ABSTRACT
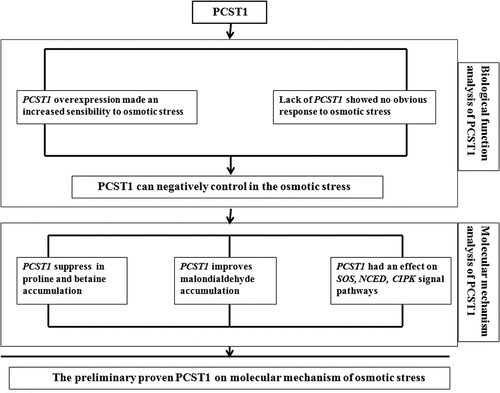
Analysis of PCST1 expression characteristics and the role of PCST1 in response to osmotic stress in Arabidopsis thaliana. The structure of PCST1 was analyzed using Bioinformatics method. Real-time PCR, GUS tissue localization and subcellular localization were adopted to analyze the expression pattern of PCST1 in Arabidopsis. To validate the transgenic positive strain of PCST1 using Real-time PCR, overexpression experiments were performed in wild type. Full-length cDNA was cloned and connected into a binary vector with 35S promoter, and the construction was transformed into wild type. With NaCl and mannitol treatments, the germination rate, green leaves rate, physiological indexes were carried out and counted in Arabidopsis with overexpression of PCST1 and T-DNA insertion mutants. The molecular mechanism of PCST1 in response to osmotic stress in Arabidopsis was analyzed. Based on the bioinformatic analysis, PCST1 is a hydrophobin with 403 amino acids, and the molecular weight is 45.3236 KDa. It contains only the START (the lipid/sterol – binding StAR – related lipid transfer protein domains) conservative domain. PCST1 possesses phosphatidylcholine binding sites and transmembrane region. Expression pattern analysis showed that expression of PCST1 increased with time. The PCST1 widely expressed in Arabidopsis, including roots, axils of stem leaves, flowers (sepal, conductive tissue of the petal, thrum, anther and stigmas), and the top and basal parts of the siliquas. It mainly localized in cell membrane. The overexpression of PCST1 enhanced the sensitivity to osmotic stress in Arabidopsis based on the germination rate. While expression of PCST1 decreased, and the sensitivity to osmotic stress had no obvious change in Arabidopsis. Its molecular mechanism study showed, that PCST1 response to osmotic stress resistance by regulating the proline, betaine synthesis, as well as the expression of key genes SOS, NCED, CIPK. PCST1 is composed of 403 amino acids. The START conservative domain, a transmembrane structure, the phosphatidyl choline binding sites are contained in PCST1. It is localized in cytoplasmic membrane. The PCST1 widely expressed in the root, leaf, flower and siliquas. NaCl and mannitol suppressed the expression of PCST1 and PCST1 can negatively control action of Arabidopsis in the osmotic stress. PCST1 regulates the synthetic pathway of proline, betaine and the expression of SOS, NCED and CIPK in response to the osmotic stress resistance.
Background
In the process of agricultural production, all kinds of drastic/harsh environment are important reasons directly affecting crop yield. Mining the critical stress-related genes is the key to improve crop yieldsCitation1. High salt, drought and low temperature seriously affect crop growth, development, directly affecting crop production.Citation2 Therefore, how to improve the tolerance of crops becomes a major problem that should be solved. In the long-term evolution of plants, a variety of mechanisms are formed to resist environmental stresses, including morphology, physiology and biochemistry adaptive mechanism. With the Arabidopsis genome sequencing completed, a large number of gene sequences accumulated in the database, while the functions of most of them are unknown. PCST1 (At1g55960) is one of possessing START domain protein family in Arabidopsis. The function of PCST1 is no clear yet.
The conserved START domain is 30 KDa and consists of approximately 200 amino acids. It belongs to a class of steroid regulatory protein and could combine and transport cholesterol to inner membrane of mitochondria.Citation3 It is defined as a protein related to combine with lipids and sterols amino acid sequence.Citation4 In animals, the ligand portion of START has been confirmed. START and metastatic lymph protein (MLN64) can combine with cholesterol,Citation5 phosphatidylcholine can transport protein-binding phosphatidylcholine,Citation6 carotenoid-binding protein can combine carotenoids in silkworm.Citation7 In addition, a protein called CERT (nephritis syndrome antibody binding proteins) have been discovered recently, ceramide can be transported by the START domain of CERT.Citation8
By using X-ray crystallography method, the structure of START domain has been determined, which belongs to three species of mammal proteins: PCTP, MLN64 and StarD4. On the basis of this structure, START domains are classified as super-helical fold proteins family and exist in everywhere of cell organism everywhere.Citation9 Researchers speculated that the members of the START superfamily bind lipids and steroids, while other fold supercoiled families can interact with a wide range of metabolites and other molecules, such as antibiotics, RNA, and antigens.Citation10 START domains may have conservative interactions with lipids and sterols in higher species such as animals and plants. The function of START domains in animals (e.g. StAR and PCTP) are sterols and phospholipids substances transportation and metabolism.
In Arabidopsis, there are 35 members in the START domain protein-family.Citation11 The function of nine of them has been determined and they all contain HD ZLZ START domain, they participate in the process of cortex development regulation (ATML1),Citation12,Citation13 floral organ formation (PDF2),Citation13 anthocyanin accumulation (ANL2, FWA),Citation10,Citation14 epidermal hair growth (GL2),Citation15 adaxial and abaxial polarity (PHV, PHB, REV)Citation16–18 and vascular bundle development (ATHB-8).Citation19 Previous studies reported that SSMP (At3g13062),Citation11 which contains START domain, related to salt resistance. The PCST1 which possessed the START domain in Arabidopsis had the highest homology with SSMP. It is consists of 403 amino acids and possessed phosphatidylcholine binding sites and transmembrane structure. The structural characteristic of PCST1 may affect the composition and permeability of cell membranes, and further involves in plant osmotic stress regulatory processes.
Osmotic stress is a condition that the plant cannot get enough water because of some adverse environmental factors. Common osmotic stress factors include salt damage, drought, etc. Osmotic adjustment substances, such as proline and betaine, can reduce plant damages caused by osmotic stress.
Salt stress is an important abiotic stress, which directly affects plant growth and development by osmotic stress, specific ion toxicity, ion imbalance, oxygen stress and interfering with absorption and transport of mineral nutrients.Citation20 Salt stress results in changes in many biochemical processes in plants, including the accumulation of low molecular weight metabolite, such as amino acid and betaine. This mechanism is commonly referred as the regulatory mechanism of compatible solutes and ion transport.Citation21 In addition, oxygen stress will occur simultaneously with salt stress. This phenomenon results from the production of reactive oxygen species (ROS),Citation22 such as super oxide radical (O2-), hydrogen peroxide (H2O2), and hydroxyl radical (OH−). In plant cells, the produced ROS interacts with some of the important cellular molecules and metabolites, which result in a number of damaging processes that cause cell damage. Excessive accumulation of ROS may cause toxic reaction of plants, such as lipid peroxidation, protein degradation and DNA mutation. It is found that some plant species protect their cellular and subcellular systems by enhancing some enzyme activity to suppress the reactive oxygen species and free radicals. The enzymes contain superoxide dismutase, catalase, peroxidase, glutathione reductase, and polyphenol oxidase. Ascorbic acid and glutathione are also important in the process of scavenging reactive oxygen species and free radicals.Citation23–25 SOS (Salt Overly Sensitive) pathway is a classical salt resistance signal pathway in previous studies.Citation26 SOS showed a critical role in regulating the ion homeostasis and improving the sodium resistance of plants under salt stress.Citation27
Under drought stress, plants need osmotic adjustment to prevent dehydration of plants to maintain photosynthesis and stomatal opening, enhance the growth of root and promote the absorption of water. Water channel protein is a membrane channel, which plays an important role in the control of water content in cells.Citation28–30 Many endogenous hormones in plants involve in drought stress. They may lead to a large number of plant signal transduction.Citation28 When plants are subjected to drought stress, stress signals are perceived firstly through the receptor system and then transmit stress stimulus signals to second messenger.Citation31 As a stress signal, ABA plays a critical role in plant stress resistance, especially in drought resistance.Citation32 ABA biosynthesis is the basis of ABA signal generation. ABA synthesis key genes of NCED (9-cis-epoxycarotenoid dioxygenase)Citation33 and CIPK (CBL-interacting protein kinase)Citation34 play an important roles in the regulation of drought stress.
Proline is one of the components of plant protein and widely existed in plants in free state. In drought, salinity and other stress conditions, a large number of proline accumulated in many plants.Citation35 Proline is the cytoplasmic osmotic adjustment substance, in addition, the accumulation of proline plays an important role in stabilizing the structure of biological macromolecules, reducing the acidity of cells, releasing ammonia and regulating cell redox potential as an energy base. The content of proline in plants reflects the stress resistance of plants to a certain extent. Proline is a response signal on osmotic substances and salt stress.Citation36 The Δ1- pyrroline-5-carboxylate synthetase (P5CS) is a key enzyme in proline biosynthesis,Citation37–39 and proline dehydrogenase (PRODH) catalyze proline degradation. Stress induces proline accumulation in plants, depending on the increase of P5CS activity.Citation40 Under drought, high salt and other stress conditions, betaine is a very large amount of accumulation of osmotic adjustment substances, which has a great effect on the resistance to adversity. Betaine is synthesized by the catalysis of choline (CMO) and betaine aldehyde dehydrogenase (BADH).Citation41 The content of malondialdehyde (MDA) reflects the peroxidation degree of cell membrane lipid in plants.Citation42–44 High content of MDA usually accompanied with high degree of cell membrane lipid peroxidation and serious cell membrane damage.Citation45,Citation46 In general, the stress conditions, such as high temperature, salinity, and strong light, tend to induce membrane lipid peroxidation.
Proteins that contain START conserved domain functioned on lipids transport and metabolism, signal transduction, transcription regulation or other processes. PCST1 has a distant homology relationship with the START gene family in Arabidopsis, while PCST1 showed a closer relationship with PCTP that belongs to START family gene in mammals. Reports about the function of protein only containing START domain are less. The studies on the function of this protein on plant growth regulation and development are significant on clarifying the position of START domain in evolution.
In this study, the method of biological information was used to analyze the structure characteristic of PCST1 in Arabidopsis. To explicit the function of PCST1 in Arabidopsis under osmotic stress, the overexpression of PCST1 and deletion mutant were obtained and used for analysis of stress resistance, then the mechanism of PCST1 participating in abiotic stress regulation maybe further explicit in Arabidopsis.
Materials and methods
Plant materials and growth conditions
The Arabidopsis thaliana (L.) Heynh. ecotype Colombia (Col-0) was used as the WT and the genetic background for transgenic plants. The T-DNA insertion mutants of PCST1 (SALK_053628, SALK_053637) were obtained from the Arabidopsis Biological Resource Center (ABRC), Columbus, OH, USA. Homozygous lines were generated from three generations for the analysis of segregation ratios. The deletion of gene in the mutant was determined by real-time reverse transcription-polymerase chain reaction (RT-PCR) using gene-specific primers and a left border T-DNA primer. The stable transformed plants were generated by floral dip method. Seeds were grown in Murashige and Skoog (MS) nutrient agar medium under continuous 70 μmol/(m2·s) light intensity at 22 ± 2°C. The basal MS medium was supplemented with 3% (w/v) sucrose, the mannitol or NaCl was added in treated groups. After transplanting Arabidopsis seedlings to soil (vermiculite: humus = 1:1), they were kept in a growth chamber with a photoperiod of 16 h/8 h (light/dark) at 22 ± 2°C. To analyze the expression of PCST1 in response to osmotic stress, the 4-wk-old plants were treated with a solution of 150 mM NaCl or 300 mM mannitol. Plants treated with fresh water were used as controls. The tissues were collected at 1, 3, 6, 9, 12 and 24 h after treatment.
Plasmid construction
Full-length cDNA of PCST1 was amplified with the following primers containing restriction sites of BamHI and SacI: forward, 5′-GGATCCATGAAGAGCGTATCAACCTGGG-3′, reverse, 5′-GAGCTCTTAAATGGTGGTGGTCTGGGTG-5′. The amplified fragments were introduced into the T-vector PMDTM19 (TaKaRa Biomedicals, Dalian, China) and cloned into pSN1301. To create the PCST1 pro::β-glucuronidase (GUS) construct, a 1.5-kb fragment upstream from the initiation codon was amplified using the forward primer with BamHI and HindIII sites 5′-GGATCCTTGCTCTCGAGAATGAACCTTTTGA-3′ and reverse primer 5′-AAGCTTCGAATTCCAAAATCTCGACGATTTG-3′, and cloned into the pCAMBIA 1381z vector. The plasmid was electroporated into Agrobacterium tumefaciens GV3101 and transformed into Col-0 by the floral dip method. Transgenic plants were selected using 50 μg·ml−1 hygromycin. T3 or T4 homozygous lines were used for further experiments.
Localization analysis
The full-length PCST1 coding region was amplified with the primers containing SacI and BamHI sites (forward primer,5′-GAGCTCATGAAGAGCGTATCAACCTGGG −3′, reverse primer, 5′-GGATCCAATGGTGGTGGTCTGGGTG-3′). The PCR products were cleaved by SacI/BamHI and cloned into Cam35S-GFP (pCAMBIA2300-35S-GFP) to generate the PCST1 expression plasmid. The plasmid (1.5 μg) was transformed into Arabidopsis and detected using a Laser scanning confocal microscope (FV1000IX81) with a wavelength of 488 nm.
Stress tolerance assays
Arabidopsis seeds were sown on 1/2 MS solid medium with 200, 250, 300, 350 mM mannitol, or 100, 125, 150, 175 mM NaCl for 15 d. The germination rates, green cotyledon rates were counted, and the root length was measured.
Expression analysis of genes related to proline and betaine biosynthesis
To analyze the expression of genes related to proline and betaine biosynthesis. OE, WT and pcst1 plants were treated with 150 mM NaCl or 300 mM mannitol for 24 h. Seedlings grown under normal conditions were control. Realtime RT-PCR was performed to analyze gene expression levels (the gene names are listed in Table S1). The expression levels of genes in each type of plants (with or without stress treatment) were calculated.
Real-time RT-PCR
Total RNA was isolated using the Trizol reagent and digested with DNaseI to remove DNA contamination. cDNA was synthesized using oligo dT as the primer. The genes of 18S rRNA and actin 3 were used as the internal controls. The primers used for RT-PCR are listed in Table S1. The reaction mixture (20 μl) contained 10 μl of SYBR Green Realtime PCR Master Mix (DBI®, Bioscience, Germany), 0.5 μM each of forward and reverse primers, and 1 μl of cDNA template (equivalent to 50 ng of total RNA). PCR was performed on an ABI 7500 system using the SYBR Premix Ex TaqTM(perfect real time) kit (TaKaRa Biomedicals, Dalian, China) under the following conditions: 94°C for 30s, 40 cycles of 94°C for 5s, 58°C for 15s, and 72°C for 10s, and 72°C for 7 min. Three biological replicates were performed. The data was calculated using the 2−ΔΔCt method.Citation47
Water loss measurement
Eight leaves per individual mutant and WT plant growing in normal conditions for 4 week were excised. The leaves were kept on the laboratory bench at 22°C and weighed at the designated time intervals. Four replicates were performed for each line.
Signaling pathway analysis
To analyze the role of PCST1 in the stress resistance signaling pathway, WT, OE and pcst1 mutant were treated with 150 mM NaCl and 300 mM mannitol and sampled at the designated time intervals. Total RNA of samples were extracted and reversed transcribed into cDNA. Real-time PCR was performed to analyze the expression of NaCl stress related gene SOS1, SOS2, SOS3, HKT1 and mannitol stress-related genes CIPK3, NCED3. Primers are shown in Table S1.
Statistical analysis
Data analysis was performed using SPSS 16.0 software (SPSS Inc, Chicago, IL, USA). ANOVA Tukey’s multiple comparison tests were used to exam significant differences (p < .05).
Results
Bioinformatic analysis of PCST1
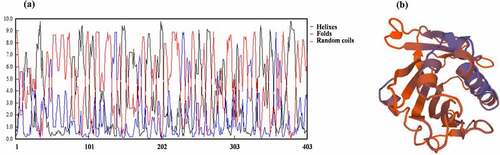
By consulting NCBI (National Center of Biotechnology Information) and TAIR (The Arabidopsis Information Resource) website, we know that PCST1 is a hydrophobin with 403 amino acids, the molecular weight is 45.3236 KDa, the isoelectric point is 9.75, only the START conservative domain (84–295) is contained. PCST1 possesses phosphatidylcholine binding sites. The function of PCST1 is unknown. Using TMHMM Server v. 2.0 to forecast the transmembrane structure of PCST1, it showed that the 21–43 amino acid position possesses the transmembrane region. Forecast PCST1 secondary structure, consists of 148 helixes, 46 folds and 199 random coils (). Tertiary structures of PCST1were predicted by SWISS – MODEL with homologous MODEL service software (). A 1.3-kb fragment received from TAIR upstream from the initiation codon was amplified, analysis PCST1 promoter contains components (), this study found 26 cis-element, participation in Arabidopsis response art defense, environmental stimuli, and hormone related components, including defense and stress response element (TC-rich repeats), drought induced MYB transcription factor binding sites (MBS); Gibberellin response element (P-box, GARE-motif) and auxin response elements (TGA-element); Anaerobic Response Element (Anaerobic Response Element, motorcycle), light Response Element (LAMP-Element, rbcS-CMA7a), etc. Which contains many relevant elements to the resistance to adversity, the response to the environmental stimulus and the hormone in the Arabidopsis. Show that PCST1 may be involved in plant response to adversity defense.
Table 1. Promoter analysis of PCST1.
Expression pattern of PCST1 in Arabidopsis
To explore the expression pattern of PCST1, Transgenic plants PCST1pro::GUS were analyzed by using GUS staining (). PCST1 was mainly expressed in hypocotyls, cotyledons and true leaves in seedlings, with more expression quantities in old parts than that in young parts. In mature strains, PCST1 was widely expressed in roots, axils of stem leaf, the flowers (sepal, conducting tissue of the petal, thrum, anther and stigmas) and the top and basal parts of the siliquas. The temporal and spatial expression of PCST1 in Arabidopsis was analyzed. The study demonstrated that expression of PCST1 increased with time; PCST1 expression was found in the root, rosette leaf, stem leaf, flower and siliqua of 6 weeks’ seedlings in Arabidopsis (). The subcellular localization of PCST1 was analyzed, showing that PCST1 was mainly localized in cell membrane ().
Figure 2. Expression pattern of PCST1 in Arabidopsis. (a) Observation the GUS staining of proPCST1::GUS transgenic plants. (1) 5-d-old seeding; (2) 10-d-old seeding; (3) flower; (4) dissect of flower; (5) silique; (6) whole plant. (b) The expression of PCST1 during different growth periods and different tissues. (1) Assay of the accumulation of PCST1 transcript in different growth periods Arabidopsis thaliana plants. Total RNA was isolated from 7, 14, 21, 28-d-old wild-type (WT) plants; (2) Detection of PCST1 transcript accumulation in different tissues of Arabidopsis. Total RNA was isolated from various tissues of 4-wk-old wild-type plants. Real-time reverse transcription-polymerase chain reaction (RT-PCR) quantifications were normalized to the expression of 18S rRNA. Values are means (±SE) of three replications. (c) Observation of location of PCST1-GFP under the laser confocal microscope. (d) Regulation of PCST1 transcripts by NaCl and mannitol treatments. Assay of the accumulation of PCST1 transcript in Arabidopsis in response to NaCl and mannitol treatments by real-time reverse transcription-polymerase chain reaction (RT-PCR). Wild-type were grown with sufficient water for 4 wk. The expression levels were normalized to that of Actin 3. Error bars represent standard errors (n = 3). Three independent replicates were performed. Statistical significance was determined by one-way ANOVA with Tukey’s post hoc test (p < .05).
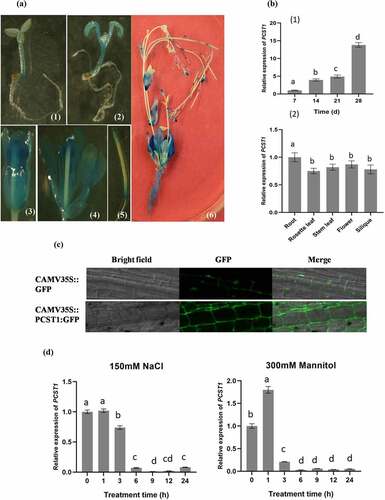
The 4 weeks’ old wild type was treated with 150 mM NaCl or 300 mM mannitol, leading to nearly 1/10 drop in expression of PCST1 along the time to 6 h (). The result indicated that NaCl and mannitol suppressed the expression of PCST1.
Analysis of the expression of PCST1 in SALK and OE plants
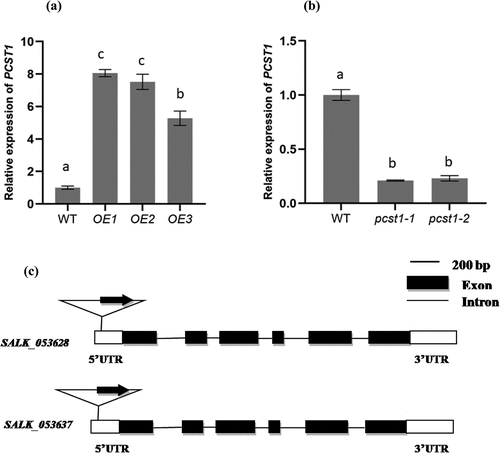
We examined the expression of PCST1 in mutant plants (SALK_053628, SALK_053637, pcst1) and overexpressing PCST1 (OE) using real-time RT-PCR. The results showed that the expression of PCST1 strongly increased in OE plants, and was downregulated more than 10-fold in SALK plants, compared with the control ().
Figure 3. Analysis of the expression of PCST1 in OE and SALK plants. The Arabidopsis plant with overexpression (a) and knockdown (b) of PCST1 were analyzed using real-time PCR. The x-axis in each figure showed the line number, and 3 lines of overexpression (OE) and 2 lines of knockdown (pcst1) of PCST1 were analyzed. The expression levels of PCST1 were log2 transformed. Data are means ± SD from three independent experiments. (c) Schematic diagram of the T-DNA insertion site in the PCST1 locus. Positions are shown for T-DNA insertions (triangle), exons (black rectangles), untranslated region (UTR) (white rectangles) and introns (lines). Data are means ± SD from three independent experiments. Statistical significance was determined by one-way ANOVA with Tukey’s post hoc test (p < .05).
Overexpression of PCST1 enhanced NaCl and mannitol stress sensitivity in Arabidopsis
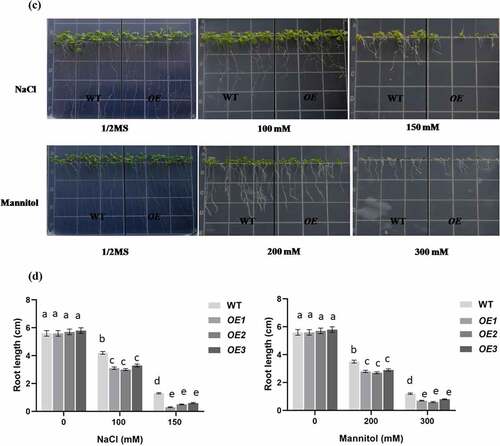
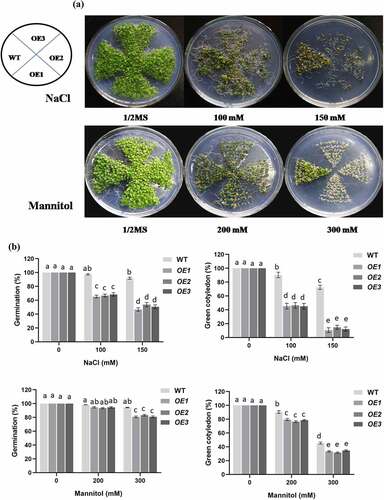
To characterize the function of PCST1 in stress tolerance, the types of plant with different PCST1 expression levels were studied, including the following: WT plants; independent T3 homozygous transgenic plants which overexpressed PCST1 (OE). After planting the wild type (Col-0) and PCST1 transgenic seeds on the one-half-strength Murashige (MS) culture medium supplemented with/without 100, 150 mM NaCl or 200, 300 mM mannitol and culturing for 15 d. Under normal growth conditions, there were no differences in germination rate, green leaves rate, root length and growth phenotype among these plants (). Under NaCl and mannitol treatment conditions, WT had significantly higher germination rate and green leaves rate than OE plants (). In addition, OE plants had significantly lower root growth rates than WT plants (). These results showed that overexpression of PCST1 made an increased sensibility to osmotic stress.
Figure 4. NaCl and mannitol responses of overexpression PCST1 transgenic Arabidopsis during germination and post-germination growth. (a) Growth of WT and OE seedlings on Murashige and Skoog (MS) medium containing 100, 150 mM NaCl and 200, 300 mM Mannitol. Seeds were germinated and grown for 15 d. The photographs show representative seedlings. (b) Effects of NaCl and mannitol on the germination and green leave rates of WT and OE plants. Error bars represent the standard errors of c. 200 seeds from three independent experiments. (c) NaCl and mannitol effects on the growth of germinated WT and OE seedlings. Seeds of different genotypes were grown in a vertical position on MS medium containing 100, 150 mM NaCl and 200, 300 mM Mannitol. The photographs show representative seedlings. (d) Root length of WT and OE plants in response to NaCl and mannitol. Error bars represent the standard errors of c. 60 plants from three independent experiments. Statistical significance was determined by one-way ANOVA with Tukey’s post hoc test (p < .05).
PCST1 mutants showed no obvious response to osmotic stress of transgenic plants
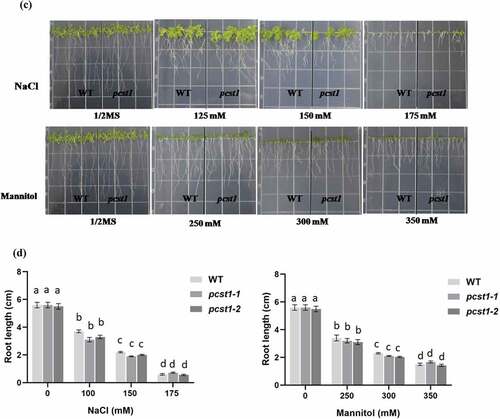
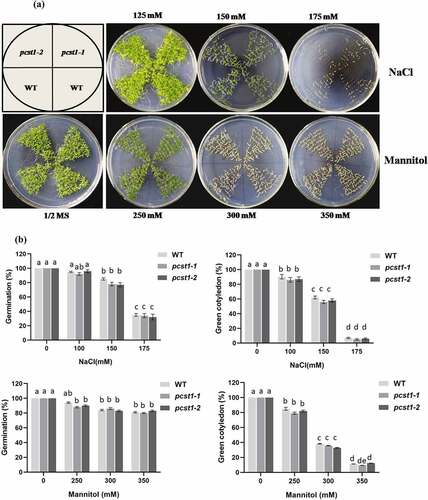
Three types of plant with different PCST1 expression levels were studied, including the following: WT plants, PCST1 mutant plants (SALK_053628, pcst1-1; SALK_053637, pcst1-2). After planting the wild type and PCST1 mutants seeds on the one-half-strength Murashige (MS) culture medium supplemented with 125, 150, 175 mM NaCl or 250, 300, 350 mM mannitol and culturing for 15 d. Under normal growth conditions, there were no differences in germination rate, green cotyledon rate, root length and growth phenotype among these plants (). Under NaCl and mannitol treatment conditions, pcst1-1 and pcst1-2 plants showed no obvious response to them and had the same phenotype with the wild type (). In addition, WT, pcst1-1 and pcst1-2 plants had generally similar root growth rates (). The results showed that PCST1 mutants had no obvious response to sensibility to osmotic stress.
Figure 5. PCST1 mutant plants are no obvious response to sensibility to salt and mannitol stresses. (a) Growth phenotype of WT and pcst1 mutants in media containing 125, 150, 175 mM NaCl and 250, 300, 350 mM mannitol. Seeds were germinated and grown for 15 d. The photographs show representative seedlings. (b) Effects of NaCl and mannitol on the germination and greening leave rate of WT and pcst1. Error bars represent the standard errors of c. 200 seeds from three independent experiments. (c) Seedling root length of WT and pcst1 mutants as affected by NaCl and mannitol. (d) The data analysis was the same as in (b). Statistical significance was determined by one-way ANOVA with Tukey’s post hoc test (p < .05).
PCST1 confers stress tolerance through suppress of proline and betaine but improves water loss rate and Malondialdehyde accumulation
Proline acts as an osmotic agent and radical scavenger to protect cells from damage caused by osmotic stress, and also helps plants to maintain sustainable growth under long-term stress conditions. As proline is quite important for osmotic stress tolerance, we examined whether PCST1 is involved in proline accumulation under osmotic stress conditions. The proline contents among OE, WT and pcst1 under salt and mannitol treatment conditions were significantly different, while there was no significant difference in proline content in these types of plants under normal growth conditions. The proline contents were lowest in OE1 and OE2, whereas pcst1-1 and pcst1-2 had generally similar proline contents with WT (). We further examined the expression of proline biosynthesis- and degradation-related genes, including P5CS (At3g55610), PRODH (At4g34590). Under NaCl or mannitol treatment conditions, the expression of these genes was significantly different. The expression of P5CS was highest in WT plants and pcst1-1, pcst1-2 plants, followed by OE1 and OE2. In contrast, under NaCl and mannitol stress conditions, the expression of PRODH was highest in both OE lines, whereas it was lowest in WT and pcst1.
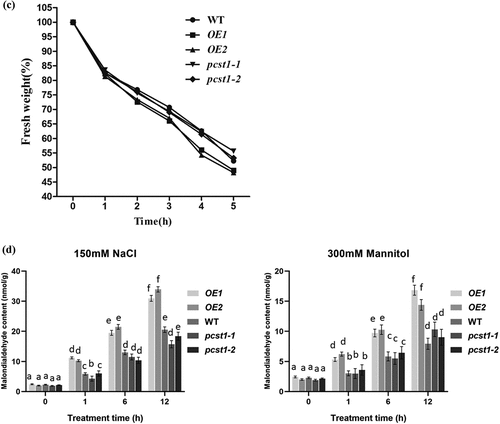
Figure 6. PCST1 confers stress tolerance through suppress of proline and betaine but improves water loss rate and Malondialdehyde accumulation. (a) Proline biosynthesis regulated by PCST1. Analysis of proline level in OE, WT and pcst1 and the expression of proline biosynthesis (P5CS) and degradation-related (PRODH) genes. The plants were treated with NaCl, mannitol for 1, 6, 12 and 24 h, and plants grown under normal conditions were harvested at the corresponding time points as controls. The relative expression level was calculated as the transcript level of a gene under NaCl or mannitol treatment for 6 or 24 h divided by the transcript level of that gene under normal conditions at the corresponding time point. Data are means ±SD from three independent experiments. (b) Analysis of betaine level in OE, WT and pcst1 and the expression of betaine biosynthesis (CMO) genes. The plants were treated with NaCl, mannitol for 1, 6 and 24 h, and plants grown under normal conditions were harvested at the corresponding time points as controls. The relative expression level was calculated as the transcript level of a gene under NaCl or mannitol treatment for 6 or 24 h divided by the transcript level of that gene under normal conditions at the corresponding time point. Data are means ±SD from three independent experiments. (c) Water loss from detached leaves of WT, OE, pcst1 transgenic plants. Water loss was expressed as the percentage of initial fresh weight. Values are means from eight leaves for each of five independent experiments. (d) Malondialdehyde content regulated by PCST1. Analysis of malondialdehyde level in OE, WT and pcst1. The plants were treated with NaCl, mannitol for 1, 6 and 12 h. Data are means ±SD from three independent experiments. Statistical significance was determined by one-way ANOVA with Tukey’s post hoc test (p < .05).
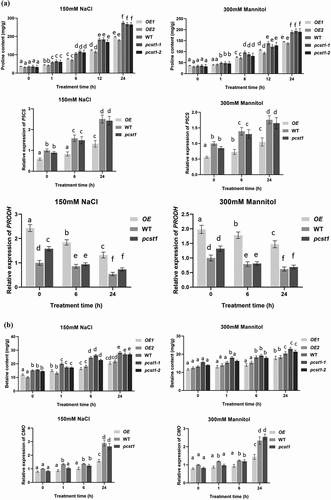
Betaine is a very large amount of accumulation of osmotic adjustment substances, which has a great effect on the resistance to adversity. There was no significant difference in betaine content among OE, WT and pcst1 under normal growth conditions; on salt and mannitol treatment, the betaine contents in these types of plant were significantly different. The betaine contents were lowest in OE1 and OE2, whereas pcst1and WT had generally similar betaine contents (). We further examined the expression of betaine that synthesized by CMO. Under salt or mannitol treatment, the expression of these genes was significantly different. The expression of CMO was highest in WT and pcst1. CMO was downregulated in OE, compared with the WT. Overexpression of PCST1 improved water loss rate response to osmotic stress (). For water loss rate treatment, the leaves of 4-wk-old WT, OE and pcst1 lines were weighed immediately (fresh weight, FW), kept on the laboratory beach at 25°C and weighed at the designated time intervals. Water loss was represented as the percentage of initial fresh weight at each time point. OE1 and OE2 had higher fresh weight decrease rate than WT and pcst1, whereas WT, pcst1-1 andpcst1-2 had generally similar fresh weight decrease rate. The results showed that the water holding capacity of OE strain was weakest, and the lowest stress tolerance. Overexpression of PCST1 improved Malondialdehyde accumulation (). The content of MDA is the embodiment of the plant cell membrane lipid peroxidation degree of expression. There was no significant difference in MDA content among OE, WT and pcst1 under normal growth conditions; on salt and mannitol treatment, the MDA contents in these types of plant were significantly different. The MDA contents were highest in OE1 and OE2, whereaspcst1and WT had generally similar MDA contents.
PCST1 had an effect on genes of adversity resistance signal pathways
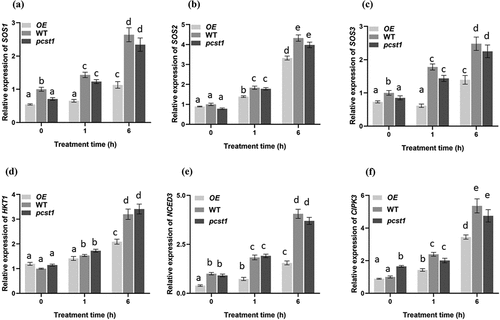
Under the treatment of osmotic stress, the samples were taken at the designated time intervals. The qRT-PCR was carried out for detecting the expression of NaCl stress related gene SOS1, SOS2, SOS3 and HKT1, and mannitol stress related gene CIPK3, NCED3.
Under normal growth conditions, there were no differences in those genes among these plants (). Under osmotic stress treatment conditions, those genes in WT had significantly higher expression than OE plants lines, whereas pcst1 plants lines showed no obvious response to it and had the same expression with the wild type.
Figure 7. Analysis of the expression of NaCl stress related gene SOS1, SOS2, SOS3 and HKT1, and mannitol stress related gene CIPK3, NCED3 regulated by PCST1. Analysis of relative expression level in OE, WT and pcst1. The plants were treated with NaCl, mannitol for 1and 6 h. The expression of (a) SOS1, (b) SOS2, (c) SOS3, (d) HKT1, (e) CIPK3, (f) NCED3. Data are means ±SD from three independent experiments. Statistical significance was determined by one-way ANOVA with Tukey’s post hoc test (p < .05).
Discussion
Expression patterns of PCST1 in Arabidopsis
PCST1 have the homology to PCTPCitation11 protein which contained mammalian START domain . Studies have shown that, PCTP can be combined with phosphatidylcholine,Citation48 whose functions are transporting and metabolizing sterols and phospholipids substances. Due to PCST1 also has a phosphatidylcholine-binding site and phosphatidylcholine is a major component of phospholipids and phospholipid metabolism has a close relationship to cell membrane permeability and fluidity. At present, the protein only containing START domain in the process of plant stress resistance were rarely reported, SSMP only contains START domain and participates in the process of stress tolerance in Arabidopsis transmembrane protein, but PCST1 consisting transmembrene structure has nearly 70% of homology to SSMP. Bioinformatics analyzes defense-related stress tolerance response elements of PCST1 upstream promoter sequence contained (TC-rich repeats) and MYB transcription factor binding sites (MBS), and the presence of these elements prompted that PCST1 may be associated with defense response resilience, this is consistent results of this study, scilicet PCST1are involved in the regulation process under osmotic stress in Arabidopsis.
In this study, by analyzing the tissue localization of PCST1 in Arabidopsis, PCST1 in the whole growing cycle of growing from just two cotyledons to bear fruit in Arabidopsis were expressed. Seedling period is mainly composed of hypocotyl gradually to new site expression, from hypocotyl to cotyledon and true leaves, older parts more than young site expression; Adult seedling were expressed in the root, the Indus leaves, cauline leaf axil, stem, flowers (sepals, filaments, anther, stigma), pods. By analyzing the expression of partial distribution and growth period in PCST1 of Arabidopsis, we found that with the increase of plant growth days, PCST1 expression quantity increased significantly, were expressed in various organizations, fully meet the GUS staining. Subcellular localization of PCST1 orientation on the cytoplasmic membrane, confirmed the bioinformatics analysis of PCST1 across a membrane structure.
PCST1 regulate the molecular mechanism of osmotic stress in Arabidopsis
Studies have reported that the excessive Na+ ion in the soil caused imbalance in the body, moisture deficiency and ion toxicity,Citation49 so some plants formed a Na+ efflux and Na+ segment processing, therefore they can maintain low intracellular Na+ concentration to adapt to the effects of salt stress on plant’s growth and development. SOS pathway in previous research is more classic salt signaling pathway.Citation26 SOS3 and SOS2 located in the cytoplasm regulate SOS1 (plasma membrane Na+/H+ antiporter) on the cytoplasmic membrane, while the high-affinity potassium transporter HKT (high-affinity potassium transporters) and Na+/H+ antiporter NHX1 control the K+, Na+ regulation.Citation50 So it will achieve intracellular balance of K+, Na+. Under drought stress, plants need osmotic adjustment to prevent plant dehydration phenomenon, maintain photosynthesis and stomatal opening, enhance root growth and promote the absorption of water. The water film is aquaporin channel. It plays an important roleCitation28–30 in terms of water content control cells.
Drought stress signaling pathways are divided into two kinds: dependent ABA and ABA not relying on two ways.Citation51 ABA, as a stress signal in particular, plays an important role in drought resistance in plants.Citation33,Citation52 Since the ABA biosynthesis is the fundamental basis for ABA signal, ABA generates the biosynthetic NCED,Citation33 and CIPKCitation34 which plays an important role on the regulation of drought stress. Osmolytes proline and betaine play important roles in the regulation of plant salt and drought. Under drought, salinity and other stress conditions, many plants have accumulated much proline.Citation35 Proline content in plants depends P5CS activity, and to some extent reflects the resistance of plants. P5CS is a key enzyme in the biosynthesis of proline, and PRODH catalyzed the proline degradation.Citation40Betaine, as osmotic adjustment substance, which has much accumulation, has a very large role in resistance to stress . Betaine synthesis by CMO catalytic synthesis.Citation41 MDA is a plant cell membrane lipid peroxidation degree of expression, and the high content of MDA, indicates higher plant plasma membrane lipid peroxidation, meanwhile the cell membrane was serious injury.Citation45,Citation46
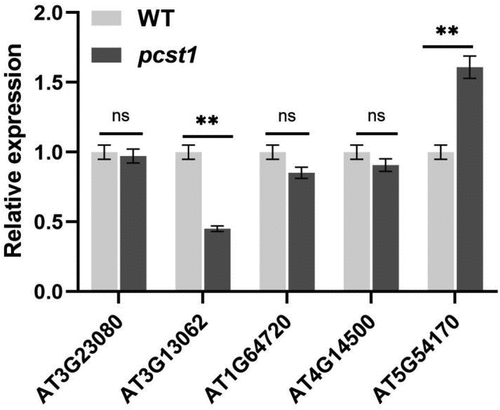
This study through the PCST1 function of gain and loss of transgenic Arabidopsis response to the stress of NaCl, mannitol, its various strains seedling germination rate, the rate of leaves and root length of phenotype observation and data statistics, and to analyze the phenotype of sprout, found expression strain sensitivity to osmotic stress increases, the T-DNA insertion mutants of osmotic stress sensitivity is not obvious; To express under osmotic stress, wild type and mutant Arabidopsis proline, betaine, malondialdehyde content determination, found expression strain proline, betaine content significantly lower than the wild type, MDA content is significantly higher than the wild type, while the mutant and wild type difference is not obvious. The expression analysis of osmotic stress gene SOS1, SOS2, SOS3, HKT1, CIPK3, NCED3 showed overexpression of these genes expression in PCST1 is significantly lower than the wild-type plant, and the difference between wild type and mutant plant is not obvious. We analyzed under osmotic stress, on the one hand, due to the excessive expression of PCST1 through regulating effect on the key enzymes of osmotic regulation substances that affect the accumulation of osmotic regulation substances and the sensitivity to osmotic stress. On the other hand, because PCST1 excess expression suppresses the osmotic stress signaling pathways of gene expression, the ionic imbalance inside and outside the cells causes more sensitivity to osmotic stress. PCST1 T-DNA insertion mutants of Arabidopsis to osmotic stress sensitivity is not obvious due to redundancy of genetic function of family, we analyzed the expression of other gene that only contains the START structure domain in PCST1 (), found that in pcst1, AT3G13062 and AT5G54170 express obvious changes have taken place, the next step we will analyze whether pcst1 due to genetic redundancy is not sensitive to osmotic stress, for each pair of interacting proteins is PCST1 screening at the same time, improve the PCST1 regulatory mechanism of osmotic stress.
Figure 8. Analysis only contains the START structure genes expressed in pcst1. Statistical significance was determined by Student’s t-test (two-tailed) and are expressed as mean ± s.d. ns > 0.05, **p < .01.
The results drawn from this study PCST1 regulation sketch (), preliminarily prove the molecular mechanism of osmotic stress regulation of PCST1 in Arabidopsis.
Conclusion
PCST1 is composed of 403 amino acids, containing only START conservative domain, across a membrane structure, and the phosphatidyl choline binding sites, Positioning on the cytoplasmic membrane, the root, leaf, flower and fruit pods are expressed in the organization, in the expression is more mature organizations. NaCl and mannitol suppress the expression of PCST1. And PCST1 can negatively control in the osmotic stress action of Arabidopsis. PCST1 regulates the synthetic pathway of proline, betaine and the expression of SOS, NCED and CIPK for responding the osmotic stress resistance.
Author contributions
J.D. and W.F. designed the experiments. H.Z., K.Z., T.L. and Y.Z. contributed equally to this work. H.Z., Y.Z., K.Z., T.L. and Z.T. carried out all of the experiments with technical support from K.Z. and J.D. and F.W. analyzed data. H.Z., K.Z. and F.W. wrote the manuscript with input from J.D.
Supplemental Material
Download MS Excel (23 KB)Acknowledgments
This research was supported by a grant (ZD2018085) from Science and technology research project of colleges and universities in Hebei province.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/15592324.2022.2134675
Additional information
Funding
References
- Maiti RK, Satya P. Research advances in major cereal crops for adaptation to abiotic stresses [J]. GM Crops Food. 2014;5(4):259–16. doi:10.4161/21645698.2014.947861.
- Urao T, Katagiri T, Mizoguchi T, Yamaguchi-Shinozaki K, Hayashida N, Shinozaki K. Two genes that encode Ca(2+)-dependent protein kinases are induced by drought and high-salt stresses in Arabidopsis thaliana [J]. Molecular and General Genetics MGG. 1994;244(4):331–340. doi:10.1007/BF00286684.
- Ponting CP, Aravind L. START: a lipid-binding domain in StAR, HD-ZIP and signalling proteins [J]. Trends Biochem Sci. 1999;24(4):130–132. doi:10.1016/S0968-0004(99)01362-6.
- Tsujishita Y, Hurley JH. Structure and lipid transport mechanism of a StAR-related domain [J]. Nat Struct Biol. 2000;7(5):408–414. doi:10.1038/75192.
- Roderick SL, CHAN WW, Agate DS, Olsen LR, Vetting MW, Rajashankar KR, Cohen DE. Structure of human phosphatidylcholine transfer protein in complex with its ligand [J]. Nat Struct Biol. 2002;9(7):507–511. doi:10.1038/nsb812.
- T H, S H, T Y, Fujii H, Banno Y, Jouni ZE, Kobayashi M, Sato R, Maekawa H, Tsuchida K, et al. Isolation, characterization, and cDNA sequence of a carotenoid binding protein from the silk gland of Bombyx mori larvae [J]. J Biol Chemi. 2002;277(35):32133–32140. doi:10.1074/jbc.M204507200.
- H K, K K, Y S, Miura Y, Kawano M, Fukasawa M, Nishijima M. Molecular machinery for non-vesicular trafficking of ceramide [J]. Nature. 2003;426(6968):803–809. doi:10.1038/nature02188.
- Romanowski MJ, Soccio RE, Breslow JL, Burley SK. Crystal structure of the Mus musculus cholesterol-regulated START protein 4 (StarD4) containing a StAR-related lipid transfer domain. Proc Natl Acad. 2002;99(10):6949–6954. doi:10.1073/pnas.052140699.
- Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures [J]. Nucleic Acids Res. 1997;25(1):236–239. doi:10.1093/nar/25.1.236.
- Kubo H, KOORNNEEF M, Aarts MGM, Pereira A, Koornneef M. ANTHOCYANINLESS2, a homeobox gene affecting anthocyanin distribution and root development in arabidopsis. Plant Cell. 1999;11(7):1217–1226. doi:10.1105/tpc.11.7.1217.
- Schrick K, Nguyen D, Karlowski WM, et al. START lipid/sterol-binding domains are amplified in plants and are predominantly associated with homeodomain transcription factors [J]. Genome Biol. 2003;5(6):50–67(18.
- Rabino I, Mancinelli AL. Light, temperature, and anthocyanin production [J]. Plant Physiol. 1986;81(3):922–924. doi:10.1104/pp.81.3.922.
- M ABE, Katsumata H, Komeda Y, Takahashi T. Regulation of shoot epidermal cell differentiation by a pair of homeodomain proteins in arabidopsis. Development. 2003;130(4):635–643. doi:10.1242/dev.00292.
- Fujimoto R, KINOSHITA Y, Kawabe A, Kinoshita T, Takashima K, Nordborg M, Nasrallah ME, Shimizu KK, Kudoh H, Kakutani T, et al. Evolution and control of Imprinted FWA genes in the genus arabidopsis [J]. PLoS Genet. 2008;4(4):162–168. doi:10.1371/journal.pgen.1000048.
- Szymanski DB, Jilk RA, Pollock SM, Marks MD. Control of GL2 expression in Arabidopsis leaves and trichomes [J]. Development. 1998;125(7):1161–1171. doi:10.1242/dev.125.7.1161.
- EMERY JF, Floyd SK, Alvarez J, Eshed Y, Hawker NP, Izhaki A, Baum SF, Bowman JL. Radial patterning of arabidopsis shoots by class III HD-ZIP and KANADI genes [J]. Current Biol. 2003;13(20):1768–1774. doi:10.1016/j.cub.2003.09.035.
- Mcconnell JR, EMERY J, ESHED Y, Bao N, Bowman J, Barton MK. Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots [J]. Nature. 2001;411(6838):709–713. doi:10.1038/35079635.
- Elhiti M, Stasolla C. Structure and function of homodomain-leucine zipper (HD-Zip) proteins [J]. Plant Signal Behav. 2009;4(2):86–88. doi:10.4161/psb.4.2.7692.
- Baima S, Possenti M, Matteucci A, Wisman E, Altamura MM, Ruberti I, Morelli G. The arabidopsis ATHB-8 HD-zip protein acts as a differentiation-promoting transcription factor of the vascular meristems [J]. Plant Physiol. 2001;126(2):643–655. doi:10.1104/pp.126.2.643.
- Brosa C, Soca L, Terricabras E, Ferrer JC, Alsina A. New synthetic brassinosteroids: a 5α-hydroxy-6-ketone analog with strong plant growth promoting activity [J]. Tetrahedron. 1998;54(40):12337–12348. doi:10.1016/S0040-4020(98)00743-1.
- Bajguz A, Tretyn A. The chemical characteristic and distribution of brassinosteroids in Plants [J]. Phytochemistry. 2003;62(7):1027–1046. doi:10.1016/S0031-9422(02)00656-8.
- XIONG X, GAN J, WEI Z, et al. Effects of oxygen and weak magnetic field on Fe 0 /bisulfite system: performance and mechanisms [J]. Environ Sci Pollut Res. 2016;1–10.
- AL-ANBAKY Q, AL-KARAKOOLY Z, Kilaparty SP, Agrawal M, Albkuri YM, RanguMagar AB, Ghosh A, Ali N. Cytotoxicity of manganese (III) complex in human breast adenocarcinoma cell line is mediated by the generation of reactive oxygen species followed by mitochondrial damage [J]. Int J Toxicol. 2016;35(6):672–682. doi:10.1177/1091581816659661.
- Huang J, Hirji R, ADAM L, Rozwadowski KL, Hammerlindl JK, Keller WA, Selvaraj G. Genetic engineering of glycinebetaine production toward enhancing stress tolerance in plants: metabolic limitations [J]. Plant Physiol. 2000;122(3):747–756. doi:10.1104/pp.122.3.747.
- Guler NS, Pehlivan N. Exogenous low-dose hydrogen peroxide enhances drought tolerance of soybean (Glycine max L.) through inducing antioxidant system [J]. Acta Biol Hung. 2016;67(2):169–183. doi:10.1556/018.67.2016.2.5.
- Chinnusamy V, Schumaker K, ZHU JK. Molecular genetic perspectives on cross-talk and specificity in abiotic stress signalling in plants [J]. J Exp Bot. 2004;55(395):225–236. doi:10.1093/jxb/erh005.
- ZHU JK. Cell signaling under salt, water and cold stresses [J]. Curr Opin Plant Biol. 2001;4(5):401–406. doi:10.1016/S1369-5266(00)00192-8.
- Dubois M, Claeys H, Broeck LV, et al. Time of day determines Arabidopsis transcriptome and growth dynamics under mild drought. Plant Cell & Environment; 2016. J.
- ZHANG L, WEI H, Yuan W, et al. The MaASR gene as a crucial component in multiple drought stress response pathways in Arabidopsis [J]. Funct Integr Genomics. 2014;15(2):1–14.
- KHAN MIR, Nazir F, Asgher M, Per TS, Khan NA. Selenium and sulfur influence ethylene formation and alleviate cadmium-induced oxidative stress by improving proline and glutathione production in wheat [J]. J Plant Physiol. 2015;173(3):9–18. doi:10.1016/j.jplph.2014.09.011.
- Ravikumar G, Manimaran P, Voleti SR, Subrahmanyam D, Sundaram RM, Bansal KC, Viraktamath BC, Balachandran SM. Stress-inducible expression of AtDREB1A transcription factor greatly improves drought stress tolerance in transgenic indica rice [J]. Transgenic Res. 2014;23(3):421–439. doi:10.1007/s11248-013-9776-6.
- XIONG L, ZHU JK. Regulation of abscisic acid biosynthesis [J]. Plant Physiol. 2003;133(1):29–36. doi:10.1104/pp.103.025395.
- Estradamelo AC, Chao REIDMS, et al. Overexpression of an ABA biosynthesis gene using a stress-inducible promoter enhances drought resistance in petunia [J]. Horticulture Res. 2015;2(1):52–60.
- Manik SM, SHI S, Mao J, Dong L, Su Y, Wang Q, Liu H. The calcium sensor CBL-CIPK Is involved in plant’s response to abiotic stresses [J]. Int J Genomics. 2014;2015(4):1–10. doi:10.1155/2015/493191.
- Jahantigh O, Najafi F, Badi HN, Khavari-Nejad RA, Sanjarian F. Changes in antioxidant enzymes activities and proline, total phenol and anthocyanine contents in Hyssopus officinalis L. plants under salt stress. Acta Biol Hung. 2016;67(2):195–204. doi:10.1556/018.67.2016.2.7.
- C LINY, Kao CH. Proline accumulation induced by excess nickel in detached rice leaves [J]. Biologia Plantarum. 2007;51(2):351–354. doi:10.1007/s10535-007-0071-3.
- Zhang B, Öy T, Zhou X, Guo CJ, Liu X, Thind A, Hu HH, Zhao S, Liu JL. The proline synthesis enzyme P5CS forms cytoophidia in Drosophila. J Genet Genomics. 2020 Mar 20;47(3):131–143. doi:10.1016/j.jgg.2020.02.005. Epub 2020 Mar 19. PMID: 32317150.
- Funck D, Baumgarten L, Stift M, von Wirén N, Schönemann L. Differential contribution of P5CS isoforms to stress tolerance in arabidopsis. Front Plant Sci. 2020 Sep 25;11. 565134. 10.3389/fpls.2020.565134. PMID: 33101333; PMCID: PMC7545825.
- Rai AN, Penna S. Molecular evolution of plant P5CS gene involved in proline biosynthesis. Mol Biol Rep. 2013 Nov;40(11):6429–6435. Epub 2013 Sep 26. PMID: 24068435. doi:10.1007/s11033-013-2757-2.
- Krishna P. Brassinosteroid-mediated stress responses [J]. J Plant Growth Regul. 2004;22(4):289–297. doi:10.1007/s00344-003-0058-z.
- Bhuiyan NH, Hamada A, Yamada N, Rai V, Hibino T, Takabe T. Regulation of betaine synthesis by precursor supply and choline monooxygenase expression in Amaranthus tricolor [J]. J Exp Bot. 2007;58(15–16):4203–4212. doi:10.1093/jxb/erm278.
- Ayala A, Muñoz MF, Argüelles S. Lipid peroxidation: production, metabolism, and signaling mechanisms of malondialdehyde and 4-hydroxy-2-nonenal. Oxid Med Cell Longev. 2014;2014:360438. Epub 2014 May 8. PMID: 24999379; PMCID: PMC4066722. doi:10.1155/2014/360438.
- Tsikas D. Assessment of lipid peroxidation by measuring malondialdehyde (MDA) and relatives in biological samples: analytical and biological challenges. Anal Biochem. 2017 May 1;524. 13–30. 10.1016/j.ab.2016.10.021. Epub 2016 Oct 24. PMID: 27789233.
- Sri Iswari R, Dafip M, Purwantoyo E. Malondialdehyde (MDA) production in atherosclerosis supplemented with steamed tomato. Pak J Biol Sci. 2021 Jan;24(3):319–325. PMID: 34486316. doi:10.3923/pjbs.2021.319.325.
- WANG Y, ZHOU M, Gong X, Liu C, Hong M, Wang L, Hong F. Influence of lanthanides on the antioxidative defense system in maize seedlings under cold stress [J]. Biol Trace Elem Res. 2011;142(3):819–830. doi:10.1007/s12011-010-8814-y.
- N-CERDA K MANQUI, Escudey M, G ZIGA, et al. Effect of cadmium on phenolic compounds, antioxidant enzyme activity and oxidative stress in blueberry (Vaccinium corymbosum L.) plantlets grown in vitro [J]. Ecotoxicol Environ Saf. 2016;133(316).
- Zhang H, Guo L, Li Y, Zhao D, Liu L, Chang W, Zhang K, Zheng Y, Hou J, Fu C, et al. TOP1α fine-tunes TOR-PLT2 to maintain root tip homeostasis in response to sugars. Nat Plants. 2022 Jul;8(7):792–801. Epub 2022 Jul 11. Erratum in: Nat Plants. 2022 Sep 8; PMID: 35817819. doi:10.1038/s41477-022-01179-x.
- Edelstein LC, Simon LM, Montoya T, Holinstat M, Chen ES, Bergeron A, Kong X, Nagalla S, Mohandas N, Cohen DE, et al. Racial differences in human platelet PAR4 reactivity reflect expression of PCTP and miR-376c [J]. Nat Med. 2013;19(12):1609–1616. doi:10.1038/nm.3385.
- R D MARKTESTER. Na+ tolerance and Na+ transport in higher plants [J]. Ann Bot. 2003;91(5):503–527. doi:10.1093/aob/mcg058.
- Camilla HILL, Jha, Deepa D, Bacic A, Tester M, Roessner U. Characterization of ion contents and metabolic responses to salt stress of different arabidopsis AtHKT1;1 genotypes and their parental strains [J]. Mol Plant. 2013;6(2):350–368. doi:10.1093/mp/sss125.
- Shinozaki K, Yamaguchi-Shinozaki K. Gene networks involved in drought stress response and tolerance [J]. J Exp Bot. 2007;58(2):221–227. doi:10.1093/jxb/erl164.
- ZHANG J, W JIA, Yang J, Ismail AM. Role of ABA in integrating plant responses to drought and salt stresses [J]. Field Crops Res. 2006;97(1):111–119. doi:10.1016/j.fcr.2005.08.018.