron 1 region, and transcription start site (TSS) of the PAD4 gene. Three biological replicates were used for the experiment. To test for significance among the dataset, a two-way analysis of variance (ANOVA) followed by Tukey’s multiple comparisons test was performed using GraphPad prism software at *p < .05. ** p < .01, *** p < .001.
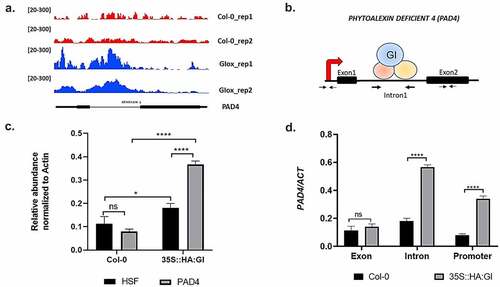
In order to test our hypothesis, we performed chromatin immunoprecipitation followed by quantitative RT-PCR (ChIP-qPCR). Chromatin was isolated from plants expressing HA-tagged GI protein (35S::HA:GI) and control Col-0 plants, followed by chromatin fragmentation, and then immunoprecipitation was performed using an anti-HA antibody. Here, Col-0 plants were used as a negative control to ensure the specific binding of proteins with anti-HA antibody only. Pull-down was further followed by qPCR from the purified DNA fragments using primers specific to the intronic sequences of the PAD4 gene. Exon-specific primers and Transcription start site (TSS)-specific primers of the PAD4 gene were used as a control to show the specific binding of GI to PAD4 intron in comparison to promoter and exon regions. Our ChIP‐qPCR result detected the highest enrichment of PAD4 in the intronic region followed by the promoter region of the PAD4 while binding to the exon region was not significant. In this experiment, heat shock protein (HSF) gene was used as a negative experimental control to show that binding of GI to PAD4 is specific and it is not binding to any random gene sequences. The samples were normalized with ACTIN8 as an internal control. Thus, our results confirmed that GI specifically binds to the intronic region of PAD4 (.
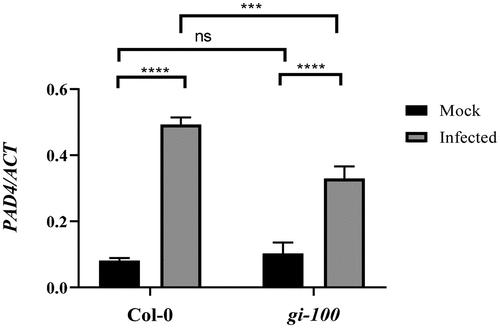
After confirming that GI can physically associate at the regulatory sequences of the PAD4 gene, we wanted to explore whether changes in GI expression can regulate PAD4 gene expression during Hpa infection. The amount of PAD4 expression was measured using quantitative RT-PCR in Col-0 and gi-100 mutant plants, 48 h after inoculation with the virulent Noco2 pathogen. As expected, pathogen‐infected WT plants displayed much higher levels of PAD4 mRNA expression in comparison to uninfected plants. However, in the case of gi-100 mutant plants, the PAD4 expression level was much reduced in comparison to wild-type Col-0 plants upon infection (). The above observation suggested that GI is required for the expression of PAD4 gene in Arabidopsis during Noco2 pathogen-induced infection.
Figure 3. Regulation of PAD4 expression by GI after Noco2 infection. Graph showing qRT-PCR result of PAD4 gene expression relative to ACTIN1 gene at 48 hpi. Three biological replicates were used for the experiments. Data represented as Mean ± SE. To test for significance among the dataset, a two-way analysis of variance (ANOVA) followed by Tukey’s multiple comparisons test was performed using GraphPad prism software at *p < .05. ** p < .01, *** p < .001.
3.3. GIGANTEA promotes SA-dependent defense genes after infection
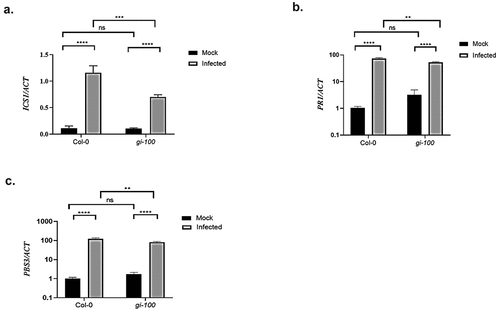
After confirming the role of GI in activating PAD4 expression to confer pathogen resistance, we further investigated the status of PAD4 downstream signaling pathways that are involved in the disease resistance. Salicylic acid (SA) is the universal component responsible for conferring resistance against biotrophic pathogens. Therefore, we checked the transcription status of SA marker gene, PR1 and genes involved in SA production, ICS1 and AVRPPHB SUSCEPTIBLE3 (PBS3). Quantitative RT-PCR was performed using Col-0 and gi-100 plant samples after 48 h post-Noco2 infection (hpi). A significant decrease in the transcript level of ICS1, PR1, and PBS3 was recorded in the gi-100 mutant in comparison to the Col-0 plants upon Noco2 infection (), indicating that in the absence of GI, pathogen-induced expression of SA pathway components is compromised after Hpa Noco2 infection.
Figure 4. Transcripts accumulation of key genes involved in the Salicylic acid pathway during Hpa Noco2 infection. Relative abundance of ICS1 (a), PR1 (b) and PBS3 (c) transcripts, derived from qRT-PCR, after 48 h of Noco2 inoculation in gi-100 as well as Col-0 lines of Arabidopsis. Values are expressed as mean ± SD. The Y-axis represents the 2−^CT of ICS1 gene and log10 (2−^^CT) of PR1 and PBS3 transcription. Transcripts levels were normalized with the internal control gene, ACTIN1. Three biological replicates were used for the experiments. To test for significance among the dataset, a two-way analysis of variance (ANOVA) followed by Tukey’s multiple comparisons test was performed using GraphPad prism software at *p < .05. ** p < .01, *** p < .001.
3.4. GIGANTEA promotes SA-independent defense genes after infection
In addition to the SA-dependent pathway, EDS1 and PAD4 cooperate to activate the expression of SA-independent signaling genes upon pathogen infection. FLAVIN-DEPENDENT MONOOXYGENASE 1 (FMO1), marker genes for SA-independent pathway, is expressed downstream of EDS1- and PAD4-mediated defense signaling and is required for basal resistance to invasive virulent pathogens.Citation41,Citation44 Here, we quantified FMO1 expressions in Col-0 and gi-100 mutant lines after infection with Noco2, by qRT-PCR in relation to a constitutive reference gene, ACTIN1. This analysis revealed that the expression level of FMO1 transcripts was significantly reduced in the gi-100 mutant in comparison to Col-0 plants upon Noco2 infection (). Thus, as a result of compromised activation of the disease-resistance genes in the absence of GI, the gi-100 mutant plants are more susceptible to the Noco2 infection. Taken together, our findings provide a strong indication for a previously unknown function of GI in conferring disease resistance during Hpa Noco2 infection.
Figure 5. Function of GIGANTEA in activation of SA-independent defense genes after Noco2 infection. Graph showing transcripts accumulation of FMO1 gene which is involved in SA-independent defense pathway after 48 h of Noco2 infection in gi-100 as well as Col-0. Values are expressed as mean ± SD. The Y-axis represents the log10 (2−^^CT) of FMO1 transcription. Transcripts levels were normalized with the internal control gene, ACTIN1. Three biological replicates were used for the experiments. To test for significance among the dataset, a two-way analysis of variance (ANOVA) followed by Tukey’s multiple comparisons test was performed using GraphPad prism software at *p < .05. ** p < .01, *** p < .001.
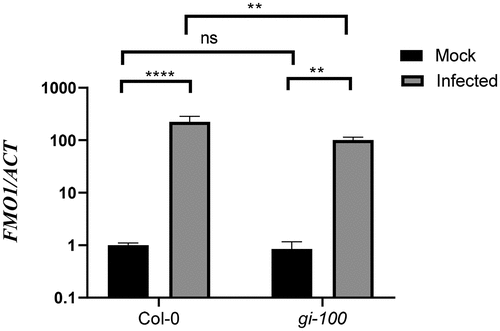
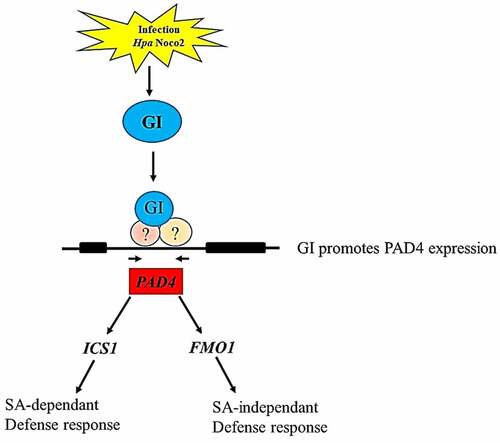
Figure 6. A concluding model depicting the role of GI in regulating PAD4 expression which is a key player involved in modulating both SA-dependent and SA-independent pathways. Our result shows that GI expression is induced in response to Hpa Noco2 infection that contributes to enhanced expression of the PAD4 gene. Graphical summary of the GI-mediated immune response against Hyaloperonospora arabidopsidis infection (). Activated PAD4 further induces its downstream signaling genes expression to initiate the immune response against biotrophic pathogens infection by activating SA-dependent and SA-independent signaling pathways.
Discussion
GIGANTEA (GI) is a plant-specific nuclear protein and has been shown to be involved in the regulation of many physiological and developmental processes in plants.Citation24 Recent studies have revealed a link between GIGANTEA and plant defense signaling.Citation20,Citation59 It was observed that late-flowering mutants are immune to the Fusarium infection and the absence of GI makes the plant more resistant to the Fusarium pathogen.Citation20 Another recent publication showed the role of GI as a negative regulator of plant defense signaling during hemibiotrophic fungal infection and confirmed that GI causes susceptibility to Bipolaris sorokiniana in Arabidopsis.Citation59 Based on our results, we proposed a new model where GI may act as a positive regulator of defense signaling and can be a resistance factor for downy mildew disease, which is caused by an obligate biotrophic pathogen, Hyaloperonospora arabidopsidis (Hpa). Our studies show an additional layer of transcriptional regulation of the plant-pathogen defense signaling pathway. In gi-100 mutant, upon Hpa Noco2 infection, the formation of aerial conidiospores is significantly higher than the Col-0 (control), eds1-2 mutant (susceptible plant to Noco2 infection) and Ws-2 (an accession resistant to Noco2 infection). This suggests that GI plays a positive role in defense signaling against Hpa Noco2 infection and the absence of GI weakens this defense against biotrophic fungus pathogens. We have also investigated the Hpa Noco2 fungal infection severity using another GI T-DNA insertional mutant line, gi-2.Citation56 Our results established that gi-2 shows a similar but comparatively weaker phenotype than gi-100 upon infection. The gi-2 mutant lines were more susceptible to Hpa Noco2 infection in comparison to Col-0 but could not surpass eds1-2 mutant phenotype. This difference in the severity of phenotypes could be because both the gi mutants, gi-2 and gi-100, have been shown to form different sizes of transcripts after T-DNA insertion,Citation24,Citation56 and there could be a difference in the functionality of truncated proteins formed in both the mutants. There are previous reports that have shown a difference in phenotype severity in different alleles of the same gene. For instance, the gi-1 mutation shortened period lengths of leaf movement, cab2::1uc luminescence and RNA transcript abundance rhythms; in contrast, gi-2 caused a gradual lengthening of the cab2::1uc luminescence and RNA transcript abundance rhythm along with shortened period lengths leaf movement.Citation21 In another example, the period of gi-3 did not display wild-type phenotype at 17°C unlike the gi-11 mutant.Citation60 Also, the gi-2 allele shows an exception in the temperature-independent flowering phenotype of gi mutants.Citation61 Thus, such variations in different gi mutant phenotypes resulting from differences in the functionality of truncated transcripts could also explain the difference in the observed pathogen-related phenotype in our current study. Another possible explanation for variations in disease severity phenotype of gi-100 and gi-2 could be that the expression of GI neighboring genes is slightly altered in gi-100 mutant seedlingsCitation52 and our data could not rule out the contributions from the altered expression of the neighboring genes in plant defense signaling.
Our results also suggest the context-dependent function of GIGANTEA in plant disease resistance. During hemibiotrophic vascular infections like Fusarium oxysporum or Bipolaris sorokiniana, GI functions as a negative regulator and makes the plant more susceptible to the disease.Citation20,Citation59 On the other hand, during downy mildew infection caused by the obligate biotrophic pathogen Hpa, GI acts as a positive regulator. To keep the host cell alive, biotrophic pathogens and hemibiotrophs in their biotrophic stage delay senescence. The host can achieve resistance by activating senescence-like processes. On the other hand, necrotrophic pathogens and hemibiotrophs in their necrotrophic stage induce senescence in the host and preventing early senescence is a resistance approach of plants.Citation62 This difference in the mode of action could explain the contrasting phenotypes of gi mutants with these two pathogens, Fusarium and Hpa.
Like humans, plants have developed a very effective system of immunity, which enables them to protect themselves from infection and to produce seeds successfully. Several studies demonstrate that PAD4 along with the EDS1 forms one of the core components to regulate plant basal immunity against virulent biotrophic pathogens.Citation42 We hypothesized that the defense responses regulated by GI might also involve a PAD4/EDS1 signaling cascade. It has been shown that the regulatory region of the PAD4 gene lies within the intron, which contains the core ACGT sequences, called G-box sequences, that are required for binding of the bZIP class of transcription regulators, such as GBF1.Citation58 GBF1 is a well-established regulator of plant defense signaling and modulates PAD4 expression.Citation58 It has been speculated that GI can also be a G-box binding protein;Citation57 however, there is no concrete evidence to date. Based on the above-mentioned findings, we hypothesized that GI can bind to the regulatory intronic sequence of the PAD4 gene to direct its expression just like GBF1. To confirm our hypothesis, we explored the ChIP-seq results of GI available online,Citation57 and to our surprise, we found out that GI can directly be involved in the transcriptional regulation of PAD4. We validated this finding with the ChIP assay followed by qPCR. We are first to confirm that GI binds to the regulatory region present in the intron of the PAD4 gene. Although there is another non‐recognized G‐box (TACGTA) present in the PAD4 promoter about 1.26 kb upstream of the transcription start site, however, we found less enrichment of the PAD4 start site than the intronic region after pull-down with GI. However, whether GI and GBF1 function together or independently to regulate PAD4 expression remains an open question. It might be possible that GI along with GBF1 forms a complex that binds to the intronic region of PAD4 to regulates the plant defense responses. Here, we have confirmed that GI is required for the upregulation of PAD4 expression during Noco2 infection as gi-100 mutant showed a reduction in the expression of PAD4 in comparison to the WT plants. Since PAD4 expression was not completely blocked in the gi-100 mutant, it led us to propose that several other alternative regulators (such as GBF1) may function together or independent of GI to fine-tune the expression of PAD4, thereby regulating plant defense response to various pathogens.
After confirming the role of GI in the regulation of PAD4 expression, we next explored the status of PAD4 downstream signaling events. EDS1 and PAD4 are important regulators of basal resistance to obligate biotrophic and certain hemibiotrophic pathogens, governing the accumulation of the phenolic signaling molecule salicylic acid.Citation43,Citation63 The EDS1-PAD4 complex promotes the expression of key genes involved in SA biosynthesis, ICS1 and PBS3 genes, and SA marker gene, PR1.Citation38 Our results demonstrated that following the reduced PAD4 expression in gi-100 plants after Noco2 infection, PAD4‐dependent defense responses were also faded. Here, we found that after infection, the absence of GI leads to reduced expression of ICS1, PBS3 and PR1. Hence, in wild-type plants, Hpa infection results in increased GI expression, which enhances the PAD4 expression, leading to the upregulation of downstream defense genes ICS1, PBS3, and PR1 genes, and hence makes the wild-type plant relatively more resistant to Noco2 infection than gi-100 plants.
EDS1 and PAD4 also function through the SA-independent mechanism that offers resistance in the presence of FLAVIN-DEPENDENT MONOOXYGENASE 1 (FMO1).Citation41–43 FMO1 is the marker gene of the EDS1-PAD4-controlled, SA-independent signaling pathway. It has been shown that its expression is being locally and systemically stimulated in Arabidopsis plants upon inoculation with virulent or avirulent Pseudomonas syringae bacteria and oomycete, Hyaloperonospora arabidopsidis pathogens, whereas fmo1 loss-of-function mutants lead to compromised resistance to virulent or avirulent P. syringae or H. arabidopsidis.Citation41,Citation44 Our results verified that the absence of GI leads to downregulation of FMO1 after Hpa Noco2 infection, hence making the plant more susceptible than Col-0 plants. Further studies in this direction are required to unravel how GI regulates pattern-triggered and the effector‐triggered immune response in Arabidopsis.
In summary, GI expression is induced in response to Hpa Noco2 infection that contributes to enhanced expression of the PAD4 gene. Activated PAD4 further induces its downstream signaling gene expression to initiate the immune response against biotrophic pathogen infection by activating SA-dependent and SA-independent signaling pathways. Thus, our results provide a framework that addresses the role of GIGANTEA in regulating plant defense response.
Author contributions
AS has designed and performed the experiments. AS has analyzed the data, written the manuscript and arranged the figures.
Supplemental Material
Download Zip (2.4 MB)Acknowledgments
We thank Dr Jane Parker for providing Ws-2 and eds1-2 mutant seeds and for her helpful discussions during the whole project. We also thank Prof. George Coupland, Max Planck Institute, for providing Col-0, 35S::HA:GI, gi-2 and gi-100 mutant seeds. Special thanks to Jaqueline Bautor for her guidance in performing pathogen experiments. A big thanks to Dr Ankita Singh, Dr Aparajita Singh and Dr Kishore Panigrahi for proofreading the manuscript.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website
Additional information
Funding
References
- Korves TM, Bergelson J. A developmental response to pathogen infection in Arabidopsis. Plant Physiol. 2003;133:339–10. doi:10.1104/pp.103.027094.
- Schiestl FP, Kirk H, Bigler L, Cozzolino S, Desurmont GA. Herbivory and floral signaling: phenotypic plasticity and tradeoffs between reproduction and indirect defense. New Phytol. 2014;203:257–266. doi:10.1111/nph.12783.
- Martínez C, Pons E, Prats G, León J. Salicylic acid regulates flowering time and links defence responses and reproductive development. Plant J. 2004;37:209–217. doi:10.1046/j.1365-313X.2003.01954.x.
- Wang GF, Seabolt S, Hamdoun S, Ng G, Park J, Lu H. Multiple roles of WIN3 in regulating disease resistance, cell death, and flowering time in Arabidopsis. Plant Physiol. 2011;156:1508–1519. doi:10.1104/pp.111.176776.
- Lee J, Nam J, Park HC, Na G, Miura K, Jin JB, Yoo CY, BaekD, Kim DH, Jeong JC, et al. Salicylic acid-mediated innate immunity in Arabidopsis is regulated by SIZ1 SUMO E3 ligase. Plant J. 2007;49:79–90. doi:10.1111/j.1365-313X.2006.02947.x.
- Li W, Ahn IP, Ning Y, Park CH, Zeng L, Whitehill JG, Lu H, Zhao Q, Ding B, Xie Q, et al. The U-Box/ARM E3 ligase PUB13 regulates cell death, defense, and flowering time in Arabidopsis. Plant Physiol. 2012;159:239–250. doi:10.1104/pp.111.192617.
- Liu L, Zhang J, Adrian J, Gissot L, Coupland G, Yu D, Turck F. Elevated levels of MYB30 in the phloem accelerate flowering in Arabidopsis through the regulation of FLOWERING LOCUS T. PLoS One. 2014;9:e89799. doi:10.1371/journal.pone.0089799.
- Song S, Qi T, Fan M, Zhang X, Gao H, Huang H, Wu D, Guo H, Xie D. The bHLH subgroup IIId factors negatively regulate jasmonate-mediated plant defense and development. PLoS Genet. 2013;9:e1003653. doi:10.1371/journal.pgen.1003653.
- Ogawara T, Higashi K, Kamada H, Ezura H. Ethylene advances the transition from vegetative growth to flowering in Arabidopsis thaliana. J Plant Physiol. 2003;160:1335–1340. doi:10.1078/0176-1617-01129.
- Zhou C, Zhang L, Duan J, Miki B, Wu K. HISTONE DEACETYLASE19 is involved in jasmonic acid and ethylene signaling of pathogen response in Arabidopsis. Plant Cell. 2005;17:1196–1204. doi:10.1105/tpc.104.028514.
- Wu K, Zhang L, Zhou C, Yu CW, Chaikam V. HDA6 is required for jasmonate response, senescence and flowering in Arabidopsis. J Exp Bot. 2008;59:225–234. doi:10.1093/jxb/erm300.
- Yu CW, Liu X, Luo M, Chen C, Lin X, Tian G, Lu Q, Cui Y, Wu K. HISTONE DEACETYLASE6 interacts with FLOWERING LOCUS D and regulates flowering in Arabidopsis. Plant Physiol. 2011;156:173–184. doi:10.1104/pp.111.174417.
- Andrés F, Coupland G. The genetic basis of flowering responses to seasonal cues. Nat Rev Genet. 2012;13:627–639. doi:10.1038/nrg3291.
- Lyons R, Iwase A, Gänsewig T, Sherstnev A, Duc C, Barton GJ, Hanada K, Higuchi-Takeuchi M, Matsui M, Sugimoto K, et al. The RNA-binding protein FPA regulates flg22-triggered defense responses and transcription factor activity by alternative polyadenylation. Sci Rep. 2013;3:2866. doi:10.1038/srep02866.
- Singh V, Roy S, Giri MK, Chaturvedi R, Chowdhury Z, Shah J, Nandi AK. Arabidopsis thaliana FLOWERING LOCUS D is required for systemic acquired resistance. Mol Plant Microbe Interact. 2013;26(9):1079–1088. doi:10.1094/MPMI-04-13-0096-R.
- Singh V, Roy S, Singh D, Nandi AK. Arabidopsis FLOWERING LOCUS D influences systemic-acquired-resistance- induced expression and histone modifications of WRKY genes. J Biosci. 2014;39:119–126. doi:10.1007/s12038-013-9407-7.
- Winter CM, Austin RS, Blanvillain-Baufumé S, Reback MA, Monniaux M, Wu MF, Sang Y, Yamaguchi A, Yamaguchi N, Parker J, et al. LEAFY target genes reveal floral regulatory logic, cis motifs, and a link to biotic stimulus response. Dev Cell. 2011;20:430–443. doi:10.1016/j.devcel.2011.03.019.
- Blazquez MA, Green R, Nilsson O, Sussman MR, Weigel D. Gibberellins promote flowering of Arabidopsis by activating the LEAFY promoter. Plant Cell. 1998;10:791–800. doi:10.1105/tpc.10.5.791.
- Navarro L, Bari R, Achard P, Lisón P, Nemri A, Harberd NP, Jones JDG. DELLAs control plant immune responses by modulating the balance of jasmonic acid and salicylic acid signaling. Curr Biol. 2008;18:650–655. doi:10.1016/j.cub.2008.03.060.
- Lyons R, Rusu A, Stiller J, Powell J, Manners JM, Kazan K. Investigating the association between flowering time and defense in the Arabidopsis thaliana-Fusarium oxysporum interaction. PloS One. 2015;10:e0127699. doi:10.1371/journal.pone.0127699.
- Park DH, Somers DE, Kim YS, Choy YH, Lim HK, Soh MS, Kim HJ, Kay SA, Nam HG. Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science. 1999;285:1579–1582. doi:10.1126/science.285.5433.1579.
- Fowler S, Thomashow MF. Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell. 2002;14:1675–1690. doi:10.1105/tpc.003483.
- Martin-Tryon EL, Kreps JA, Harmer SL. GIGANTEA acts in blue light signaling and has biochemically separable roles in circadian clock and flowering time regulation. Plant Physiol. 2007;143:473–486. doi:10.1104/pp.106.088757.
- Mishra P, Panigrahi KC. GIGANTEA - an emerging story. Front Plant Sci. 2015;6:8. doi:10.3389/fpls.2015.00008.
- Cao S, Ye M, Jiang S. Involvement of GIGANTEA gene in the regulation of the cold stress response in Arabidopsis. Plant Cell Rep. 2005;24:683–690. doi:10.1007/s00299-005-0061-x.
- Fornara F, de Montaigu A, Sánchez-Villarreal A, Takahashi Y, Ver Loren van Themaat E, Huettel B, Davis SJ, Coupland G. The GI – CDF module of Arabidopsis affects freezing tolerance and growth as well as flowering. Plant J. 2015;81:695–706. doi:10.1111/tpj.12759.
- Kurepa J, Smalle J, Van Montagu M, Inzé D. Oxidative stress tolerance and longevity in Arabidopsis: the late-flowering mutant gigantea is tolerant to paraquat. Plant J. 1998;14:759–764. doi:10.1046/j.1365-313x.1998.00168.x.
- Riboni M, Galbiati M, Tonelli C, Conti L. GIGANTEA enables drought escape response via Abscisic acid-dependent activation of the florigens and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS. Plant Physiol. 2013;162:1706–1719. doi:10.1104/pp.113.217729.
- Kim WY, Ali Z, Park HJ, Park SJ, Cha JY, Perez-Hormaeche J, Quintero FJ, Shin G, Kim MR, Qiang Z, et al. Release of SOS2 kinase from sequestration with GIGANTEA determines salt tolerance in Arabidopsis. Nat Commun. 2013;4:1352. doi:10.1038/ncomms2357.
- Dodds PN, Rathjen JP. Plant immunity: towards an integrated view of plant-pathogen interactions. Nat Rev Genet. 2010;11:539–548. doi:10.1038/nrg2812.
- Macho AP, Zipfel C. Targeting of plant pattern recognition receptor-triggered immunity by bacterial type-III secretion system effectors. Curr Opin Microbiol. 2015;23:14–22. doi:10.1016/j.mib.2014.10.009.
- Cui H, Tsuda K, Parker JE. Effector-triggered immunity: from pathogen perception to robust defense. Annu Rev Plant Biol. 2015;66:487–511. doi:10.1146/annurev-arplant-050213-040012.
- Tsuda K, Somssich IE. Transcriptional networks in plant immunity. New Phytol. 2015;206:932–947. doi:10.1111/nph.13286.
- Zhou N, Tootle TL, Tsui F, Klessig DF, Glazebrook J. PAD4 functions upstream from salicylic acid to control defense responses in Arabidopsis. Plant Cell. 1998;10:1021–1030. doi:10.1105/tpc.10.6.1021.
- Wagner S, Stuttmann J, Rietz S, Guerois R, Brunstein E, Bautor J, Niefind K, Parker J. Structural basis for signaling by exclusive EDS1 heteromeric complexes with SAG101 or PAD4 in plant innate immunity. Cell Host Microbe. 2013;14:619–630. doi:10.1016/j.chom.2013.11.006.
- Rietz S, Stamm A, Malonek S, Wagner S, Becker D, Medina-Escobar N, Corina Vlot A, Feys BJ, Niefind K, Parker JE, et al. Different roles of ENHANCED DISEASE SUSCEPTIBILITY1 (EDS1) bound to and dissociated from PHYTOALEXIN DEFICIENT4 (PAD4) in Arabidopsis immunity. New Phytol. 2011;191:107–119. doi:10.1111/j.1469-8137.2011.03675.x.
- Seyfferth C, Tsuda K. Salicylic acid signal transduction: the initiation of biosynthesis, perception and transcriptional reprogramming. Front Plant Sci. 2014;5:697. doi:10.3389/fpls.2014.00697.
- Fu ZQ, Dong X. Systemic acquired resistance: turning local infection into global defense. Annu Rev Plant Biol. 2013;64:839–863. doi:10.1146/annurev-arplant-042811-105606.
- Zhang X, Chen S, Mou Z. Nuclear localization of NPR1 is required for regulation of salicylate tolerance, ISOCHORISMATE SYNTHASE1 expression and salicylate accumulation in Arabidopsis. J Plant Physiol. 2010;167:144–148. doi:10.1016/j.jplph.2009.08.002.
- Coates ME, Beynon JL. Hyaloperonospora arabidopsidis as a pathogen model. Annu Rev Phytopathol. 2010;48:329–345. doi:10.1146/annurev-phyto-080508-094422.
- Bartsch M, Gobbato E, Bednarek P, Debey S, Schultze JL, Bautor J, Parker JE . Salicylic acid-independent ENHANCED DISEASE SUSCEPTIBILITY1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the Nudix hydrolase NUDT7. Plant Cell. 2006;18:1038–1051.
- Cui H, Gobbato E, Kracher B, Qiu J, Bautor J, Parker JE. A core function of EDS1 with PAD4 is to protect the salicylic acid defense sector in Arabidopsis immunity. New Phytol. 2017;213:1802–1817. doi:10.1111/nph.14302.
- Joglekar S, Suliman M, Bartsch M, Halder V, Maintz J, Bautor J, Zeier J, Parker JE, Kombrink E. Chemical activation of EDS1/PAD4 signaling leading to pathogen resistance in Arabidopsis. Plant Cell Physiol. 2018;59:1592–1607. doi:10.1093/pcp/pcy106.
- Mishina TE, Zeier J. The Arabidopsis flavin-dependent monooxygenase FMO1 is an essential component of biologically induced systemic acquired resistance. Plant Physiol. 2006;141:1666–1675. doi:10.1104/pp.106.081257.
- Chen Y-C, Holmes EC, Rajniak J, Kim J-G, Tang S, Fischer CR, Mudgett MB, Sattely ES. N-hydroxy-pipecolic acid is a mobile metabolite that induces systemic disease resistance in Arabidopsis. Proc Natl Acad Sci USA. 2018;115:E4920–E9. doi:10.1073/pnas.1805291115.
- Hartmann M, Zeier T, Bernsdorff F, Reichel-Deland V, Kim D, Hohmann M, Scholten N, Schuck S, Bräutigam A, Hölzel T, et al. Flavin monooxygenase-generated N-hydroxypipecolic acid is a critical element of plant systemic immunity. Cell. 2018;173:456–69. e16. doi:10.1016/j.cell.2018.02.049.
- Feys BJ, Wiermer M, Bhat RA, Moisan LJ, Medina-Escobar N, Neu C, Cabral A, Parker JE. Arabidopsis SENESCENCE-ASSOCIATED GENE101 Stabilizes and Signals within an ENHANCED DISEASE SUSCEPTIBILITY1 Complex in Plant Innate Immunity. Plant Cell. 2005;17(9):2601–2613. doi:10.1105/tpc.105.033910.
- Cabral A, Stassen JH, Seidl MF, Bautor J, Parker JE, Van den Ackerveken G. Identification of Hyaloperonospora arabidopsidis transcript sequences expressed during infection reveals isolate-specific effectors. PLoS One. 2011;6:e19328. doi:10.1371/journal.pone.0019328.
- Aarts N, Metz M, Holub E, Staskawicz BJ, Daniels MJ, Parker JE. Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene-mediated signaling pathways in Arabidopsis. Proc Natl Acad Sci USA. 1998;95:10306–10311. doi:10.1073/pnas.95.17.10306.
- Ó’Maoiléidigh DS, van Driel AD, Singh A, Sang Q, Le Bec N, Vincent C, de Olalla EBG, Vayssières A, Romera Branchat M, Severing E, et al. Systematic analyses of the MIR172 family members of Arabidopsis define their distinct roles in regulation of APETALA2 during floral transition. PLoS Biol. 2021;19:e3001043.
- Kaufmann K, Muino JM, Østerås M, Farinelli L, Krajewski P, Angenent GC. Chromatin immunoprecipitation (ChIP) of plant transcription factors followed by sequencing (ChIP-SEQ) or hybridization to whole genome arrays (ChIP-CHIP). Nat Protoc. 2010;5:457–472. doi:10.1038/nprot.2009.244.
- Huq E, Tepperman JM, Quail PH. GIGANTEA is a nuclear protein involved in phytochrome signaling in Arabidopsis. Proc Natl Acad Sci U S A. 2000;97:9789–9794. doi:10.1073/pnas.170283997.
- Botella MA, Parker JE, Frost LN, Bittner-Eddy PD, Beynon JL, Daniels MJ, Holub EB, Jones JDG. Three genes of the Arabidopsis RPP1 complex resistance locus recognize distinct peronospora parasitica avirulence determinants. Plant Cell. 1998;10:1847–1860. doi:10.1105/tpc.10.11.1847.
- Asai S, Rallapalli G, Piquerez SJ, Caillaud MC, Furzer OJ, Ishaque N, Wirthmueller L, Fabro G, Shirasu K, Jones JDG, et al. Expression profiling during Arabidopsis/downy mildew interaction reveals a highly-expressed effector that attenuates responses to salicylic acid. PLoS Pathog. 2014;10:e1004443. doi:10.1371/journal.ppat.1004443.
- Asai S, Yoshioka H. Nitric oxide as a partner of reactive oxygen species participates in disease resistance to necrotrophic pathogen Botrytis cinerea in Nicotiana benthamiana. Mol Plant Microbe Interact. 2009;22:619–629. doi:10.1094/MPMI-22-6-0619.
- Fowler S, Lee K, Onouchi H, Samach A, Richardson K, Morris B, Coupland G, Putterill, J, . GIGANTEA: a circadian clock‐controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane‐spanning domains. The EMBO J. 1999;18:4679–4688. doi:10.1093/emboj/18.17.4679.
- Nohales MA, Liu W, Duffy T, Nozue K, Sawa M, Pruneda-Paz JL, Maloof JN, Jacobsen SE, Kay SA. Multi-level modulation of light signaling by GIGANTEA regulates both the output and pace of the circadian clock. Dev Cell. 2019;49:840–51. e8. doi:10.1016/j.devcel.2019.04.030.
- Giri MK, Singh N, Banday ZZ, Singh V, Ram H, Singh D, Chattopadhyay S, Nandi AK. GBF1 differentially regulates CAT2 and PAD4 transcription to promote pathogen defense in Arabidopsis thaliana. Plant J. 2017;91:802–815. doi:10.1111/tpj.13608.
- Kundu P, Sahu R. GIGANTEA confers susceptibility to plants during spot blotch attack by regulating salicylic acid signalling pathway. Plant Physiol Biochem. 2021. doi:10.1016/j.plaphy.2021.02.006.
- Gould PD, Locke JC, Larue C, Southern MM, Davis SJ, Hanano S, Moyle R, Milich R, Putterill J, Millar AJ, et al. The molecular basis of temperature compensation in the Arabidopsis circadian clock. Plant Cell. 2006;18:1177–1187. doi:10.1105/tpc.105.039990.
- Araki T, Komeda Y. Analysis of the role of the late‐flowering locus, GI, in the flowering of Arabidopsis thaliana. Plant J. 1993;3:231–239. doi:10.1046/j.1365-313X.1993.t01-15-00999.x.
- Häffner E, Konietzki S, Diederichsen E. Keeping control: the role of senescence and development in plant pathogenesis and defense. Plants. 2015;4:449–488. doi:10.3390/plants4030449.
- Wiermer M, Feys BJ, Parker JE. Plant immunity: the EDS1 regulatory node. Curr Opin Plant Biol. 2005;8:383–389. doi:10.1016/j.pbi.2005.05.010.
