ABSTRACT
Different abiotic stresses induce OsTZF1, a tandem CCCH-type zinc finger domain gene, in rice. Here, we report that transgenic rice plants overexpressing OsTZF1 under own promoter (POsTZF1:OsTZF1-OX [for overexpression]) transferred to soil showed normal growth similar to vector control plants. The POsTZF1:OsTZF1-OX produced normal leaves without any lesion mimic phenotype and exhibited normal seed setting. The POsTZF1:OsTZF1-OX plants showed significantly increased tolerance to salt and drought stresses and enhanced post stress recovery. Microarray analysis revealed a total of 846 genes up-regulated and 360 genes down-regulated in POsTZF1:OsTZF1-OX salt-treated plants. Microarray analysis of POsTZF1:OsTZF1-OX plants showed the regulation of many abiotic stress tolerance genes. These results suggest that OsTZF1-OX under own promoter show abiotic stress tolerance and produces no pleiotropic effect on phenotype of transgenic rice plant.
Introduction
Zinc finger proteins are the most abundant and diverse families of plant regulatory proteins. Zinc finger proteins play an important role in certain cellular functions, containing transcriptional regulation, RNA binding, programmed cell death and protein–protein interactions.Citation1 Various kinds of zinc finger proteins are classified on the basis of cysteine (Cys) and histidine (His) residues that binds to zinc ion. Different types of these proteins are C2H2, C2C2, C2HC, C2C2C2C2, C2HCC2C2, and CCCH.Citation2–4
Tandem zinc finger proteins (TZF) are characterized by two similar C-x8-C-x5-C-x3-H motifs parted by 18 amino acids.Citation5 Three Cys residues and one His is linked to the zinc ion and each CCCH zinc finger has the ability to bind to the 5`-UAUU-3` half site of the class II AU-rich element (ARE) 5`-UAUUUAUU-3`.Citation6–8 In humans, ZFP36, ZFP36L1, and ZFP36L2 belong to TZF gene family. Insulin, serum and other growth factors cause the expression of ZFP36 also known as tristetraprolin (TTP).Citation7 TTP binds to several important regulators such as tumor necrosis factor at the 3`-UTR of AREs and cause mRNA degradation.Citation9 Tandem zinc finger proteins are also involved in epigenetic mRNA silencing by activating mRNA decay enzymesCitation10 as well as nucleate processing bodies (PBs).Citation11,Citation12
A genome-wide survey of CCCH-type zinc finger genes in maize (Zea mays L.) identified 68 CCCH genes [ZmC3H1-68; Citation13]. Similarly, 34 and 91 CCCH family genes have been identified in Medicago truncatula and popular plants, respectively.Citation14,Citation15 Through computational analysis 68 and 67 CCCH family genes were identified in Arabidopsis and rice, respectively.Citation16 These CCCH family genes were categorized into 11 sub-families in Arabidopsis and 8 subfamilies in rice.Citation16 Arginine-rich (RR) tandem CCCH zinc finger (RR-TZF) group comprise of a large subfamily IX.Citation17 Members in RR-TZF have been functionally characterized to function in seed maturation, germination and abiotic stress tolerance. AtTZF1 regulates abscisic acid (ABA), gibberellin and sugar-mediated growth and stress responses.Citation18 On the other hand, AtTZF2 and AtTZF3 are involved in jasmonate, ABA and oxidative stress responses.Citation19–21 Other TZF genes such as AtTZF4 (SOMNUS), AtTZF6 (PEI1) and AtTZF11/ AtTZF10 (AtSZF1/AtSZF2) function in light-dependent seed germination, embryogenesis and salt stress response, respectively.Citation22–24 OsDOS (also known as OsTZF2) was reported to delay leaf senescence in rice.Citation25 OsTZF5 confers drought tolerance and increased grain yield under drought stress.Citation26 OsTZF1 was reported to be induced by drought and salt stress conditions and resulted in delayed leaf senescence.Citation27
In this study the phenotype of OsTZF1 overexpression plants driven by own promoter (POsTZF1:OsTZF1-OX) were analyzed to examine if the OsTZF1 own promoter could overcome the pleiotropic effects previously reported in Ubi:OsTZF1-OX rice.Citation27 The POsTZF1:OsTZF1-OX rice plants were also subjected to salt and drought stress to test if OsTZF1 under own promoter confers tolerance against abiotic stresses.
Material and methods
Plant material and growth conditions
Rice (Oryza sativa ssp. japonica ‘Nipponbare’) was used in the present study. Initially, Seeds incubation was carried out at 42°C for 3 d in oven. Seeds were washed with sterilized distilled water and then sterilized with 0.2% HgCl2 solution. Seeds were rinsed five times with sterilized distilled water and then incubated on Murashige and Skoog (MS) media containing 25 mg/ml hygromycin. The germinated seedlings were transferred to soil for establishment and acclimatization under 12 hours light and dark cycles, flooded water and at 28°C.
Senescence analysis of POsTZF1:OsTZF1 transgenic rice
For leaf senescence testing, leaf fragments (6 cm) from 6 weeks old OsTZF1 transgenic (Ubi:OsTZF1-OX, POsTZF1:OsTZF1-OX) and vector control plants were taken. The leaf fragments were incubated at 27°C for 4 d in petri plates containing water, different hormones or NaCl solution. Hormones included 10 µM methyl jasmonate (MeJA), 100 µM salicylic acid (SA) and 10 µM ABA solutions. The salt solution used was 250 mM NaCl. After 4 d, the difference in leaf coloring of different transgenic plants was observed.Citation27
Stress tolerance of POsTZF1:OsTZF1 transgenic rice
For high-salt stress, 4 weeks old POsTZF1:OsTZF1 transgenic plants and vector control grown in soil were subjected to 250 mM sodium chloride (NaCl) solution for 5 d. After salt stress treatment, plants were watered with fresh water for 2 weeks to check the survival rate. The surviving and continuously growing plants were counted and examined.Citation27 For drought stress, 4 weeks old POsTZF1:OsTZF1 transgenic plants and vector control grown in soil were subjected to dehydration stress until the appearance of symptoms. After treatment, plants were watered for 2 weeks. The Plants maintaining continuous and suitable growth were counted and examined.Citation27
Transcriptomic analysis of POsTZF1:OsTZF1 transgenic rice
Microarray analysis was performed on 14 d old seedlings of the transgenic rice plants exposed to 2 days (2d) salt stress. Total RNAs were extracted from the harvested seedlings by TRIzole method/protocol. Cy3 and Cy5-labeled cDNA probes were made from the isolated total RNA. The probes were hybridized using the 44 K rice oligo microarray (Agilent Technologies). Briefly, two independent POsTZF1:OsTZF1 transgenic lines (OX#1 and OX#3) and one vector control were used. In each experiment, the reproducibility of the microarray analysis was measured by dye swapping. Using Feature Extraction software (version 10.10.1.1, Agilent Technologies), microarray slides were scanned and the data was analyzed after hybridization. Data analyses were carried out according to the Agilent methodology. Raw data was analyzed by Gene Spring GX software (version 11.5.1, Agilent Technologies). Lowess normalization method was used to normalize raw data.
Real-time quantitative PCR (qRT-PCR)
Real-time quantitative PCR was performed for four (two up-regulated and two down-regulated) randomly selected genes identified in microarray experiment. RNA was isolated from transgenic rice leaves using TRIzole method. Synthesis of cDNA was performed from RNA through Superscript II reverse transcriptase using oligo dT primer. The qRT-PCR was done for three independent biological replicates of salt-treated POsTZF1:OsTZF1 transgenic rice and vector control with specific primers, using ABI 3700/ABI Studio quant and SYBR Green fluorescent dye chemistry (Takara, Japan). The primers used for up-regulated (Os01g0871600 & Os03g0587200) and down-regulated genes (Os07g014755 & Os10g0409400) are given in the following table.
Statistical analysis
Statistical analysis was performed on the data obtained from salt and drought stress experiments. Statistical t-test was applied keeping level of significance less than 0.05 (P < .05).
Results
Phenotype of POsTZF1:OsTZF1 transgenic rice plants
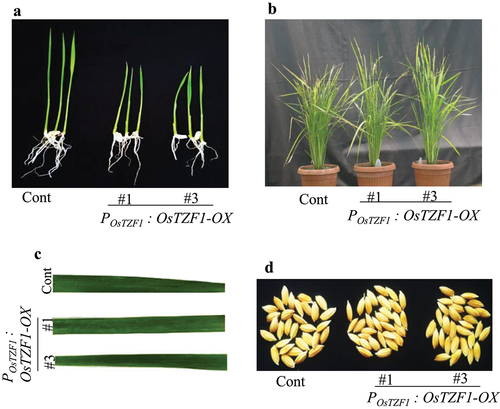
Ten days after seed germination, the growth of POsTZF1:OsTZF1-OX seedlings was relatively slow compared to vector control (). The POsTZF1:OsTZF1-OX seedlings transferred to soil showed normal growth and were similar in growth to vector control plants (). The POsTZF1:OsTZF1-OX showed no difference in stature, number of tillers and number of panicle in comparison to vector control (). However, 8 weeks after heading (WAH) leaf yellowing phenotype appeared in vector control while POsTZF1:OsTZF1-OX plants remained relatively green (data not shown). The leaf phenotype in vector control and POsTZF1:OsTZF1-OX was similar and no brown lesions/spots were observed in case of POsTZF1:OsTZF1-OX (). Furthermore, no difference in seed color of POsTZF1:OsTZF1-OX and vector control plants was observed at the time of harvest (). It is concluded that OsTZF1 gene driven under own promoter has no pleiotropic effect on the phenotype of rice.
Figure 1. Phenotype of POsTZF1:OsTZF1-OX and control plants. A) Rice seedling growth of control and POsTZF1:OsTZF1-OX after 10 d of germination. B) Transgenic rice POsTZF1:OsTZF1-OX (#1 and #3), and control plants grown in soil exhibited similar phenotype. C) Leaves from control and POsTZF1:OsTZF1-OX (#1 and #3) plants at seed setting stage. No brown lesions were observed. D) Phenotype of seeds harvested from control and POsTZF1:OsTZF1-OX (#1 and #3) at the time of harvest.
Delayed senescence exhibited by POsTZF1:OsTZF1 transgenic rice
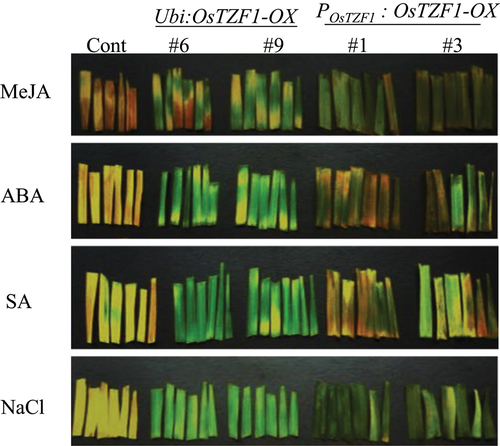
To study the role of OsTZF1 gene in leaf senescence, leaf fragments (6 cm) from 6-week old OsTZF1 transgenic (Ubi:OsTZF1-OX, POsTZF1:OsTZF1-OX) and vector control were taken and examined for senescence. For 10 µM MeJA treatment, the leaf fragments of Ubi:OsTZF1-OX (#6 and #9) and POsTZF1:OsTZF1-OX (#1 and #3) were compared to vector control. Under the treatment of 100 µM salicylic acid for 4 d in dark, the vector control leaf fragment appeared yellow while the leaf fragments of Ubi:OsTZF1-OX and POsTZF1:OsTZF1-OX remained green (). Similar results of delayed leaf senescence in case of OsTZF1-OX were observed under 10 µM ABA and 250 mM NaCl treatment (). These results showed that OsTZF1 driven under own promoter also delayed leaf senescence in rice compared to vector control.
Figure 2. Delayed leaf senescence phenotype of Ubi:OsTZF1-OX and POsTZF1:OsTZF1-OX compared to controls under different senescence inducing conditions. Leaf fragments from the OsTZF1-OX and control plants were incubated in 10 µM MeJA, 10 µM ABA, 100 µM SA and 250 mM NaCl solutions in the dark for 4 d.
Salt and drought stress tolerance of POsTZF1:OsTZF1 transgenic rice
Four week old plants grown in soil pots were irrigated with 250 mM NaCl solution for 5 d. The survival rates of POsTZF1:OsTZF1-OX were significantly higher than vector control. About 67% POsTZF1:OsTZF1-OX#1 and 69% POsTZF1:OsTZF1-OX#3 plants survived, whereas the survival rate of vector control plants was 30% ().
Figure 3. A) Salt-stress tolerance of POsTZF1:OsTZF1-OX OsTZF1-RNAi and control plants. The results are the average of three independent experiments with 12 plants per experiment. Asterisks indicate statistical significance (*, P < .050). B) Dehydration stress tolerance of POsTZF1:OsTZF1-OX, OsTZF1-RNAi and control plants. The results are the average of three independent experiments with 12 plants per experiment. Asterisks indicate statistical significance (*, P < .050).
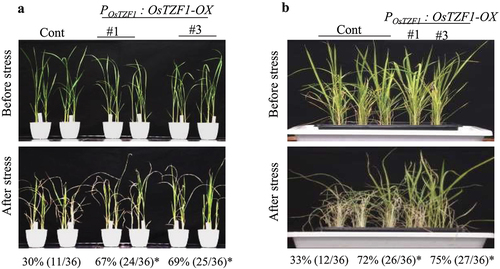
Four week old plants grown in pots were exposed to drought stress. The plants were revived from drought stress by re-watering. Symptoms like leaf rolling and wilting appeared earlier in vector control compared to POsTZF1:OsTZF1-OX. The survival rates of POsTZF1:OsTZF1-OX plants were significantly higher than vector control. The survival rate for POsTZF1:OsTZF1-OX#1 was 72% and POsTZF1:OsTZF1-OX#3 was 75% compared to 33% in vector control ().
Expression of stress related genes in POsTZF1:OsTZF1 transgenic rice
The POsTZF1:OsTZF1-OX plants showed increased tolerance to salt stress as shown in . Transcriptomic analysis was carried out using two POsTZF1:OsTZF1-OX lines (OX#1 and OX#3) and one vector control. Microarray analysis revealed that 1206 genes were regulated in POsTZF1:OsTZF1-OX transgenic lines compared to vector control by two-fold or greater than two-fold under 2 d salt stress. Among 1206 genes, 846 were up-regulated and 360 were down-regulated in POsTZF1:OsTZF1-OX salt stress treated plants. Previously, the microarray analysis of Ubi:OsTZF1-OX revealed the regulation of 4192 genes where 2051 genes were up-regulated and 2141 were down-regulated under 2 d salt stress.Citation27 Comparative transcriptome analysis of up-regulated genes in POsTZF1:OsTZF1-OX and Ubi:OsTZF1-OX revealed that 148 genes were co-expressed while 1903 genes and 598 genes were uniquely up-regulated in Ubi:OsTZF1-OX and POsTZF1:OsTZF1-OX, respectively (). Analysis of down-regulated genes in POsTZF1:OsTZF1-OX and Ubi:OsTZF1-OX showed that 134 genes were co-expressed while 2,007 and 226 genes were specifically down-regulated in Ubi:OsTZF1-OX and POsTZF1:OsTZF1-OX, respectively ().
Figure 4. Comparative transcriptome analysis of Ubi:OsTZF1-OX and POsTZF1:OsTZF1-OX up-regulated and downregulated genes under high salt stress. A) Venn diagram of Ubi:OsTZF1-OX and POsTZF1:OsTZF1-OX up- regulated genes, where 148 genes were co-expressed. B) Venn diagram of Ubi:OsTZF1-OX and POsTZF1:OsTZF1- OX down-regulated genes, where 134 genes were co-expressed. C) Heat map of 148 upregulated genes co- expressed in Ubi:OsTZF1-OX and POsTZF1:OsTZF1-OX under 250 mM NaCl stress. D) Heat map of 134 downregulated genes co-expressed in Ubi:OsTZF1-OX and POsTZF1:OsTZF1-OX under 250 mM NaCl stress.
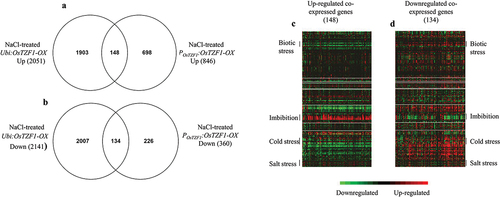
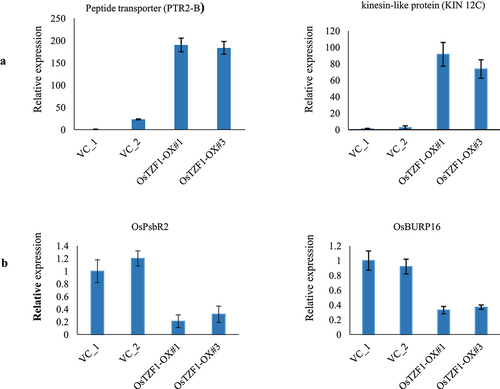
The heat map of co-expressed up-regulated genes (148) in Ubi:OsTZF1-OX and POsTZF1:OsTZF1-OX revealed that genes for biotic stress and cold stress were down-regulated while genes for imbibition and salt stress were up-regulated (). Similarly, heat map for co-expressed down-regulated genes (134) in Ubi:OsTZF1-OX and POsTZF1:OsTZF1-OX revealed that genes for biotic stress and cold stress were up-regulated while genes for imbibition and salt stress were down-regulated (). To validate the microarray, two genes each from top 30 up or down-regulated genes were selected randomly. Results showed that peptide transporter (PTR2-B) and kinesin-like protein (KIN-12C) genes were up-regulated and OsPsbR2 and OsBURP16 were down-regulated in POsTZF1:OsTZF1-OX comparison to vector control ().
Figure 5. qRT-PCR analysis of up-regulated genes identified in POsTZF1:OsTZF1-OX plants by microarray analysis. A) The analyzed upregulated genes were peptide transporter (Os01g0871600) and kinesin-like protein (Os03g0587200). B) The downregulated genes analyzed were OsPsbR2 (Os07g0147550) and OsBURP16 (Os10g0409400).
Discussion
Zinc finger proteins are involved in plant growth, development and stress responses through transcriptional regulation, RNA binding and protein–protein interactions.Citation1,Citation28 There are 67 CCCH zinc finger genes in rice divided into 8 subfamilies.Citation16 Recently, functional studies on some CCCH zinc finger genes have been performed, but much remains to be revealed. Here, we report the effect of OsTZF1 driven by own promoter in rice.
Previously, Citation27,reported that Ubi:OsTZF1-OX transgenic plants exhibited pleiotropic effects. Ubi:OsTZF1-OX plants showed reduced seed setting, delayed seed germination, retarded seedling growth, delayed leaf senescence, brown lesions on leaves and brownish seeds.Citation27 In similarity to Ubi:OsTZF1-OX plants, the seedling growth in POsTZF1:OsTZF1-OX plants was slow compared to vector control (). However, at subsequent stages such as mature vegetative or reproductive stages, the POsTZF1:OsTZF1-OX transgenic and vector control plants showed similar phenotype (). Neither any difference was observed in seeds or leaves phenotype between POsTZF1:OsTZF1-OX and vector control nor any brown lesions were observed on the leaves of POsTZF1:OsTZF1-OX plants (). The ectopic overexpression of OsDOS showed several pleiotropic developmental phenotypes such as delayed growth, shorter stature, abnormally developed panicles, deferred heading and severe sterility, as well as delayed leaf senescence.Citation25 Extremely stunted growth and reduced seed setting was observed in Ubi:OsTZF5-OX transgenic rice plants. However, expression of OsTZF5 under stress-inducible OsNAC6 promoter resulted in drought tolerance without negatively affecting growth in rice plants.Citation26 Maize ubiquitin promoter is considered a constitutive promoter and has broad spectrum expression patterns. Putative OsTZF1 promoter fragment is a stress inducible promoter. Previously, Jan et al., 2013, reported various cis-acting elements involved in the response to abiotic stresses in the 1,417-bp promoter region of OsTZF1.Citation27 The identified cis-acting elements were five ABA responsive elements [ACGTG; Citation29], three MYB core sequences [CNGTTR; Citation30] and four recognition sites for MYC [CANNTG; Citation31]. The OsTZF1 promoter also contained some putative cis-acting elements involved in the response to biotic stresses, including three WRKY71OS sequences [TGAC-containing W-box; Citation32] and seven W-boxes of different types, which are known as recognition sites for WRKY transcription factors. With the above information, putative promoter of OsTZF1was considered to be a good stress inducible promoter. Our results show that though OsTZF1 expressed under own promoter has slow seedling growth in the initial 10 d after germination, it has no other negative effect on the phenotype of rice plants and confers abiotic stress tolerance ().
Under different phytohormones such as ABA, MeJA, SA or NaCl stress, delayed leaf senescence was observed in POsTZF1:OsTZF1-OX and Ubi:OsTZF1-OX compared to vector control (). Previously, it was reported that delayed leaf senescence was associated with tolerance to oxidative stressCitation33. Ubi:OsTZF1-OX plants exhibited delayed leaf senescence under ABA, JA, SA, hydrogen peroxide and several abiotic stresses, showing that delayed senescence might be due to tolerance to oxidative stress.Citation27 As described earlier, the putative promoter of OsTZF1 is a stress inducible promoter, POsTZF1:OsTZF1-OX also exhibited delayed senescence phenotype under hormones tested or NaCl. Further characterization of POsTZF1:OsTZF1-OX transgenic plants may help to reveal its function under other stresses.
The POsTZF1:OsTZF1-OX plants exhibited enhanced salt and drought tolerance compared to vector control plants. The survival rates of OsTZF1-OX plants were significantly higher than vector control plants after recovery from salt and drought stress (). It was reported that overexpression of OsDRZ1 increased drought tolerance in rice seedling. The transgenic plants had accumulated high free proline and less reactive oxygen species and had enhanced antioxidant enzymes activity.Citation34 OsTZF5 over expression could confer drought tolerance and increased grain yield under drought stress.Citation26 Different transcription factors like OSISAP1, TFIIIA-type ZFP252, ZFP179, OsDREB1A, OsDREB1F and OsDREB2A have been reported that showed improved salinity and other abiotic stress tolerance in plants.Citation16,Citation35–39 Consistent with the previous results, our finding suggests that overexpression of OsTZF1 under own promoter could enhance salt and drought stress tolerance in rice.
Transcriptomic analysis revealed that a total of 1206 genes with two-fold change were regulated in POsTZF1:OsTZF1-OX, among which 846 genes were up-regulated and 360 genes were down-regulated under high salt stress. A total of 2,051 and 2,141 genes were up- and down-regulated, respectively, under salt stress in Ubi:OsTZF1-OX compared to vector control.Citation27 Comparative transcriptome analysis of salt-treated POsTZF1:OsTZF1-OX and Ubi:OsTZF1-OX revealed that 148 genes were co-expressed in up-regulated genes while 134 genes were co-expressed in down-regulated genes (). The heat map of co-expressed up-regulated genes revealed that genes for biotic stress and cold stress were down-regulated while genes belonging to biotic stress and cold stress were up-regulated in co-expressed down-regulated genes (). Jan et al., 2013, reported a contrasting expression pattern of genes to salt and drought stress response. The contrasting results indicate that OsTZF1-OX may show attenuated response to abiotic stresses (salt and drought stress).Citation27 For example, the atszf1-1 atszf2-1 double mutant also exhibited higher induction of stress-inducible genes compared to vector control and were less tolerant to high salt.Citation24 Further detailed transcriptomic analysis may help to shed light on the attenuated stress response of OsTZF1-OX transgenic plants to abiotic stresses.
Conclusion
POsTZF1:OsTZF1-OX exhibited no pleiotropic effects previously observed in Ubi:OsTZF1-OX transgenic plants. Delayed leaf senescence was also observed in POsTZF1:OsTZF1-OX compared to vector control under different stress inducing conditions. POsTZF1:OsTZF1-OX plants exhibited high tolerance to salt and drought stress. The expression of OsTZF1 under other stress-inducible promoters such as Oshox24 is suggested to generate stress tolerant plants without any affect on plant growth and development.
Acknowledgments
We thank K. Shinozaki (RIKEN) for their valuable advice; M. Kishimoto, K. Amano, E. Kishi, and K. Murai (JIRCAS) for their technical support.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Ciftci-Yilmaza S, Mittler R. The zinc finger network of plants. Cell Mol Life Sci. 2008;65(7–8):1150–7. doi:10.1007/s00018-007-7473-4.
- Klug A, Schwabe JW. Protein motifs 5. Zinc fingers. FASEB J. 1995;9(8):597–604. doi:10.1096/fasebj.9.8.7768350.
- Mackay JP, Crossley M. Zinc fingers are sticking together. Trends Biochem Sci. 1998;23(1):1–4. doi:10.1016/S0968-0004(97)01168-7.
- Sanchez-Garcia I, Rabbitts TH. The LIM domain: a new structural motif found in zinc-finger-like proteins. Trends Genet. 1994;10(9):315–320. doi:10.1016/0168-9525(94)90034-5.
- Blackshear PJ, Phillips RS, Lai WS. Tandem CCCH zinc finger proteins in mRNA binding. In: Iuchi S, Kuldell N, editors. Zinc finger proteins: from atomic contact to cellular function. London: Kluwer Academic/ Plenum Publishers; 2005. p. 80–90.
- Barreau C, Paillard L, Osborne HB, Paillard L, Osborne HB, Osborne HB, Paillard L, Osborne HB, Osborne HB, Osborne HB. AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res. 2005;33(22):7138–7150. doi:10.1093/nar/gki1012.
- Carrick DM, Lai WS, Blackshear PJ. The tandem CCCH zinc finger protein tristetraprolin and its relevance to cytokine mRNA turnover and arthritis. Arthritis Res Ther. 2004;6(6):248–264. doi:10.1186/ar1441.
- Hudson BP, Martinez-Yamout MA, Dyson HJ, Wright PE. Recognition of the mRNA AU-rich element by the zinc finger domain of TIS11d. Nat Struct Mol Biol. 2004;11(3):257–264. doi:10.1038/nsmb738.
- Carballo E, Lai WS, Blackshear PJ. Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science. 1998;281:1001–1005. doi:10.1126/science.281.5379.1001.
- Lykke-Andersen J, Wagner E. Recruitment and activation of mRNA decay enzymes by two ARE-mediated decay activation domains in the proteins TTP and BRF-1. Genes Dev. 2005;19(3):351–361. doi:10.1101/gad.1282305.
- Franks TM, Lykke-Andersen J. TTP and BRF proteins nucleate processing body formation to silence mRNAs with AU-rich elements. Genes Dev. 2007;21(6):719–735. doi:10.1101/gad.1494707.
- Stoecklin G, Anderson P. In a tight spot: ARE-mRNAs at processing bodies. Genes Dev. 2007;21(6):627–631. doi:10.1101/gad.1538807.
- Peng X, Zhao Y, Cao J, Zhang W, Jiang H, Li X, et al. CCCH type zinc finger family in maize: genome-wide identification, classification and expression profiling under abscisic acid and drought treatments. PLoS One. 2012;7:401–420.
- Chai G, Hu R, Zhang D, Qi G, Zuo R, Cao Y, Chen P, Kong Y, Zhou G. Comprehensive analysis of CCCH zinc finger family in poplar (Populus trichocarpa). BMC Genomics. 2012;13(1):1471–2164. doi:10.1186/1471-2164-13-253.
- Zhang C, Zhang H, Zhao Y, Jiang H, Zhu S, Cheng B, Xiang Y. Genome-wide analysis of the CCCH zinc finger gene family in Medicago truncatula. Plant Cell Rep. 2013;32(10):1543–1555. doi:10.1007/s00299-013-1466-6.
- Wang D, Guo Y, Wu C, Yang G, Li Y, Zheng C. Genome-wide analysis of CCCH zinc finger family in Arabidopsis and rice. BMC Genomics. 2008;9(1):44. doi:10.1186/1471-2164-9-44.
- Pomeranz MC, Hah C, Lin PC, Kang SG, Finer JJ, Blackshear PJ, Jang JC. The Arabidopsis tandem zinc finger protein AtTZF1 traffics between the nucleus and cytoplasmic foci and binds both DNA and RNA. Plant Physiol. 2010;152(2):151–165. doi:10.1104/pp.109.145656.
- Lin P, Pomeranz MC, Jikumaru Y, Kang SG, Hah C, Fujioka S, Kamiya Y, Jang JC. The Arabidopsis tandem zinc finger protein AtTZF1 affects ABA- and GA-mediated growth, stress and gene expression response. Plant J. 2011;65(2):253–268. doi:10.1111/j.1365-313X.2010.04419.x.
- Huang P, Chung MS, Ju HW, Na HS, Lee DJ, Cheong HS, Kim CS. Physiological characterization of the Arabidopsis thaliana oxidation-related zinc finger 1, a plasma membrane protein involved in oxidative stress. J Plant Res. 2011;124(6):699–705. doi:10.1007/s10265-010-0397-3.
- Huang P, Ju HW, Min JH, Zhang X, Chung JS, Cheong HS, Kim CS. Molecular and physiological characterization of the Arabidopsis thaliana oxidation-related zinc finger 2, a plasma membrane protein involved in ABA and salt stress response through the ABI2-mediated signalling pathway. Plant Cell Physiol. 2012;53(1):193–203. doi:10.1093/pcp/pcr162.
- Lee SJ, Jung HJ, Kang H, Kim SY. Arabidopsis zinc finger proteins AtC3H49/AtTZF3 and AtC3H20/AtTZF2 are involved in ABA and JA responses. Plant Cell Physiol. 2012;53(4):673–686. doi:10.1093/pcp/pcs023.
- Kim DH, Yamaguchi S, Lim S, Oh E, Park J, Hanada A, Kamiya Y, Choi G. SOMNUS, a CCCH-type zinc finger protein in arabidopsis, negatively regulates light-dependent seed germination downstream of PIL5. Plant Cell. 2008;20(5):1260–1277. doi:10.1105/tpc.108.058859.
- Li Z, Thomas TL. PEI1, an embryo-specific zinc finger protein gene required for heart-stage embryo formation in arabidopsis. Plant Cell. 1998;10(3):383–398. doi:10.1105/tpc.10.3.383.
- Sun J, Jiang H, Xu Y, Li H, Wu X, Xie Q, Li C. The CCCH-type zinc finger proteins AtSZF1 and AtSZF2 regulate salt stress responses in Arabidopsis. Plant Cell Physiol. 2007;48(8):1148–1158. doi:10.1093/pcp/pcm088.
- Kong Z, Li M, Yang W, Xu W, Xue Y. A novel nuclear-localized CCCH-type zinc finger protein, OsDOS, is involved in delaying leaf senescence in rice. Plant Physiol. 2006;141(4):1376–1388. doi:10.1104/pp.106.082941.
- Selvaraj MG, Jan A, Ishizaki T, Valencia M, Dedicova B, Maruyama K, Ogata T, Todaka D, Yamaguchi-Shinozaki K, Nakashima K, et al. Expression of the CCCH-tandem zinc finger protein gene OsTZF5 under a stress-inducible promoter mitigates the effect of drought stress on rice grain yield under field conditions. Plant Biotech J. 2020;18(8):1711–1721. doi:10.1111/pbi.13334.
- Jan A, Maruyama K, Todaka D, Kidokoro S, Abo M, Yoshimura E, Shinozaki K, Nakashima K, Yamaguchi-Shinozaki K. OsTZF1, a CCCH-tandem zinc finger protein, confers delayed senescence and stress tolerance in rice by regulating stress-related genes. Plant Physiol. 2013;161(3):1202–1216. doi:10.1104/pp.112.205385.
- Bogamuwa SP, Jang JC. Tandem CCCH zinc finger proteins in plant growth, development and stress response. Plant Cell Physiol. 2014;55(8):1367–1375. doi:10.1093/pcp/pcu074.
- Simpson SD, Nakashima K, Narusaka Y, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Two different novel cis -acting elements of erd1, a clpA homologous Arabidopsis gene function in induction by dehydration stress and dark-induced senescence. Plant J. 2003;33(2):259–270. doi:10.1046/j.1365-313X.2003.01624.x.
- Urao T, Yamaguchi-Shinozaki K, Urao S, Shinozaki K. An Arabidopsis myb homolog is induced by dehydration stress and its gene product binds to the conserved MYB recognition sequence. Plant Cell. 1993;5(11):1529–1539. doi:10.1105/tpc.5.11.1529.
- Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell. 2003;15(1):63–78. doi:10.1105/tpc.006130.
- Eulgem T, Rushton PJ, Robatzek S, Somssich IE. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000;5(5):199–206. doi:10.1016/S1360-1385(00)01600-9.
- Woo HR, Kim JH, Nam HG, Lim PO The Delayed Leaf Senescence Mutants of Arabidopsis, ore1, ore3, and ore9 are Tolerant to Oxidative Stress. Plant Cell Physiol. 2004;45(7): 923–932. 10.1093/pcp/pch110
- Yuan X, Huang P, Wang R, Li H, Lv X, Duan M, Tang H, Zhang H, Huang J. A zinc finger transcriptional repressor confers pleiotropic effects on rice growth and drought tolerance by down‐regulating stress‐responsive genes. Plant Cell Physiol. 2018;59(10):2129–2142. doi:10.1093/pcp/pcy133.
- Dubouzet JG, Sakuma Y, Ito Y, Kasuga M, Dubouzet EG, Miura S, Seki M, Shinozaki K, Shinozaki KY. OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, highsalt- and cold-responsive gene expression. Plant J. 2003;33(4):751–763. doi:10.1046/j.1365-313X.2003.01661.x.
- Mallikarjuna G, Mallikarjuna K, Reddy MK, Kaul T. Expression of OsDREB2A transcription factor confers enhanced dehydration and salt stress tolerance in rice (Oryza sativa L.). Biotechnol Lett. 2011;33(8):1689–1697. doi:10.1007/s10529-011-0620-x.
- Mukhopadhyay A, Vij S, Tyagi AK. Overexpression of a zinc-finger protein gene from rice confers tolerance to cold, dehydration, and salt stress in transgenic tobacco. Proc Natl Acad Sci USA. 2004;101(16):6309–6314. doi:10.1073/pnas.0401572101.
- Sun SJ, Guo SQ, Yang X, Bao YM, Tang HJ, Sun H, Huang J, Zhang HS. Functional analysis of a novel Cys2/His2-type zinc finger protein involved in salt tolerance in rice. J Exp Bot. 2010;61(10):2807–2818. doi:10.1093/jxb/erq120.
- Xu DQ, Hang J, Gao SQ, Yang X, Bao YM, Tang HJ, Zang HS. Overexpression of a TFIIIA type Zinc finger protein ZFP252 enhances drought and salt tolerance in rice (Oryza sativa L.). FEBS. Letters. 2008;582:1037–1043.
