ABSTRACT
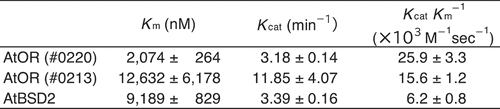
Orange protein (OR) is known to interact with phytoene synthase (PSY) that commits the first step in carotenoid biosynthesis, and functions as a major post-transcriptional regulator on PSY. We here tried to reveal enzymatic characteristics of OR, that is, protein disulfide reductase (PDR) activity of the Arabidopsis thaliana OR protein (AtOR) was analyzed using dieosin glutathione disulfide (Di-E-GSSG) as a substrate. The AtOR part containing only the zinc (Zn)-finger motif was found to show PDR activity, with an apparent Km of 12,632 nM, Kcat of 11.85 min−1, and KcatKm−1 of 15.6 × 103 M−1sec−1. To evaluate the significance of the N-terminal region of AtOR, we examined the kinetic parameters of a fusion protein composed of the N-terminal region and the Zn-finger motif from AtOR. Consequently, the fusion protein had lower values for Km (2,074 nM) and Kcat (3.18 min−1) and higher catalytic efficiency (25.9 × 103 M−1sec−1) than that of only the Zn-finger motif part, suggesting that the N-terminal region of AtOR should be important for substrate affinity and catalytic efficiency of PDR activity. Complementation experiments with E. coli further demonstrated that AtOR containing the N-terminal region and the Zn-finger motif increases phytoene synthase activity of AtPSY especially under reduced circumstances retaining a NADPH- and H+-regeneration system.
Introduction
Carotenoids are diverse color pigments naturally occurring in plants and algae, as well as parts of fungi, bacteria, and archaea. They play important roles in development, photosynthesis, root-mycorrhizal interactions, and the production of phytohormones, such as abscisic acid and strigolactones.Citation1 Phytoene synthase (PSY) catalyzes the formation of phytoene (15-cis-phytoene) from geranylgeranyl diphosphate (GGPP) as the first committed step in carotenoid biosynthesis and is one of the most important regulators in the carotenoid biosynthesis.Citation2,Citation3 The Orange (OR) gene was isolated from an orange cauliflower mutant (Brassica olerancea var. botrytis) that accumulates ß-carotene in organs that normally do not contain carotenoids.Citation4 It was found that PSY and the OR protein (OR) physically interacted with each other in plastids, and OR served as the major post-transcriptional regulators on PSY.Citation2 It was furthermore shown that the N-terminal region of the BoOR protein interacted with PSY.Citation2 The Arabidopsis thaliana OR gene (At5g61670) encodes a protein (AtOR) of 307 amino acids with a putative molecular weight of 33,789. It contains 11 cysteine residues and has two predicted zinc (Zn) finger-like motifs (CXXCXGXG), Zn1 and Zn2 (Supplementary data). The two Zn finger motifs of AtOR are similar to the C4-type Zn finger motifs of the E. coli DnaJ and AtBSD2 proteins.Citation5–8 In chloroplasts, four other proteins likewise contain the Zn-finger motifs: CYO1/SCO2,Citation9–11 LQY1Citation12 and HCF222Citation13 in/at the thylakoid membrane, and PSA2 in the thylakoid lumen.Citation14 They all exhibit protein-disulfide reductase (PDR) activity like DnaJ.Citation15 We thus tested whether AtOR retains PDR activity with a protein disulfide reductase (PDR) assay.Citation8,Citation10,Citation16 In this assay, fluorescence increases if the reduction of disulfide bonds is mediated by dieosin glutathione (Di-E-GSSG). The present study reveals the enzymatic characteristics of AtOR as PDR.
Materials and methods
Expression and purification of truncated AtOR proteins
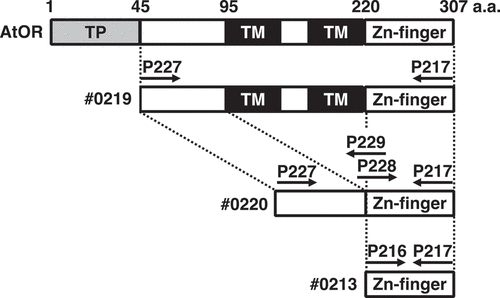
An expression vector (pET-#0219) that contains the cDNA fragment encoding amino acids between 45 and 307 of the wild-type AtOR (At5g61670) protein from Arabidopsis thaliana was amplified by RT-PCR using primers, P217 and P227 (). The amplified DNA fragments were ligated with the expression vector pET24a (+) digested with NdeI and XhoI using In-Fusion HD cloning kit (Takara bio, Japan). cDNA fragments encoding amino acids between 45 and 95, and between 220 and 307 of AtOR were amplified by RT-PCR using primer sets of P227 and P229, and of P228 and P217, respectively. These DNA fragments were ligated with the expression vector pET24a(+) digested with NdeI and XhoI using In-Fusion HD cloning kit, designated pET-#0220. The amplified DNA fragments using primers P206 and P217 were ligated with pET24a (+), designated pET-#0213. The sequences of the DNA fragments were confirmed. The E. coli BL21-CodonPlus (DE3; Agilent Technologies, U.S.A.) was used to express the OR fusion proteins with 6× His tags. The expression and purification of these proteins were performed as described.Citation9
Preparation of Di-E-GSSG and assay for PDI-dependent disulfide reduction
Di-E-GSSG was prepared as described.Citation16 PDI disulfide reduction activity was monitored in PDI assay buffer (100 mM potassium phosphate, pH 7.0) containing OR protein (100 nM) and Di-E-GSSG (188–4700 nM) with or without DTT (5 μM) based on the increased fluorescence at 545 nm with excitation at 525 nm.Citation16 The kinetic parameters were calculated using KaleidaGraph software (Synergy software, U.S.A.).
Bacterial strains and plasmid construction
The plasmid pAC-HIEI was constructed for the expression of the Pantoea ananatis crtE and crtI genes in addition to the Haematococcus pluvialis IDI gene by eliminating crtB encoding phytoene synthase from the plasmid pAC-HIEBI that produces lycopene in E. coli.Citation17 The plasmids pCDF-AtPSY, pCDF-gdh-AtPSY and pCDF-zwf-AtPSY were constructed by inserting the AtPSY gene fragment into pCDF, pCDF-gdh, and pCDF-zwf, respectively.Citation18 The #0213, #0219, #0220 fragments were inserted into pET24a creating pET-#0213, pET-#0219, pET-#0220, respectively.
As E. coli strain as the host, we used BL21(DE3). The transformed E. coli cells were cultured in 2YT medium (1.6% Bactotryptone, 1.0% yeast extract, 0.5% NaCl) containing antibiotics, ampicillin (75 mg L−1), tetracycline (12.5 mg L−1) and spectinomycin (100 mg L−1), at 37°C. Then, we inoculated this preculture into the new 2YT medium with the antibiotics and 0.05 mM IPTG, and cultured at 20°C for 2 days.
Analysis of carotenoids accumulated in recombinant E. coli
We extracted and analyzed the carotenoids as described.Citation19
Results and discussion
To elucidate the functional differences in each region of AtOR, we generated three expression vectors encoding truncated AtOR proteins. #0219 is the expression vector encoding amino acid 45–307 of the full length of AtOR except for the putative transit peptide, the putative mature AtOR protein (Supplemental data). #0220 and #0213 are the expression vectors from #0219 without the putative transmembrane region, “95–220 amino acids” and encoding only Zn-finger motif of 220–307 amino acids, respectively. We tried to synthesize the putative mature AtOR protein (#0219) in E. coli, but it was not observed under all E. coli growth/IPTG concentration conditions we tried (Supplemental data). Therefore, we expressed the truncated AtOR genes that coding for the putative mature protein deleting the transmembrane region (#0219) and only the Zn-finger motif (#0213). These two truncated proteins were soluble and could be used for protein disulfide reductase assay using Di-E-GSSG at a substrate.Citation8,Citation10 In the sample incubated with Di-E-GSSG and dithiothreitol (DTT), the close proximity of the eosin moieties resulted in self-quenching,Citation16 and the molecule had relatively low fluorescence at an excitation of 525 nm.Citation10 In the sample incubated with Di-E-GSSG, DTT and the AtOR protein (#0213), the two eosin moieties were spatially separated because of the reduction of disulfide bonds of Di-E-GSSG, and fluorescence increased.Citation10 Time course analyses of the reduction of Di-E-GSSG with AtOR protein (#0213) and DTT, but not with DTT alone ()), suggesting that AtOR protein has PDR activity. To estimate Km and Kcat of AtOR proteins, we incubated AtOR proteins with DTT and 500–7,000 nM Di-E-GSSG at 25°C and monitored initial rates of eosin glutathione (E-GSSG) formation ()). The reductase activity of AtOR (#0213) increased as a function of Di-E-GSSG, with a Km of 12,632 ± 6,178 nM and Vmax of 1185 ± 407 nM min−1. Therefore, we estimated the Kcat and catalytic efficiency (KcatKm−1) as11.85 ± 4.07 min−1 and 15.6 ± 1.2 × 103 M−1sec−1, respectively (). To analyze the significance of the N-terminal region of AtOR, we estimated the kinetic parameters of AtOR (#0220)()). The Km, Kcat and KcatKm−1 of AtOR (#0220) were 2,074 ± 246 nM, 3.18 ± 0.14 min−1, and 25.9 ± 3.3 × 103 M−1sec−1, respectively (). The Km of AtOR (#0220) was about 6-fold lower than that of AtOR (#0213), suggesting the N-terminal region of AtOR should be important for substrate affinity. The catalytic efficiency of AtOR (#0220) was higher than that of AtOR (#0213). The results indicated that the N-terminal region of AtOR should promote the catalytic efficiency of PDR activity.
Figure 2. PDR activity of AtOR protein. (a) Di-E-GSSG was incubated with DTT at 25°C in the presence (black line) or absence (gray line) of AtOR (#0213). The fluorescence was recorded with excitation at 525 nm and emission at 545 nm. (b) Michaelis-Menten curves for PDR kinetics with 5 µM DTT of AtOR (#0213)(open circles and broken line) and AtOR (#0220)(closed circles and solid line). AtOR and DTT were incubated with varying concentrations of Di-E-GSSG at 25°C, and initial rates of E-GSH formation were monitored as a function of the concentration of Di-E-GSSG. Values represent the mean ± S.D. (n= 3).
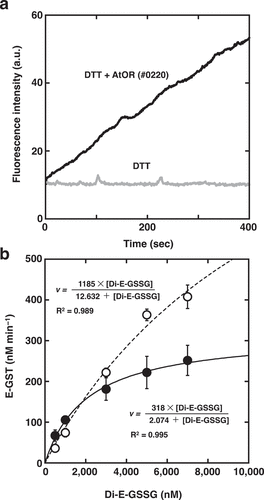
Figure 3. Kinetic properties of AtOR and AtBSD2. Data for AtBSD2 was obtained in the previous study.8
The holdase activity of sweetpotato (Ipomoea batatas) OR protein (IbOR) protected the stability of IbPSY protein and enhanced heat and oxidative stress tolerance in plants.Citation3 IbOR protein inhibited oxidative stress-induced aggregation of IbPSY in vitro. The treatment of IbPSY with H2O2 induced aggregation but the presence of IbOR protein inhibits the aggregation of IbPSY protein.Citation3 Since AtPSY protein has six cysteine residues, PSY protein should form disulfide bound under oxidative stress and the enzymatic activity is reduced, however the PDR activity of OR protein suppresses the oxidative inactivation of PSY. This hypothesis requires further investigation.
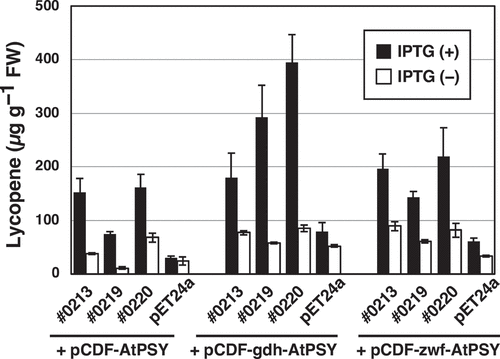
We further performed complementation experiments with E. coli to examine the effect of AtOR toward phytoene synthase activity of the AtPSY protein (). When AtOR coexisted with AtPSY in the pAC-HIEI-carrying E. coli, which further includes pET-#0213, pET#0219 or pET-#0220 and pCDF-AtPSY, lycopene content was increased several times. As for the E. coli transformants possessing pET#0219 and pET-#0220, lycopene amounts were further elevated in the presence of the Bacillus subtilis glucose dehydrogenase (gdh) gene to supply sufficient levels of NADPH and H+. This result indicated that AtOR containing the N-terminal region and the Zn-finger motif increases phytoene synthase activity of AtPSY especially under reduced circumstances having a NADPH- and H+-regeneration system. We also reported that when the gdh and zwf genes, as the NADPH-regenerating enzyme genes, were introduced into the ß-amyrin-producing E. coli, the productivity was increased only in the case of gdh.Citation18
Supplemental Material
Download MS Word (871.1 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/15592324.2022.2072094
Additional information
Funding
References
- Cazzonelli CI, Pogson BJ. Source to sink: regulation of carotenoid biosynthesis in plants. Trends Plant Sci. 2010;15:266–5. doi:10.1016/j.tplants.2010.02.003.
- Zhou X, Welsch R, Yang Y, Alvarez D, Riediger M, Yuan H,Fish, T., Liu, J., Thannhauser, T.W. and Li, L. Arabidopsis OR proteins are the major posttranscriptional regulators of phytoene synthase in controlling carotenoid biosynthesis. Proceedings of the National Academy of Sciences of the United States of America; 2015; 112:3558–3563.
- Park S, Kim HS, Jung YJ, Kim SH, Ji CY, Wang Z, Jeong JC, Lee H-S, Lee SY, Kwak -S-S, et al. Orange protein has a role in phytoene synthase stabilization in sweetpotato. Sci Rep. 2016;6:33563. doi:10.1038/srep33563.
- Lu S, Van Eck J, Zhou X, Lopez AB, O’Halloran DM, Cosman KM, Conlin BJ, Paolillo DJ, Garvin DF, Vrebalov J, et al. The cauliflower or gene encodes a DnaJ cysteine-rich domain-containing protein that mediates high levels of beta-carotene accumulation. Plant Cell. 2006;18:3594–3605. doi:10.1105/tpc.106.046417.
- Brutnell TP, Sawers RJ, Mant A, Langdale JA. BUNDLE SHEATH DEFECTIVE2, a novel protein required for post-translational regulation of the rbcL gene of maize. Plant Cell. 1999;11:849–864. doi:10.1105/tpc.11.5.849.
- Wostrikoff K, Stern D. Rubisco large-subunit translation is autoregulated in response to its assembly state in tobacco chloroplasts. Proceedings of the National Academy of Sciences of the United States of America; 2007; 104:6466–6471.
- Doron L, Segal N, Gibori H, Shapira M. The BSD2 ortholog in Chlamydomonas reinhardtii is a polysome-associated chaperone that co-migrates on sucrose gradients with the rbcL transcript encoding the Rubisco large subunit. Plant J. 2014;80:345–355. doi:10.1111/tpj.12638.
- Busch FA, Tominaga J, Muroya M, Shirakami N, Takahashi S, Yamori W, Kitaoka T, Milward SE, Nishimura K, Matsunami E, et al. Overexpression of BUNDLE SHEATH DEFECTIVE 2 improves the efficiency of photosynthesis and growth in Arabidopsis. Plant J Cell Mol Biol. 2020;102:129–137. doi:10.1111/tpj.14617.
- Shimada H, Mochizuki M, Ogura K, Froehlich JE, Osteryoung KW, Shirano Y, Shibata D, Masuda S, Mori K, Takamiya K-I, et al. Arabidopsis cotyledon-specific chloroplast biogenesis factor CYO1 is a protein disulfide isomerase. Plant Cell. 2007;19:3157–3169. doi:10.1105/tpc.107.051714.
- Muranaka A, Watanabe S, Sakamoto A, Shimada H. Arabidopsis cotyledon chloroplast biogenesis factor CYO1 uses glutathione as an electron donor and interacts with PSI (A1 and A2) and PSII (CP43 and CP47) subunits. J Plant Physiol. 2012;169:1212–1215. doi:10.1016/j.jplph.2012.04.001.
- Albrecht V, Ingenfeld A, Apel K. Snowy cotyledon 2: the identification of a zinc finger domain protein essential for chloroplast development in cotyledons but not in true leaves. Plant Mol Biol. 2008;66:599–608. doi:10.1007/s11103-008-9291-y.
- Lu Y, Hall DA, Last RL. A small zinc finger thylakoid protein plays a role in maintenance of photosystem II in Arabidopsis thaliana. Plant Cell. 2011;23:1861–1875. doi:10.1105/tpc.111.085456.
- Hartings S, Paradies S, Karnuth B, Eisfeld S, Mehsing J, Wolff C, Levey T, Westhoff P, Meierhoff K. The DnaJ-Like Zinc-finger protein HCF222 is required for thylakoid membrane biogenesis in plants. Plant Physiol. 2017;174:1807–1824. doi:10.1104/pp.17.00401.
- Fristedt R, Williams-Carrier R, Merchant SS, Barkan A. A thylakoid membrane protein harboring a DnaJ-type Zinc finger domain is required for photosystem I accumulation in plants. J Biol Chem. 2014;289(44):30657–30667. doi:10.1074/jbc.M114.587758.
- de Crouy-Chanel A, Kohiyama M, Richarme G. A novel function of Escherichia coli chaperone DnaJ. Protein-disulfide isomerase. J Biol Chem. 1995;270:22669–22672. doi:10.1074/jbc.270.39.22669.
- Raturi A, Mutus B. Characterization of redox state and reductase activity of protein disulfide isomerase under different redox environments using a sensitive fluorescent assay. Free Radical Bio Med. 2007;43:62–70. doi:10.1016/j.freeradbiomed.2007.03.025.
- Takemura M, Maoka T, Koyanagi T, Kawase N, Nishida R, Tsuchida T, Hironaka M, Ueda T, Misawa N. Elucidation of the whole carotenoid biosynthetic pathway of aphids at the gene level and arthropodal food chain involving aphids and the red dragonfly. BMC Zool. 2021;6. doi:10.1186/s40850-021-00082-w.
- Takemura M, Tanaka R, Misawa N. Pathway engineering for the production of beta-amyrin and cycloartenol in Escherichia coli-a method to biosynthesize plant-derived triterpene skeletons in E. coli. Appl Microbiol Biotechnol. 2017;101:6615–6625. doi:10.1007/s00253-017-8409-z.
- Takemura M, Kubo A, Higuchi Y, Maoka T, Sahara T, Yaoi K, Umeno D, Misawa N. Pathway engineering for efficient biosynthesis of violaxanthin in Escherichia coli. Appl Microbiol Biotechnol. 2019;103:9393–9399. doi:10.1007/s00253-019-10182-w.