ABSTRACT
Potassium (K+) plays a role in enzyme activation, membrane transport, and osmotic regulation processes. An increase in potassium content can significantly improve the elasticity and combustibility of tobacco and reduce the content of harmful substances. Here, we report that the expression analysis of Nt GF14e, a 14-3-3 gene, increased markedly after low-potassium treatment (LK). Then, chlorophyll content, POD activity and potassium content, were significantly increased in overexpression of Nt GF14e transgenic tobacco lines compared with those in the wild type plants. The net K+ efflux rates were severely lower in the transgenic plants than in the wild type under LK stress. Furthermore, transcriptome analysis identified 5708 upregulated genes and 2787 downregulated genes between Nt GF14e overexpressing transgenic tobacco plants. The expression levels of some potassium-related genes were increased, such as CBL-interacting protein kinase 2 (CIPK2), Nt CIPK23, Nt CIPK25, H+-ATPase isoform 2 a (AHA2a), Nt AHA4a, Stelar K+ outward rectifier 1(SKOR1), and high affinity K+ transporter 5 (HAK5). The result of yeast two-hybrid and luciferase complementation imaging experiments suggested Nt GF14e could interact with CIPK2. Overall, these findings indicate that NtGF14e plays a vital roles in improving tobacco LK tolerance and enhancing potassium nutrition signaling pathways in tobacco plants.
Introduction
Potassium is the most important and abundant cation in plant cells. Potassium plays an important role in enzyme activation, membrane transport, charge balance, osmotic regulation and other physiological processes.Citation1 Potassium is an activator of photosynthesis-related enzymes, oxidoreductases, and transferases in tobacco plants. Potassium participates in the metabolism of substances such as sugars, proteins, and nucleic acids; photosynthesis; energy metabolism; substance synthesis and transportation within tobacco plants; and the regulation of tobacco plant growth.Citation2 , Citation3 The potassium content in tobacco plants is an important indicator of tobacco quality. An increase in potassium content can significantly improve the elasticity and combustibility of tobacco, reduce the content of harmful substances such as tar and CO in smoke, and improve the quality of tobacco.Citation4 , Citation5
Potassium uptake by tobacco plants occurs mainly through an active absorption process with a reverse chemical gradient, which is carried out by a specialized K+ carrier protein (potassium pump) for transmembrane transport, as well as for providing sufficient organic acids and carbonic acid. K+ enters root hair epidermal cells from the soil solution, then enters small leaf vein cells and finally reaches mesophyll cells.Citation6 Previous studies have shown that the absorption of potassium by plant roots is an active process that requires energy. ATPase is a key enzyme in energy metabolism during the absorption and transport of substances in the root system. The activity of ATPase is a decisive factor in the exchange of nutrients and metabolites between the extracellular space and endosomes. The activity of ATPase is closely related to the absorption of potassium.Citation7
14-3-3 proteins are widely present and highly conserved in plant tissues, and in most higher plants, 14-3-3 proteins are typically encoded by 13–15 genes. These proteins exist in homologous or heterodimeric forms. 14-3-3 proteins can regulate plant growth and development by recognizing specific phosphorylation sequences and interacting with signaling proteins.Citation8 These signaling proteins include kinases, phosphatases, and transmembrane receptors. The binding of 14-3-3 proteins to phosphorylated proteins affects their activity, stability, conformation, and localization, thereby transmitting phosphorylation signals to target proteins for regulatory purposes.Citation9 , Citation10 The 14-3-3 protein can also bind to signal proteins through conformational modification and subsequently regulate physiological and biochemical reactions in plants.Citation10 , Citation11
The 14-3-3 protein, a highly conserved regulatory protein response to many
abiotic and biotic stress.Citation12 Overexpressing the chickpea Ca14-3-3 gene increased drought tolerance in Arabidopsis.Citation13 similar, overexpressing HaFT-1, a 14-3-3 protein from Haloxylon ammodendron, enhanced thermotolerance in Arabidopsis.Citation14 The overexpression At14-3-3 induces strong activation of K+ channels in the plasmalemma of mesophyll protoplasts.Citation15 Further research suggested that 14-3-3 proteins (14–3-3s) might regulate seed germination by controlling the movement of both the K+ in and K+ out of channels.Citation16 In K-deficient environments, the overexpression of 14-3-3κ can promote the growth of main roots in Arabidopsis.17. Currently, a total of 17 14-3-3 genes have been found in tobacco, one of which is involved in salt tolerance.Citation17 However, there have been no reports on the role of the 14-3-3 gene in tobacco potassium nutrition.
In the present study, we found that the overexpression of Nt GF14e in tobacco can enhance the chlorophyll content, POD activity and potassium contents under low potassium (LK) stress. Moreover, the net K efflux rates were severely lower in the transgenic tobacco plants than in the HD plants under LK stress. Under low-potassium treatment, 8495 genes were significantly differentially expressed (DEGs) between Nt GF14e overexpressing transgenic tobacco plants and HD plants, with 5708 upregulated genes and 2787 downregulated genes. The DEGs were enriched mainly in plant metabolic pathways such as carbon metabolism, oxidative phosphorylation, RNA transport, transcriptional regulation, and amino acid synthesis after analysis. The result of yeast two-hybrid and luciferase complementation imaging experiments suggested Nt GF14e could interact with CIPK2.In summary, our results revealed the molecular mechanism through which Nt GF14e increased LK tolerance in tobacco plants.
Materials and methods
NtGf14e cloning and tobacco transformation
Total RNA was isolated from tobacco plants (Nicotiana tabacum) that had been growing for one month and subsequently transferred into cDNA. Nt GF14e was cloned from cDNA via PCR according to the coding region sequence (accession XM_009590008.3). A plant transformation plasmid (pBI121- Nt GF14e) was constructed using the XbaI and SmaI restriction enzymes. Then, the cells were transformed into Agrobacterium GV3101. For tobacco tissue culture, tobacco seeds were sown in Murashige and Skoog (MS) agar media after surface sterilization and subsequently put into a growth chamber with a 16/8 h (day/night) photoperiod. The temperature was 26°C, and the light intensity was 200 μmol/mCitation2/s. The methods used for tobacco genetic transformation are described.Citation18
Tobacco growth conditions
Tobacco plants that had grown in the soil for 30 days were transferred to Hoagland culture media for 7 days. For the LK treatment, the tobacco plants were placed in a plastic box filled with LK nutrient solution (lacking K2SO4). For the control samples, the plants were grown in solution supplemented with 1 mM K+. The nutrient solution was prepared according to previous methods.Citation19 After 14 days of LK treatment, experimental samples were obtained for physiological measurements.
Subcellular localization analysis
For the subcellular localization experiment, a recombinant plasmid (pC2300-35S- Nt GF14e-GFP) was constructed. First, the complete coding sequence (CDS) of Nt GF14e was amplified with specific primers, and restriction enzymes (BamHI and SpeI) were used. The recombinant plasmid was subsequently transformed into Nicotiana benthamiana leaves with infiltration buffer as described previously.Citation20
Noninvasive microtest technique
Tobacco plants were grown in the soil for 30 days and subsequently transferred to Hoagland culture media for 7 days to adapt to the hydroponic conditions. Then, the tobacco seedling roots were rinsed after 3 hours of LK treatment with redistilled water, and the root hair zone was chosen for ion flux measurements; one example was measured for 5 min.The measurements were carried out using the SIET system BIO-003A (Younger USA Science and Technology). Net fluxes were calculated using JCal V3.2.2 (xuyue.net).
Quantitative real-time PCR
Nine genes closely related to low-potassium stress were selected for transcriptome data validation. For qRT‒PCR analysis, total RNA from tobacco samples was prepared using TRIzol reagent (Invitrogen, Carlsbad, USA), and cDNA synthesis was conducted utilizing the PrimeScript RT Kit with gDNA Eraser (TaKaRa) following the manufacturer’s protocols. The iTaq Universal SYBR Green Supermix (Bio-Rad) was used for the qRT‒PCR analysis. Elongation Factor 1 alpha-like (EF1αL) was used as an internal control, and the 2 −∆∆Ct method was used to calculate the relative gene expression level. The primer sequences for the nine genes are listed in Supplementary Table 2.
Yeast two-hybrid and luciferase complementary experiment
First, the full-length CDS of Nt GF14e was fused to the vector pGBKT7 with NcoI and BamHI restriction enzymes, and three CPKS were fused to pGADT7. Next, both pairs of plasmids (Nt GF14e-BD/CPK-AD) were cotransformed into the yeast strain AH109. PCR was used to identify positive clones. Then, the positive clones were placed on SD/-Leu-Trp-His plates to determine the extent of protein – protein interactions. AtCBL1 and AtCIPK23 were used as positive controls.Citation20 The recombinant vector of NtGF14e-JW772 and CIPKs- JW772 were transformed into Agrobacterium tumefaciens GV3101. The tobacco (Nicotiana benthamiana about 4 to 6 weeks) infected with Agrobacterium tumefaciens,then cultured in darkness at 25°C for 24 hours to check the luciferase complementary fluorescence signal.
Statistical data analysis
For statistical data analysis, the data presented here were obtained from three biological replicates. The significance of differences between samples was assayed using the F test and one-way ANOVA at p ≤ 0.01 and/or p ≤ 0.05. For the data analysis, statistical software programs (Excel and SPSS 14.0) were used.
Results
Expression analysis of the nt GF14e Gene
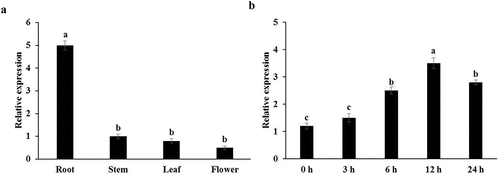
To study the function of the Nt GF14e gene, first, the expression level of the Nt GF14e gene in roots, stems, leaves, and flowers was analyzed via qRT‒PCR. The results showed that the expression level of the Nt GF14e gene was highest in the roots, followed by the stems, leaves, and flowers. The expression level of this gene in the roots of Nt GF14e was 17.67 times higher than that in the flowers (). Further analysis showed that the expression level of the Nt GF14e gene began to increase at 6 h and reached its highest level at 12 h after LK treatment, which was 8.75 times greater than that of the 0 h (). These findings indicate that Nt GF14e response to LK stress in tobacco.
Figure 1. Expression level analysis of the Nt GF14e gene. (a) Analysis of Nt GF14e expression in different tissues. (b) Analysis of Nt GF14e expression under LK treatment. The data are presented as the means ± SDs of three biological replicates. Different letters indicate significant differences at p < 0.05.
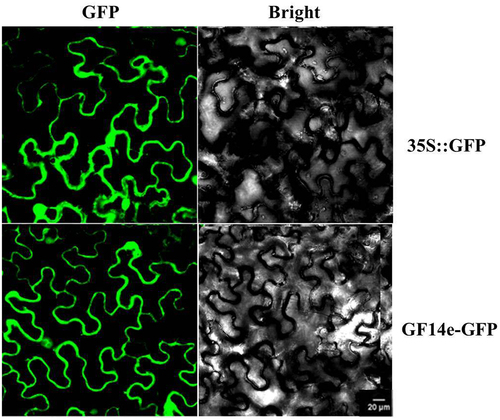
Subcellular localization analysis of nt GF14e
The subcellular localization and function of proteins are closely related. Nt GF14e-GFP was expressed in the mesophyll of Nicotiana benthamiana leaves.The subcellular localization results indicated that the green fluorescent protein (GFP) was not specific and distributed in various components of the cell (cell membrane, cytoplasm, and nucleus) in the tobacco mesophyll. Moreover, Nt GF14e-GFP was localized to the cell membrane and nucleus of tobacco mesophyll cells (), indicating that the protein may play a role in the cell membrane and nucleus.
GFP, Green fluorescent protein, bright, bright light.
Overexpression of nt GF14e in tobacco increases low-potassium tolerance
The qRT- PCR was used to measure the expression level of overexpressing
Nt GF14e tobacco plants, and the results showed that the expression level of Nt GF14e in the three transgenic tobacco lines was higher than that wild type (HD) (Figure S1). We subsequently evaluated the growth of the transgenic tobacco plants after LK treatment for 14 days, and the results showed that under normal cultivation conditions, there was almost no difference in growth between the HD plants and the three transgenic tobacco plant species (OE1, OE2, OE3). After LK treatment for 14 days, the three Nt GF14e overexpressing tobacco plants were more tolerant to LK stress than the WT plants, as the former had greener leaves and more roots (). Further experiments also showed that under normal cultivation conditions, there was almost no difference in the fresh weight of leaves or roots between the HD and the three transgenic tobacco plants. After LK treatment for 14 days, the fresh weight of the three tobacco plants overexpressing the Nt GF14e gene significantly increased compared to that of the HD (). Further research demonstrated that, after LK treatment for 14 days, the chlorophyll content and POD activity were significantly increased in Nt GF14e tobacco compared to those in HD (). At the same time, the potassium contents in the leaves and roots were also significantly increased in the three transgenic tobacco lines compared with those in the HD under LK stress (), The potassium contents increased by 1.60 and 1.76 -fold in the Nt GF14e -overexpressing tobacco leaves and roots. Indicating that overexpressing the Nt GF14e gene in tobacco plants results in greater tolerance to LK via increases in the chlorophyll content, POD activity and potassium content.
Figure 3. Analysis of transgenic Nt GF14e tobacco growth. (a) Observation of the growth phenotype. (b) Fresh weight of leaf and root. The data are presented as the means ± SDs of three biological replicates. Different letters indicate significant differences at p < 0.05. HD, control. OE1- OE3, overexpressing Nt GF14e tobacco.
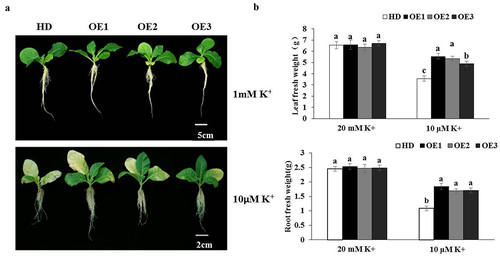
Figure 4. Determination of physiological indices (a) Chlorophyll content. (b) POD activity. (c) Potassium content in leaves. (d) Potassium content in roots. The data are presented as the means ± SDs of three biological replicates. Different letters indicate significant differences at p < 0.05. HD, wild type. OE1- OE3, overexpressing Nt GF14e tobacco.
Net K+ flux rate analysis of tobacco roots
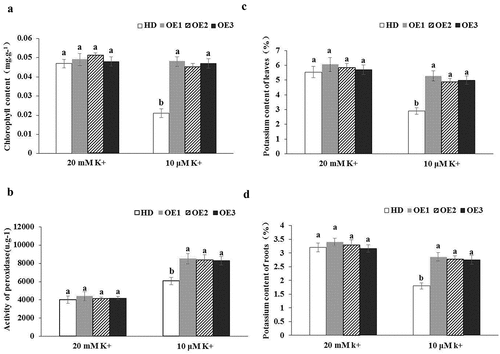
The tobacco plants were grown for 30 days in soil and subsequently transferred to Hoagland culture media for 7 days. The plants were subsequently incubated in LK solution for 3 h to measure the root net K+ flux for 5 minutes. The results suggested that the net K flux rates between the HD and OE1 roots were similar under normal nutrient solution (20 mM K+) (). Under LK stress (10μ M K+), the net K+ efflux rates were significantly lower in the OE1 line than in the WT ().
Figure 5. Measurement of the K+ flux rate in roots. (a) Net K+ flux rate analysis under control conditions. (b) Net K+ flux rate analysis under LK conditions. A positive value indicates K+ efflux. The data are presented as the means ± SDs of three biological replicates. HD, control. OE1, overexpressing Nt GF14e tobacco.
These results indicated that the overexpressing Nt GF14e tobacco absorb more K+ than the HD plants to adapt to LK stress.
Transcriptome analysis of transgenic tobacco plants
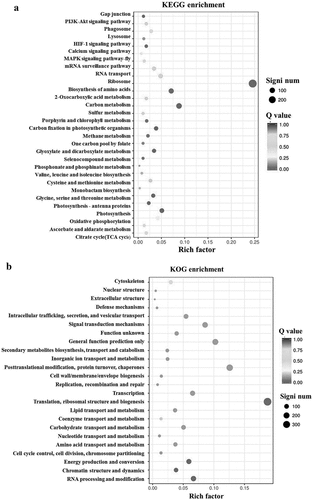
The transgenic tobacco plants that had been growing for 30 days in soil were subsequently transferred to Hoagland culture media for 7 days, after which one portion of the plants was cultured in LK culture media for 24 h for transcriptome analysis. There were 8495 upregulated DEGs in the Nt GF14e-overexpressing tobacco lines after analysis compared with those in HD. Among these DEGs, 5708 genes had elevated expression levels, and 2787 genes had decreased expression levels (Supplementary Table 1). KEGG and KOG pathway enrichment analyses of the DEGs were performed by bioinformatics analysis, and the results showed that ribosome (441), carbon metabolism (192), and biosynthesis of amino acids (156) were the three pathways with the most enriched genes in KEGG analysis (). At the same time, posttranslational modification, protein turnover, chaperones (666), translation, ribosomal structure and biogenesis (634), general function prediction only (520),signal transduction mechanisms (391) were the four pathways with the most enriched genes in KOG analysis ().
Analysis of potassium related gene expression levels
Based on the results of the transcriptome analysis, 9 genes closely related to low-potassium stress were selected for transcriptome data validation. qRT‒PCR results showed that the expression patterns of 7 genes, namely, Nt CPK2, Nt CPK23, Nt CPK25, Nt AHA2a, Nt AHA2a, Nt SKOR1, and Nt HAK5 were significantly upregulated after LK treatment, these results were consistent with the transcriptome data, and only the expression of TPK1 and KC1 were not consistent with the transcriptome data. In summary, 7 out of the 9 genes were consistent with the transcriptome data. Therefore, the transcriptome data were reliable ().
Figure 7. qRT‒PCR analyses of vital genes under potassium stress HD, control. OE1- OE3, overexpressing Nt GF14e tobacco.
Nt GF14e interacts with CIPK2 on protein level
Previous studies have indicated that the 14-3-3 protein interacts with CIPK in Arabidopsis, and the expression of Nt CIPK2, Nt CIPK23, and Nt CIPK25 increases according to the transcriptome analysis; therefore, we used a yeast two-hybrid assay to study the interaction between Nt GF14e and three CIPKs. The results of yeast two-hybrid assays indicated that Nt GF14e interacts with three CIPKs (CIPK2, CIPK23, and CIPK25) (). Subsequent luciferase complementation imaging experiment was used to study the interaction between Nt GF14e and three CIPKs.
Figure 8. Yeast two-hybrid and luciferase complementation assay. (a) Yeast two-hybrid; (b) luciferase complementation assay. AtCBL1 and AtCIPK23 were used as positive control, AD and BD were used as negative control. nLUC+cLUC were used as negative control.
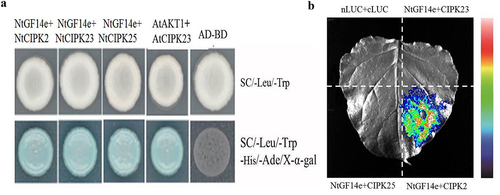
The results showed Nt GF14e could interact with CIPK2 not CIPK23 and CIPK25 (). Therefore, it is guessed that Nt GF14e and CIPK2 formed a complex to work together under LK stress.
Discussion
Overexpression Nt GF14e enhances tolerance to low potassium stress
According to the similarity of amino acid sequences in 14-3-3 proteins, these proteins can generally be separated into two categories: ε class and nonε class. The ε class contains ε, π, ρ, σ and μ type; all others are nonε class. In contrast to those of the ε class, the 14-3-3 genes of the ε class all have the same exon structure.Citation21 The Nt GF14e gene in this study has 5 exons and 4 introns and belongs to the ε group. Similarly, the number of exons and introns in the ε group of 14-3-3 genes is higher than that in the nonε group. Similar results were obtained for plants such as common kidney beansCitation22 and alfalfa.Citation23 The qRT‒PCR results in this study showed that the Nt GF14e gene was expressed mainly in the roots (). Similar results were found in hexaploid Brassica plants. The 14-3-3 gene of Brassica was also expressed mainly in the roots.Citation24 Further research has suggested that the Nt GF14e protein is distributed within the cell membrane, cytoplasm, and nucleus of tobacco epidermal cells (), similar to what has been observed for the At14-3-3 protein in Arabidopsis.Citation25 Therefore, the expression of the Nt GF14e protein is not specific.
In Arabidopsis, overexpression of 14-3-3κ promoted main root growth plants under potassium starvation stress.Citation26 Similarly, in our study, overexpression of Nt GF14e in tobacco plants enhanced potassium tolerance and increased root length; moreover, in the present study, the net K efflux rates were markedly lower in the Nt GF14e transgenic OE1 roots than those in the HD roots under LK stress (). Similar results were also reported in previous research; for example, overexpression of OsHAK5 improved the net K influx rate in rice roots.Citation27
Transcriptome analysis reveals key genes responsive to low potassium stress
To further explore the mechanism through which the Nt GF14e protein regulates tobacco adaptation to low-potassium stress, a transcriptome analysis of Nt GF14e transgenic tobacco was conducted after low-potassium treatment for 24 hours. Several studies have shown that 14-3-3 proteins regulate K+ channel activity in plants. Patch clamp experiments have shown that 14-3-3 proteins can increase the activity of outwardly rectifying K+ channels in the plasma membrane of tomato leaves and barley embryonic root cells.Citation28 After transcriptome data analysis, one voltage-gated potassium channel was downregulated in the Nt GF14e overexpression line compared with that the HD line in roots. In Arabidopsis, many related studies have elucidated the function of voltage-gated potassium channels, such as AKT1, AtKC1, GORK, and KAT2. ,Citation29 , Citation30 meanwhile, plant 14-3-3 proteins enhance GORK channel activity by interacting with the C-terminus of GORK.Citation31
Localizing of potassium ion channels in vacuoles is crucial for maintaining the stability of K ions in vivo.Citation32 Experiments have shown that TPK1 is a K+ channel protein that is independent of potential differences and activated by Ca2+. The interaction between 14-3-3 and TPK1 increases its activity and regulates the potassium ion balance in the cytoplasm.Citation33 In Arabidopsis, 14-3-3 proteins were found to colocalize on the vacuole membrane by binding to TPK1. So 14-3-3 proteins can migrate to different regions by binding to different target proteins to play function.Citation34 According to these transcriptome data, the expression level of two-pore potassium channel 5 (TPK5) in the roots is higher in the Nt GF14e overexpression line than in the HD line. Overexpressing Nt GF14e can promote TPK5 expression, enhance channel activity, and improve low potassium tolerance. We also found that HAK5 was downregulated in the Nt GF14e overexpression line compared with that in the HD line through root transcriptome analysis. Because AtCIPK1 and AtCIPK9 can regulate AtHAK5-mediated K+ uptake.Citation35 Therefore, we guessed that NtHAK5 was regulated mainly by posttranslational regulation in our study.
The Arabidopsis Na+(K+)/H+ reverse transporter proteins KEA1 and KEA2 belong to the K+ efflux antiporter (KEA) gene family. KEA4, KEA5, and KEA6 can regulate the pH and K+ homeostasis of the endometrial system and play important roles in protein transport to vacuoles.Citation36 K+ efflux antiporter 2 (KEA2) and KEA6 were upregulated in the Nt GF14e overexpression line compared with those in the HD, so we hypothesize that high expression levels of KEA2 and KEA6 can regulate endoplasmic reticulum pH and auxin homeostasis to enhance low potassium tolerance, similar to the functions of NHX5 and NHX6 in Arabidopsis.Citation37
Syntaxin (a synaptic fusion protein) can mediate membrane fusion between transport vesicles and target membranes, which play important roles in ion transport, development, geotropism, hormone signaling, defense activities, etc.Citation38 In tobacco, the mutant NtSYR1 protein affects K+ channel function to block stomatal closure.Citation39 Further research found that syp121 suppresses K+ channels to mediate K+ uptake during the stomatal opening process under high light intensities and low relative humidity conditions.Citation40 According to our transcriptome data, the expression levels of 5 syntaxins changed. In Arabidopsis, syntaxin (VAMP721) interacted with KAT1 and the KCI channel to regulate the K+ current and affected root growth.Citation41 The next step was to study 5 syntaxin-interacting proteins function and analyze the underlying mechanisms involved in low-potassium nutrition in tobacco plants.
In maize roots,the 14-3-3 protein activates proton pumps, leading to K+ influx and promoting water absorption, thereby participating in high-salt and high-drought stress responses.Citation42 The tomato 1TFT4 protein regulates H+-ATPase-mediated H+ output and indole-3-acetic acid (IAA) transport in the plasma membrane, helping primary root elongation to cope with alkaline stress under alkaline stress conditions.Citation43 In summary, 14-3-3 proteins are a complex class of regulatory proteins that interact with different target proteins to generate different regulatory mechanisms to adapt to low-potassium stress.Citation44
H+-ATPase plays a crucial role in cellular metabolism, participating in physiological processes such as water transport, nutrient uptake, stomatal opening and closing, etc. ATPase activity is regulated by environmental factors and hormone levels.Citation45 In our transcriptome study, the expression level of H+-ATPase changed, and the activity of ATPase in the roots was closely related to the rate of potassium absorption.Citation46 14-3-3 protein binds to H+- ATPase to form a complex that activates H+- ATPase.Citation47 In Arabidopsis, Further research has suggested interactions between the 14-3-3 protein and plasma membrane H+- ATPase to induce adventitious rooting in cucumber.Citation48
Protein kinase plays an hub role in low potassium stress
CPKs act as sensors for fluctuations in calcium signaling and are significant in signal transduction processes.Citation49 In our study, the expression levels of five CPKs were upregulated in the Nt GF14e overexpression line compared with those in the HD line after transcriptome analysis. Silencing the TaCPK7-D mutant improved K + uptake in wheat. Overall, these findings suggest that TaCPK7-D is a negative regulatory factor in the context of LK stress.Citation50 in tobacco,14-3-3 interacted with NtCDPK1 by identifying the unphosphorylated motif in NtCDPK1 using a new mode.Citation51 In Arabidopsis, three proteins (CPK1, CPK24 and CPK28) contain a canonical 14-3-3-binding motif; among them, the autophosphorylation site (Ser43) of CPK28 is in the 14-3-3-binding motif, which can increase the activity of 14-3-3 proteins.Citation52 So we guessed that high expression level of five CPKs in the Nt GF14e overexpression line could improve LK tolerance by phosphorylating target protein.
The CIPK family play vital roles during plant development and in response to environmental changes. AtCIPK23 complexes increased root K+ uptake through phosphorylation of HAK5 or AKT1 under different K+ concentrations in Arabidopsis.Citation20–25,Citation27–53 Similarly, in rice roots, OsCIPK23 complex also improved K+ uptake.Citation54 In tomato, SlCIPK23-LKT1 (AKT1 ortholog) pathway played vital roles in K+ accumulation in the cytosol.Citation55 Subsequently, the result of yeast two-hybrid and luciferase complementation imaging experiments suggested Nt GF14e could interact with CIPK2 in our study (). Because CIPK regulates target proteins primarily through phosphorylation,Citation56 Therefore, our subsequent research will focus on the mechanism underlying CIPK2 phosphorylation in Nt GF14e under LK stress.
Conclusion
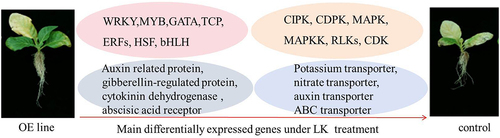
In this study, we reported that the expression level of Nt GF14e was highest in the roots. Overexpression of Nt GF14e in tobacco increased the fresh weight of the roots and leaves after LK treatment. Similarly, the chlorophyll content, potassium content, and POD activity were also significantly greater in Nt GF14e-overexpressing plants than in HD plants under LK stress. Transcriptome analysis showed 5708 upregulated genes and 2787 downregulated genes were identified in Nt GF14e-overexpressing tobacco.These genes were associated with kinases, transcription factors, reactive oxygen species, hormonal signaling and transporters. A genes work model in Nt GF14e overexpression line was summarized after LK stress (). The expression levels of several potassium-related genes, such as NtCIPK2, NtCIPK23, NtCIPK25, NtAHA2a, NtAHA4a, NtSKOR1, and NtHAK5, increased after LK stress according to the results of transcriptome qRT‒PCR analysis. The result of yeast two-hybrid and luciferase complementation imaging experiments suggested Nt GF14e could interact with CPK2. Overall, our findings provide a good reference for the study of the network through which Nt GF14e regulates LK stress in tobacco and other important crops.
OE line, Nt GF14e-overexpressing tobacco. Control, HD plant. MAPK, mitogen-activated protein kinase, MAPKK, mitogen-activated protein kinase kinase kinase; LKs, receptor-like serine/threonine-protein kinase, CDK, cyclin-dependent kinase.
Supplementary table 2.xlsx
Download MS Excel (9.9 KB)Supplementary table 1.xlsx
Download MS Excel (444.5 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/15592324.2024.2359257.
Additional information
Funding
References
- Ashley MK, Grant M, Grabov A. Plant responses to potassium deficiencies: a role for potassium transport proteinsm. Exp Bot. 2006;57(2):425–11. doi:10.1093/jxb/erj034.
- Evansh HJ. In potassium in biochemistry and physiology. Proc Colloq Int Potach Inst. 1971;8:13–29.
- Ramage CM, Williams RR. Mineral uptake in tobacco leaf discs during different developmental stages of shoot organogenesis. Plant Cell Rep. 2003;21(11):1047–1053. doi:10.1007/s00299-003-0628-3.
- Wang XQ, Wang BW, Song ZB, Zhao L, Ruan WY, Gao YL, Jia XQ, Yi KK. A spatial–temporal understanding of gene regulatory networks and NtARF-mediated regulation of potassium accumulation in tobacco. Planta. 2021;255(1):9. doi:10.1007/s00425-021-03790-2.
- Pi K, Luo W, Mo ZJ, Duan LL, Ke YZ, Wang PS, Zeng SB, Huang Y, Liu RX. Over dominant expression of related genes of ion homeostasis improves K+ content ad-vantage in hybrid tobacco leaves. BMC Plant Biol. 2022;22(1):335. doi:10.1186/s12870-022-03719-1.
- Epstein E, Rains WD, Elzam OE. Resolution of dual mechanisms of potassium absorption by barley roofs. Proc Natl Acad Sci USA. 1963;49(5):684–692. doi:10.1073/pnas.49.5.684.
- Memon AR, Saccomani M, Glass ADM. Efficiency of potassium utilization by barley varieties: The role of subcellular compartmentation. Exp Bot. 1985;36(12):1869–1876. doi:10.1093/jxb/36.1.79.
- Huang Y, Wang WS, Yu H, Peng JH, Hu ZR, Chen L. The role of 14-3-3 proteins in plant growth and response to abiotic stress. Plant Cell Rep. 2022;41(4):833–852. doi:10.1007/s00299-021-02803-4.
- Denison FC, Paul AL, Zupanska AK, Ferl RJ. 14-3-3 proteins in plant physiology. Semin Cell Dev Biol. 2011;22(7):720–727. doi:10.1016/j.semcdb.2011.08.006.
- Qi H, Lei X, Wang Y, Yu S, Liu T, Zhou SK, Chen JY, Chen QF, Qiu RL, Jiang LW. Xiao S.14-3-3 proteins contribute to autophagy by modulating SINAT-mediated of ATG13. Plant Cell. 2022;34(12):4857–4876. doi:10.1093/plcell/koac273.
- Janicka S, Augustyniak H. Multifunctional 14-3-3 proteins of plant cell. Postepy Biochemii. 2006;52(3):303–312.
- Ma Y, Wu ZY, Dong JF, Zhang SH, Zhao JL, Yang TF, Yang W, Zhou L, Wang J, Chen JS. et al. The 14-3-3 protein OsGF14f interacts with OsbZIP23 and enhances its activity to confer osmotic stress tolerance in rice. Plant Cell. 2023;35(11):4173–4189. doi:10.1093/plcell/koad211.
- Gupta S, Misra S, Kumar M, Mishra SK, Tiwar S, Anshu SN, Agrawal L, Chauhan PS, Chauhan PS. Enhancement of drought tolerance in transgenic Arabidopsis thaliana plants overexpressing Chickpea Ca14-3-3 gene. J Plant Growth Regul. 2023;42(3):1544–1557. doi:10.1007/s00344-022-10639-9.
- Pan R, Ren WJ, Liu SS, Zhang H, Deng X, Wang B. Ectopic over-expression of HaFT-1,a 14-3-3 protein from haloxylon ammodendron, enhances acquired thermotolerance in transgenic Arabidopsis. Plant Mol Biol. 2023;112(4–5):261–277. doi:10.1007/s11103-023-01361-5.
- Saalbach G, Schwerdel M, Natura G, Buschmann P, Dahse I, Dahse I. Over-expression of plant 14-3-3 proteins in tobacco: enhancement of the plasmalemma K + conductance of mesophyll cells 1 2. FEBS Lett. 1997;413(2):294–298. doi:10.1016/S0014-5793(97)00865-X.
- den Wijngaard Pw V, Sinnige MP, Roobeek I, Reumer A, Schoonheim PJ, Mo JN, Wang M, De Boer AH. Abscisic acid and 14-3-3 proteins control K+ channel activity in barley embryonic root. Plant Journal. 2005;41(1):43–55. doi:10.1111/j.1365-313X.2004.02273.x.
- He Y, Zhang Y, Chen L, Wu C, Luo Q, Zhang F, Wei Q, Li K, Chang J, Yang G. et al. A member of the 14-3-3 gene family in Brachypodium distachyon,BdGF14d,confers salt tolerance in transgenic tobacco plants. Front Plant Sci. 2017;8:340. doi:10.3389/fpls.2017.00340.
- Lu LM, Yang SY, Liu L, Lu YF, Yang SM, Liu F, Ni S, Zeng FC, Ren B, Wang XY. et al. Physiological and quantitative proteomic analysis of NtPRX63-overexpressing tobacco plants revealed that NtPRX63 functions in plant salt resistance. Plant Physiol Bioch. 2020;154:30–42. doi:10.1016/j.plaphy.2020.04.022.
- Li LQ, Lyu CC, Li JH, Wan CY, Liu L, Xie MQ, Zuo RJ, Ni S, Liu F, Zeng FC. et al. Quantitative proteomic analysis of alligator weed leaves reveals that cationic peroxidase 1 plays vital roles in the potassium deficiency stress response. Int J Mol Sci. 2020;21(7):2537. doi:10.3390/ijms21072537.
- Xu J, Li HT, Chen LQ, Wang Y, Liu LL, He L, Wu LH. A protein kinase, interacting with two calcineurin B-like proteins, regulates K+ transporter AKT1 in Arabidopsis.Cell. Cell. 2006;125(7):1347–1360. doi:10.1016/j.cell.2006.06.011.
- Sehnke PC, Delille JM, Ferl RJ. Consummating signal transduction: the role of 14-3-3 proteins in the completion of signal-induced transitions in protein activity. Plant Cell. 2002;14(1):339–354. doi:10.1105/tpc.010430.
- Tian FX, Wang T, Xie YL, Zhang J, Hu JJ, Pandey GK. Genome-wide identification, classification, and expression analysis of 14-3-3 gene family in Populus. PLoS One. 2015;10(4):e0123225. doi:10.1371/journal.pone.0123225.
- Qin C, Cheng LM, Shen JQ, Zhang YH, Cao HM, Lu D, Shen CJ. Genome-wide identification and expression analysis of the 14-3-3 family genes in Medicago truncatula. Front Plant Sci. 2016;7:320. doi:10.3389/fpls.2016.00320.
- Chandna R, Augustine R, Kanchupati P, Kumar R, Kumar P, Arya GC, Bisht NC. Class specific evolution and transcriptional differentiation of 14-3-3 family members in mesohexaploid Brassica rapa. Front Plant Sci. 2016;7:1–10. doi:10.3389/fpls.2016.00012.
- Paul AL, Sehnke PC, Ferl RJ. Isoform-specific subcellular localization among 14-3-3 proteins in Arabidopsis seems to be driven by client interactions. Mol Biol Cell. 2005;16(4):1735–1743. doi:10.1091/mbc.e04-09-0839.
- Shin R, Alvarez S, Burch AY, Jez JM, Schachtman DP. Phosphoproteomic identification of targets of the Arabidopsis sucrose nonfermenting-like kinase SnRK2.8 reveals a connection to metabolic processes. Proc Natl Acad Sci USA. 2007;104(15):6460–6465. doi:10.1073/pnas.0610208104.
- Yang TY, Zhang S, Hu YB, Wu FC, Hu QD, Chen G, Cai J, Wu T, Moran N, Yu L. et al. The role of a potassium transporter OsHAK5 in potassium acquisition and transport from roots to shoots in rice at low potassium supply levels. Plant Physiol. 2014;166(2):945–959. doi:10.1104/pp.114.246520.
- Paul WJ, Wijngaard VD, Mark PS, Roobeek I, Reumer A, Peter JS, Jos NMM, Wang M, Albertus HDB. Abscisic acid and 14-3-3 proteins control K channel activity in barley embryonic root. Plant. 2004;41(1):43–55. doi:10.1111/j.1365-313X.2004.02273.x.
- Wang Y, Wu WH. Regulation of potassium transport and signaling in plants. Curr Opin Plant Biol. 2017;39:123–128. doi:10.1016/j.pbi.2017.06.006.
- Elsa RZ, Claire CF, Frédéric SC, Christian B, Tou CH. Ca2+-dependent protein kinase 6 enhances KAT2 shaker channel activity in Arabidopsis thaliana. Int J Mol Sci. 2021;22(4):1596. doi:10.3390/ijms22041596.
- Kleeff PJMV, Gao J, Mol S, Zwart N, Zhang H, Li KW, Boer AHD. The Arabidopsis, GORK K+-channel is phosphorylated by calcium-dependent protein kinase 21 (CPK21), which in turn is activated by 14-3-3 proteins. Plant Physiol Biochem. 2018;125:219–231. doi:10.1016/j.plaphy.2018.02.013.
- Isayenkov S, Isner JC, Frans JMM. Rice two-pore K+ channels are expressed in different types of vacuoles. Plant Cell. 2011;23(2):756–768. doi:10.1105/tpc.110.081463.
- Latz A, Becker D, Hekman M, Müller T, Beyhl D, Marten IE, Fischer C, Dunkel A, Bertl M, A RU. et al. TPK1, a Ca2+-regulated Arabidopsis vacuole two-pore K+ channel is activated by 14-3-3 proteins. Plant Journal. 2007;52(3):449–459. doi:10.1111/j.1365-313X.2007.03255.x.
- Takeo K, Ito T. Subcellular localization of VIP1 is regulated by phosphorylation and 14-3-3 proteins. FEBS Lett. 2017;591(13):1972–1981. doi:10.1002/1873-3468.12686.
- Lara A, Ródenas R, Andrés Z, Martínez V, Quintero FJ, Nieves-Cordones M, Botella MA, Rubio F, Dietz K-J. Arabidopsis K+ transporter HAK5-mediated high-affinity root K+ uptake is regulated by protein kinases CIPK1 and CIPK9. J Exp Bot. 2020;71(16):5053–5060. doi:10.1093/jxb/eraa212.
- Zhu X, Pan T, Zhang X, Fan L, Quintero FJ, Zhao H, Su X, Li X, Villalta I, Mendoza I. et al. K+ efflux antiporters 4, 5, and 6 mediate pH and K+ homeostasis in endomembrane compartments. Plant Physiol. 2018;178(4):1657–1678. doi:10.1104/pp.18.01053.
- Fan L, Zhao L, Hu W, Li W, Novák O, Strnad M, Simon S, Friml J, Shen J, Jiang L. et al. Na+, K+/H+ antiporters regulate the pH of endoplasmic reticulum and auxin-mediated development. Plant, Cell & Environ. 2018;41(4):850–864. doi:10.1111/pce.13153.
- Ebine K, Okatani Y, Uemura T, Goh T, Shoda K, Niihama M, Morita MT, Spitzer C, Otegui MS, Ueda NT. A snare complex unique to seed plants is required for protein storage vacuole biogenesis and seed development of Arabidopsis thaliana. Plant Cell. 2008;20(11):3006–3021. doi:10.1105/tpc.107.057711.
- Leyman B, Geelen D, Quintero FJ, Blatt MR. A tobacco syntaxin with a role in hormonal control of guard cell ion channels. Science. 1999;283(5401):537–540. doi:10.1126/science.283.5401.537.
- Eisenach C, Chen ZH, Grefen C, Blatt MR. The trafficking protein SYP121 of Arabidopsis connects programmed stomatal closure and K+ channel activity with vegetative growth. Plant Journal. 2012;69(2):241–251. doi:10.1111/j.1365-313X.2011.04786.x.
- Zhang B, Karnik R, Wang YZ, Wallmeroth N, Blatt MR, Grefenb C. The Arabidopsis R-SNARE VAMP721 interacts with KAT1 and KC1 K+ channels to moderate K+ current at the plasma membrane. Plant Cell. 2015;27(6):1697–1717. doi:10.1105/tpc.15.00305.
- Shanko AV, Mesenko MM, Klychnikov OI, Nosov AV, Ivanov VB. Proton pumping in growing part of maize root: its correlation with 14-3-3 protein content and changes in response to osmotic stress. Biochemistry (Mosc). 2003;68:1320–1326. doi:10.1023/B:BIRY.0000011653.46422.c3.
- Xu WF, Jia LG, Shi WM, Balujka F, Kronzucker HJ, Liang JS, Zhang JH. The tomato 14-3-3 protein’TFT4 modulates Heflux, basipetal auxin transport, and the PKS5-J3 pathway in the root growth response to alkaline stress. Plant Physiol. 2013;163(4):1817–1828. doi:10.1104/pp.113.224758.
- Xiao B, Smerdon SJ, Jones DH, Dodson GG, Soneji Y, Aitken A, Gamblin SJ. Structure of a 14-3-3 protein and implications for coordination of multiple signaling pathways. Nature. 1995;376(6536):188–191. doi:10.1038/376188a0.
- Olsson A, Svennelid F, Ek B, Sommarin M, Larsson C. A phosphothreonine residue at the C-terminal end of the plasma membrane H+-ATPase is protected by fusicoccin-induced 14-3-3 binding. Plant Physiol. 1998;118(2):551–555. doi:10.1104/pp.118.2.551.
- Alamgir ANM. Effect of transpiration on the rate of absorption of potassium in some crop plants. Indian J Plant Physiol. 1997;2(3):239–241.
- Jahn T, Fuglsang AT, Olsson A, Brüntrup IM, Collinge DB, Volkmann D, Sommarin M, Palmgren MG, Larsson C. The 14-3-3 protein interacts directly with the C-terminal region of the plant plasma membrane H+-ATPase. Plant Cell. 1997;9(10):1805–1814. doi:10.1105/tpc.9.10.1805.
- Li CX, Huang DJ, Wang CL, Wang N, Yao YD, Li WF, Liao WB. NO is involved in H2-induced adventitious rooting in cucumber by regulating the expression and interaction of plasma membrane H+-ATPase and 14-3-3. Planta. 2020;252(1):9. doi:10.1007/s00425-020-03416-z.
- Chen YX, Zhou XJ, Chang S, Chu ZL, Wang HM, Han SC, Wang YD. Calcium-dependent protein kinase 21 phosphorylates 14-3-3 proteins in response to ABA signaling and salt stress in rice. Biochem Bioph Res Co. 2017;493:1450–1456. doi:10.1016/j.bbrc.2017.09.166.
- Lei XT, Chen MM, Xu K, Sun RX, Zhao SH, Wu NJ, Zhang SH, Yang HJ, Xiao K, Zhao Y. The miR166d/tacpk7-D signaling module is a critical mediator of wheat (Triticum aestivum L.) tolerance to K+ deficiency. Int J Mol Sci. 2023;24(9):7926. doi:10.3390/ijms24097926.
- Ito T, Nakata M, Fukazawa J, Ishida S, Takahashi Y. Phosphorylation-independent binding of 14-3-3 to NtCDPK1 by a new mode. Plant Signal Behav. 2014;9(12):e977721. doi:10.4161/15592324.2014.977721.
- Swatek KN, Wilson RS, Ahsan N, Tritz RL, Thelen JJ. Multisite phosphorylation of 14-3-3 proteins by calcium-dependent protein kinases. Biochem (Moscow). 2014;459(1):15–25. doi:10.1042/BJ20130035.
- Ragel P, Ródenas R, García-Martín E, Andrés Z, Villalta I, Nieves-Cordones M, Rivero RM, Martínez V, Pardo JM, Quintero FJ. CIPK23 regulates HAK5-mediated high-affinity K+ uptake in Arabidopsis roots. Plant Physiol. 2015;169:2863–2873. doi:10.1104/pp.15.01401.
- Li J, Long Y, Qi GN, Li J, Xu ZJ, Wu WH, Wang Y. The Os-AKT1 channel is critical for K+ uptake in rice roots and Is modulated by the rice CBL1-CIPK23 complex. Plant Cell. 2014;26(8):3387–3402. doi:10.1105c.114.123455.
- Amo J, Lara A, Martinez-Martinez A, Martinez V, Rubio F, Nieves-Cordones M. The protein kinase SlCIPK23 boosts K+ and Na+ uptake in tomato plants. Plant, Cell Environment. 2021;44(12):3589–3605. doi:10.1111/pce.14189.
- Saito S, Uozumi N. Calcium-regulated phosphorylation systems controlling uptake and balance of plant nutrients. Front Plant Sci. 2020;11:44. doi:10.3389/fpls.2020.00044.