ABSTRACT
The CLAVATA pathway plays a key role in the regulation of multicellular shoot and root meristems in flowering plants. In Arabidopsis, CLAVATA 3-like signaling peptides (CLEs) act via receptor-like kinases CLAVATA 1 and CRINKLY 4 (CR4). In the moss Physcomitrium patens, PpCLAVATA and PpCR4 were previously studied independently and shown to play conserved roles in the regulation of cell proliferation and differentiation. The plant calpain DEFECTIVE KERNEL 1 (DEK1) has been identified as another key regulator of cell division and cell fate in vascular plants and bryophytes. The functional interaction between CLAVATA, CR4, and DEK1 remains unknown. Here, we show that P. patens crinkly4 and dek1 mutants respond differently to CLE peptide treatments suggesting their distinct roles in the CLAVATA pathway. Reduced CLAVATA-mediated suppression of leafy shoot growth in Δcr4 mutants indicates that PpCR4 is involved in CLV3p perception, most likely as a receptor. The CLV3p strongly suppressed leaf vein development in Δcr4 mutants, suggesting that other receptors are involved in these processes and indicating a potential role of PpCR4 in organ sensitization to CLEs.
Introduction
Cell-to-cell communication plays a critical role in the differentiation and maintenance of distinct cell layers, consequently affecting multicellular body size and shape. In plants, the CLAVATA pathway represents a receptor kinase:ligand-based intercellular signaling mechanism that governs multiple developmental programs, including the organization of multicellular shoot and root apical meristems.Citation1–6 Arabidopsis CLAVATA signaling involves CLAVATA 1 (CLV1) and CLV2 leucine-rich repeats kinases that act as receptors to the CLV3 signal peptide. The family of CLV3-like peptides comprises 32 members in Arabidopsis thaliana,Citation7,Citation8 acting through diverse receptor kinases.Citation2,Citation9,Citation10 For instance, CLV3 peptides signal via RECEPTOR-LIKE PROTEIN KINASE 2/TOADSTOOL 2 (RPK2/TOAD2) homodimers and heteromultimers of CLV1 with its homologs BARELY ANY MERISTEM 1 (BAM1) and BAM2.Citation11–13 CLAVATA signaling controls spatial restriction of the WUSCHEL transcription factor (WUS), one of the key regulators of stem cell activity in plant shoot apices.Citation14,Citation15 Beyond the shoot apical meristem, roles of CLV1, CLV2, and CLEs in leaf and fruit development have been reported.Citation16 In roots, CLEs act via CLV1, CLV2, and the tumor necrosis factor-like receptor kinase CRINKLY 4 (CR4) to confine WUS-related homeobox transcription factor expression and thus spatially control root apical meristem organization.Citation2,Citation4,Citation17 Originally, CR4 function was identified in maize, where it affects the epidermis and endosperm aleurone layer differentiation.Citation18,Citation19 Following studies in monocots and dicots shown that CR4 kinase indeed plays an important role in epidermal (L1) layer formation in diverse organs.Citation20–23 In A. thaliana, heterocomplexes of CLV1 and CR4 (also known as Arabidopsis CRINKLY 4, ACR4) have been described to control stem cell specification in root apical meristem.Citation17 ACR4, and its homologs in monocots, have been implicated in surrounding tissue formation such as ovule integuments, endosperm aleurone layer, and leaf epidermis.Citation20,Citation22,Citation24–27 The membrane-anchored calpain protease DEFECTIVE KERNEL 1 (DEK1) has been identified as a key regulator of division plane orientation, stem cell formation, and epidermal cell fate specification.Citation23,Citation26,Citation28–30 DEK1 and CR4 colocalized to plasma membrane and endosomes together with a class E vacuolar sorting protein SAL1.Citation26
Genetic analyses in the moss Physcomitrium patens, a bryophyte sister of vascular plants, shed more light on the evolutionary developmental aspects of CLAVATA signaling. Unlike flowering plant sporophytes that have multilayered meristems, single apical stem cells contribute to body formation in moss gametophytes.Citation31 In P. patens, the life cycle begins with a haploid spore that germinates to produce filamentous protonemata. Protonemata form branched filaments composed of caulonema and chloronema cells.Citation32,Citation33 Distinct protonemal side-branched initial cells acquire different identitiesCitation34 and after a series of precisely oriented asymmetric divisions form a bud with an apical stem cell giving rise to a leafy gametophore.Citation35 P. patens phyllids (for simplicity hereafter called leaves) are composed of a single cell-layered lamina, and a more complex midrib composed of specialized supporting and water-conducting cells.Citation36,Citation37 Reproductive organs (gametangia) are formed at the gametophore apex, where after fertilization, a sporophyte develops, producing haploid spores to close the life cycle.Citation38,Citation39
The P. patens CLAVATA pathway involves CLV3-like peptides encoded by PpCLE1–9 genes and PpCLV1a, PpCLV1b, and PpRPK2 receptor-like kinases.Citation40 Based on genetic analyses, it was proposed that PpRPK2 regulates the distribution of the plant hormone auxin, thereby affecting stem cell activity and growth in filamentous protonemata.Citation41 CLAVATA plays a critical role during the transition from filamentous to complex three-dimensional growth by controlling oriented cell divisions leading to gametophore stem cell formation.Citation40 Downregulation of genes encoding PpCLE peptides as well as loss-of-function of PpCLV1 and PpRPK2 led to uncontrolled cell proliferation and developmental defects during vegetativeCitation40,Citation42 and reproductiveCitation43 development. The role of CLAVATA in the regulation of cell division plane orientation and cell proliferation is conserved between P. patens and Arabidopsis thaliana as revealed by comparative analyses of clv/bam mutants and cross-sensitivity to applied CLE peptides from both species.Citation40 A recent gene regulatory network analysis in P. patens identified a PpDEK1-controlled regulon affecting the CLAVATA pathway.Citation44 In P. patens, deletion of PpCR4 caused developmental defects in leafy gametophores, reproductive organs, and sporophytes.Citation45 Despite its critical role as a vital component of the CLAVATA pathway within root apices of Arabidopsis, the functional association between CR4 and CLEs in upper ground organs of angiosperms and in bryophytes remains unclear.
Here, we investigated whether PpCR4 and PpDEK1 act with CLAVATA to regulate leafy gametophore development in P. patens. We tested the effect of applied CLV3 and PpCLE peptides on P. patens plants lacking PpCR4 (Δcr4) and a mutant with modified PpDEK1 function (dek1_Δlg3), each with distinctly affected leaf development. The results indicate that CLE peptides specifically affect gametophore development in the absence of PpCR4 kinase, causing developmental arrest of leaves at their juvenile (basal) state.
Material and methods
The Gransden wild-type (WT) strain of Physcomitrium patens,Citation46 the Δcr4 mutant strain,Citation45 and dek1_ Δlg3 mutant strainCitation47 were used (all strains available in V.D. lab from previous works). Plant material was cultivated on sterile BCDAT media at 23°C in continuous light at 30–50 µmol m−2 s−1 photosynthetically active radiation in Sanyo MLR-351 growth cabinets. Synthetic CLE peptides (Genecust, >95% purity, CLV3: RTVPSGPDPLHH, PpCLE1,2,3: RMVPTGPNPLHN, and Random peptides: PHHLPGPSTRDV)Citation40 were dissolved in phosphate buffer (50 μM, pH 6, 8) to stock concentration of 10 mM and supplemented to BCDAT media to final concentration of 10 μM. To assess whole plant and gametophore phenotypes, 4-week-old spot cultures were imaged using a Keyence VHX-1000E digital microscope with a 20–50× or 50–200× objective. Procreate and ImageJ 1.54f softwares allowed cell counting, area quantification, and heatmapping. Biorender.com Beta version was used for data representation. Analysis of variance was performed using the STATGRAPHICS CENTURION V. 15 software. Boxplots show interquartile range with the median at the center and delimited by minimum and maximum values that are not outliers. The bar graphs represent the mean and the standard error of the mean (SE).
Results and discussion
crinkly4 and dek1 mutants are sensitive to CLV3-like peptides
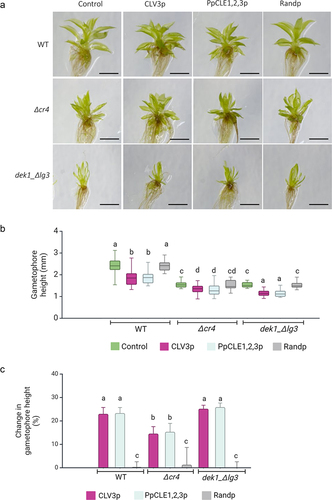
P. patens WT plants, mutants with a deleted PpCR4 gene (Δcr4), and mutants lacking the LG3-like domain in PpDEK1 (dek1_ Δlg3) were treated with CLV3 peptide (CLV3p), PpCLE1,2,3p, and random peptides, respectively (). In WT plants, the growth of gametophores was reduced in response to CLV3p (23.01%) and PpCLE1,2,3p (23.32%) (), while no significant change in the overall morphology and gametophore height was determined in the presence of control random peptides. Δcr4 mutant gametophores are smaller than WT gametophores but showed a further growth reduction upon CLV3p treatment (14.57%) as well as PpCLE1,2,3 treatment (15.34%). No significant change in gametophore height was determined in the presence of random peptides (). Gametophore development in dek1_ Δlg3 mutant is severely affected compared to WT (for details, see Johansen et al., 2016),Citation47 with reduced stem height and narrow leaves. Nevertheless, CLE3p and PpCLE1,2,3p caused further 25.19% and 25.88% gametophore height reduction respectively, and these mutants thus showed a normal response to CLEs (). The relative reduction of CLAVATA-mediated suppression of gametophore height in Δcr4 mutants () indicates that PpCR4 is required in peptide signaling, most likely acting as a receptor to CLEs.
Figure 1. The reduced gametophore height response to CLV3 and PpCLE1,2,3 peptides depends on PpCR4.
CLV3 peptides differently affect leaf morphogenesis in Δcr4 and dek1_Δlg3 mutants
The PpDEK1 calpain protease is essential for early asymmetric cell divisions that give rise to the gametophore apical stem cell.Citation30,Citation48 Based on recent gene regulatory network analyses, it has been proposed that DEK1 mediates positional signaling that involves CLAVATA regulon to control phase transition from filamentous growth to three-dimensional growth. Here, PpDEK1 seems to act in a feedback control of PpCLV1b expression separately from a PpRPK2-mediated pathway.Citation44 Previously, it has been shown that calpain activity in dek1_ Δlg3 is sufficient to maintain gametophore apical stem cell; however, it fails to maintain cell proliferation during leaf development.Citation47 Morphologically, the leaves of dek1_ Δlg3 mutant were not significantly changed after CLV3p treatment, which supports upstream-acting and fine-tuning role of PpDEK1 during leaf morphogenesis. However, CLV3 and PpCLE1,2,3 peptide treatments caused specific morphological changes in Δcr4 leaves.
In WT P. patens plants, the leaves are morphologically different depending on their position along the apical-basal axis of gametophore, a phenomenon known as heteroblasty.Citation49,Citation50 The earliest developed juvenile leaves at the gametophore base are morphologically simpler compared to successively formed leaves positioned toward the apex. They are oblong in shape and mostly formed of a single-layered lamina, lacking a vein. The transition to leaves positioned toward the middle of gametophores is associated with the formation of a vein and development of elongated marginal serrated cells. Cell size along the longitudinal axis of mature leaves also varies with elongated cells at the base and isodiametric cells toward the apex. Newly formed apical leaves contain small proliferating cells at the base and elongated cells toward the tip. Closer examination of isolated leaves from Δcr4 and dek1_Δlg3 mutants revealed distinct responses to exogenous peptides when compared to WT plants. As the effects of both CLV3p and PpCLE1,2,3p were similar, we focused on CLV3p-treated plants for more detailed observations.
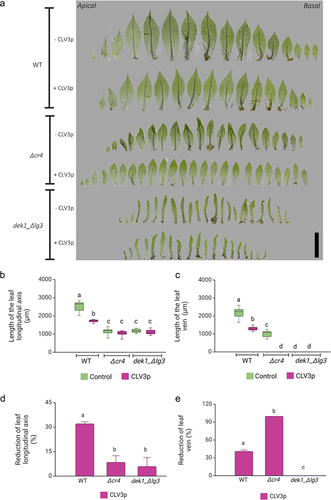
While the CLV3p treatment reduced the leaf size by 30% in WT plants and there was a 41% reduction of vein length in treated leaves, overall heteroblasty was not affected (). However, leaf development was strongly affected by CLV3p treatment in Δcr4 mutants when compared to untreated plants. The leaves of Δcr4 mutants lack elongated marginal cells, which likely disturb mechanical cues along the leaf axis leading to characteristic crinkliness, but veins form similarly in mutants to WT leaves (for details, see Demko et al., 2016).Citation45 CLV3p treatment caused a complete loss of crinkliness in Δcr4 leaves (). There was a c. 7% decrease in leaf size following treatment () and all treated Δcr4 leaves lacked defined midribs, resembling basal juvenile leaves (). This suggests that PpCR4 is required for a normal response to CLE3p treatment in leaves, and that CLEs signal through another receptor to suppress vein development.
Figure 2. CLV3p treatment suppresses vein development Δcr4 mutants.
Cell proliferation is suppressed by CLV3p in Δcr4 but not in dek1_ Δlg3 mutant’s leaves
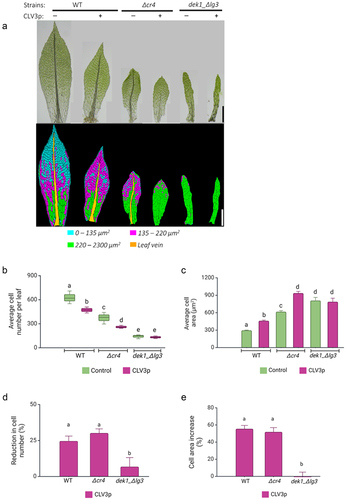
Normal heteroblastic leaf development requires spatially controlled cell proliferation and elongation along the axes of leaf symmetry.Citation49,Citation51 Therefore, we next determined the effect of CLV3p treatment on leaf cell number and cell area (, S1). In WT plants, the overall cell number was reduced upon the treatment as in previous workCitation40 (). The number of cell files along the medial-lateral axis decreased, and there were fewer cells along the apical-basal axis, especially within the distal parts of the leaves (Figure S2). In contrast, the cell area increased toward the tip of the leaves (; S1). Leaf development is already strongly affected in dek1_ Δlg3 mutant () as previously described by Johansen et al. 2016,Citation47 and CLV3p treatment had no significant effect on leaf size in dek1_ Δlg3 mutant when compared to untreated plants (). In the Δcr4 mutant, overall leaf cell numbers decreased following CLV3p treatment (). The number of cell files along the medial-lateral and apical-basal axis decreased significantly (Figure S2). In addition, CLV3p-treated Δcr4 leaves lost a typical distribution of cells with different cell size ranges along the apical-basal axis (), and cell area significantly increased throughout the midrib-less leaves (; Figure S1). Feeding WT and Δcr4 with CLV3p resulted in a similar rate of overall cell number reduction (approx. 25%; ) and cell area increase (approx. 50%; ). While gametophore length decreased upon CLV3p treatment in dek1_ Δlg3 mutant, the overall leaf size was not significantly changed (). The average cell number and cell area in mature dek1_ Δlg3 leaves were not significantly affected by added CLV3p (; Figure S1, S2). This suggests that CLAVATA pathway requires sufficient DEK1 activity to control cell proliferation during leaf development.
Figure 3. CLV3 peptides suppress cell proliferation in WT and Δcr4 mutant plants.
As previously demonstrated, P. patens CLV3-like peptides (PpCLEs) can act via conserved receptor kinases in A. thaliana.Citation40 Protein–protein interactions between PpCR4 with PpCLV1 and PpRPK2 remain to be investigated. As we show here, the CLV3p-mediated repression of cell proliferation is exacerbated in the Δcr4 mutant leading to loss of vein development. Hypothetically, the lack of PpCR4 might affect the assembly of interacting receptor kinase complexes and thereby their sensitivity to CLEs. In A. thaliana roots, the ACR4 forms heterodimers with CLV1Citation17 and contributes to spatial restriction of asymmetric cell divisions in columella stem cells as well as in pericycle during lateral root initiation.Citation27 In addition, the ACR4 expression domain is expanded by CLE40 peptide treatment.Citation16 In A. thaliana embryos, both TOAD2 (RPK2) and ACR4 localize in protodermal cells, and both play important role in epidermal identity maintenance.Citation52,Citation53 In addition, TOAD2 is required for PIN1 expression and auxin distribution during early embryogenesis in A. thaliana.Citation52 The physical interaction between PpCR4 and PpCLV1 has not been demonstrated yet. It would be also interesting to test whether PpCR4 expression is affected by CLEs in different tissues. Altogether, based on the data presented here, we propose that PpCR4 is likely a receptor for CLV3-like peptides in P. patens and together with other receptor kinases contributes to CLAVATA signaling during gametophyte development.Citation45
Authors contribution
VD and CJH conceived and designed the analysis. VD performed the experiment and collected data. AS analyzed the data, generated all figures, and wrote the manuscript. VD and CJH reviewed and edited the manuscript.
Corrected_Sup_Figure_2.tif
Download TIFF Image (2.7 MB)Corrected_Sup_Figure_1_TIFF.tif
Download TIFF Image (1.8 MB)Acknowledgments
We thank Martin Šafranek for help with figure graphics. We thank Jim Fouracre for a helpful discussion.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
All data generated during this study are included in this article and associated supplementary files. The mutant lines presented in this study are available upon request.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/15592324.2024.2386502
Additional information
Funding
References
- Fletcher JC, Brand U, Running MP, Simon R, Meyerowitz EM. Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science. 1999;283(5409):1911–8. doi:10.1126/science.283.5409.1911.
- Fiers M, Golemiec E, Xu J, van der Geest L, Heidstra R, Stiekema W, Liu C-M. The 14–amino acid CLV3, CLE19, and CLE40 peptides trigger consumption of the root meristem in Arabidopsis through a CLAVATA2-dependent pathway. Plant Cell. 2005;17(9):2542–2553. doi:10.1105/tpc.105.034009.
- Hobe M, Müller R, Grünewald M, Brand U, Simon R. Loss of CLE40, a protein functionally equivalent to the stem cell restricting signal CLV3, enhances root waving in Arabidopsis. Dev Genes Evol. 2003;213(8):371–381. doi:10.1007/s00427-003-0329-5.
- Stahl Y, Wink RH, Ingram G, Simon R. A signaling module controlling the stem cell niche in Arabidopsis root meristems. Curr Biol. 2009;19(11):909–914. doi:10.1016/j.cub.2009.03.060.
- Narasimhan M, Simon R. Spatial range, temporal span, and promiscuity of CLE-RLK signaling. Front Plant Sci. 2022;13:13. doi:10.3389/fpls.2022.906087.
- Demesa-Arevalo E, Narasimhan M, Simon R. Intercellular communication in shoot meristems. Annu Rev Plant Biol. 2024;75(1):319–344. doi:10.1146/annurev-arplant-070523.
- Cock JM, Mccormick S. A large family of genes that share homology with CLAVATA3. Plant Physiol. 2001;126(3):939–942. doi:10.1104/pp.126.3.939.
- Fletcher JC. Recent advances in Arabidopsis CLE peptide signaling. Trends Plant Sci. 2020;25(10):1005–1016. doi:10.1016/j.tplants.2020.04.014.
- Trotochaud AE, Hao T, Wu G, Yang Z, Clark SE. The CLAVATA1 receptor-like kinase requires CLAVATA3 for its assembly into a signaling complex that includes KAPP and a rho-related protein. Plant Cell. 1999;11(3):393–405. doi:10.1105/tpc.11.3.393.
- Müller R, Bleckmann A, Simon R. The receptor kinase CORYNE of Arabidopsis transmits the stem cell-limiting signal CLAVATA3 independently of CLAVATA1. Plant Cell. 2008;20(4):934–946. doi:10.1105/tpc.107.057547.
- DeYoung BJ, Bickle KL, Schrage KJ, Muskett P, Patel K, Clark SE. The CLAVATA1-related BAM1, BAM2 and BAM3 receptor kinase-like proteins are required for meristem function in Arabidopsis. Plant J. 2006;45(1):1–16. doi:10.1111/j.1365-313X.2005.02592.x.
- Kinoshita A, Betsuyaku S, Osakabe Y, Mizuno S, Nagawa S, Stahl Y, Simon R, Yamaguchi-Shinozaki K, Fukuda H, Sawa S, et al. RPK2 is an essential receptor-like kinase that transmits the CLV3 signal in Arabidopsis. Development. 2010;137(22):3911–3920. doi:10.1242/dev.061747.
- Shimizu N, Ishida T, Yamada M, Shigenobu S, Tabata R, Kinoshita A, Yamaguchi K, Hasebe M, Mitsumasu K, Sawa S, et al. BAM 1 and receptor-like protein kinase 2 constitute a signaling pathway and modulate CLE peptide-triggered growth inhibition in Arabidopsis root. New Phytol. 2015;208(4):1104–1113. doi:10.1111/nph.13520.
- Brand U, Fletcher JC, Hobe M, Meyerowitz EM, Simon R. Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science. 2000;289(5479):617–619. doi:10.1126/science.289.5479.617.
- Schoof H, Lenhard M, Haecker A, Mayer KFX, Jürgens G, Laux T. The stem cell population of Arabidopsis shoot meristems is maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell. 2000;100(6):635–644. doi:10.1016/S0092-8674(00)80700-X.
- Somssich M, Il JB, Simon R, Jackson D. CLAVATA-WUSCHEL signaling in the shoot meristem. Development. 2016;143(18):3238–3248. doi:10.1242/dev.133645.
- Stahl Y, Grabowski S, Bleckmann A, Kühnemuth R, Weidtkamp-Peters S, Pinto K, Kirschner G, Schmid J, Wink R, Hülsewede A, et al. Moderation of Arabidopsis root stemness by CLAVATA1 and ARABIDOPSIS CRINKLY4 receptor kinase complexes. Curr Biol. 2013;23(5):362–371. doi:10.1016/j.cub.2013.01.045.
- Becraft P, Stinard P, McCarty D. CRINKLY4: a TNFR-Like receptor kinase involved in maize epidermal differentiation. Science. 1996;273(5280):1406–1409. doi:10.1126/science.273.5280.1406.
- Becraft PW, Asuncion-Crabb Y. Positional cues specify and maintain aleurone cell fate in maize endosperm development. Development. 2000;127(18):4039–4048. doi:10.1242/dev.127.18.4039.
- Gifford ML, Dean S, Ingram G. The Arabidopsis ACR4 gene plays a role in cell layer organisation during ovule integument and sepal margin development. Development. 2003;130(8):4249–4258. doi:10.1242/dev.00634.
- Ingram G. Epidermal signalling and the control of plant shoot growth. Plant Cell Monogr. 2007;10127–10153. doi:10.1007/7089_2007_140.
- Watanabe M, Tanaka H, Watanabe D, Machida C, Machida Y. The ACR4 receptor-like kinase is required for surface formation of epidermis-related tissues in Arabidopsis thaliana. Plant J. 2004;39(3):298–308. doi:10.1111/j.1365-313X.2004.02132.x.
- Roeder AHK, Cunha A, Ohno CK, Meyerowitz EM. Cell cycle regulates cell type in the Arabidopsis sepal. Development. 2012;139(23):4416–4427. doi:10.1242/dev.082925.
- Cao X, Li K, Suh SG, Guo T, Becraft PW. Molecular analysis of the CRINKLY4 gene family in Arabidopsis thaliana. Planta. 2005;220(5):645–657. doi:10.1007/s00425-004-1378-3.
- Tanaka H, Watanabe M, Watanabe D, Tanaka T, Machida C, Machida Y. ACR4, a putative receptor kinase gene of Arabidopsis thaliana, that is expressed in the outer cell layers of embryos and plants, is involved in proper embryogenesis. Plant Cell Physiol. 2002;43(4):419–428. doi:10.1093/PCP/PCF052.
- Tian Q, Olsen L, Sun B, Lid SE, Brown RC, Lemmon BE, Fosnes K, Gruis D, Opsahl-Sorteberg H-G, Otegui MS, et al. Subcellular localization and functional domain studies of DEFECTIVE KERNEL1 in maize and Arabidopsis suggest a model for aleurone cell fate specification involving CRINKLY4 and SUPERNUMERARY ALEURONE LAYER1. Plant Cell. 2007;19(10):3127–3145. doi:10.1105/tpc.106.048868.
- De Smet I, Vassileva V, De Rybel B, Levesque MP, Grunewald W, Van Damme D, Van Noorden G, Naudts M, Van Isterdael G, De Clercq R, et al. Receptor-like kinase ACR4 restricts formative cell divisions in the Arabidopsis root. Science. 2008;322(5901):594–597. doi:10.1126/science.1160158.
- Johnson KL, Degnan KA, Ross Walker J, Ingram GC. AtDEK1 is essential for specification of embryonic epidermal cell fate. Plant J. 2005;44(1):114–127. doi:10.1111/j.1365-313x.2005.02514.x.
- Galletti R, Johnson KL, Scofield S, San-Bento R, Watt AM, Murray JAH, Ingram GC. DEFECTIVE KERNEL 1 promotes and maintains plant epidermal differentiation. Development. 2015;142(11):1978–1983. doi:10.1242/dev.122325.
- Perroud P, Demko V, Johansen W, Wilson RC, Olsen O-A, Quatrano RS. Defective Kernel 1 (DEK1) is required for three-dimensional growth in Physcomitrella patens. New Phytol. 2014;203(3):794–804. doi:10.1111/nph.12844.
- Kofuji R, Hasebe M. Eight types of stem cells in the life cycle of the moss Physcomitrella patens. Curr Opin Plant Biol. 2014;17:13–21. doi:10.1016/j.pbi.2013.10.007.
- Moody LA. The 2D to 3D growth transition in the moss Physcomitrella patens. Curr Opin Plant Biol. 2019;47:88–95. doi:10.1016/j.pbi.2018.10.001.
- Moody LA, Kelly S, Rabbinowitsch EH, Langdale JA. Genetic regulation of the 2D to 3D growth transition in the moss Physcomitrella patens. Curr Biol. 2018;28(3):473–478.e5. doi:10.1016/j.cub.2017.12.052.
- Aoyama T, Hiwatashi Y, Shigyo M, Kofuji R, Kubo M, Ito M, Hasebe M. AP2-type transcription factors determine stem cell identity in the moss Physcomitrella patens. Development. 2012;139(17):3120–3129. doi:10.1242/dev.076091.
- Harrison C, Roeder AH, Meyerowitz E, Langdale JA. Local cues and asymmetric cell divisions underpin body plan transitions in the moss Physcomitrella patens. Curr Biol. 2009;19(6):461–471. doi:10.1016/j.cub.2009.02.050.
- Ishikawa M, Fujiwara A, Kosetsu K, Horiuchi Y, Kamamoto N, Umakawa N, Tamada Y, Zhang L, Matsushita K, Palfalvi G, et al. GRAS transcription factors regulate cell division planes in moss overriding the default rule. Proc Natl Acad Sci USA. 2023;120(4). doi:10.1073/pnas.2210632120.
- Xu B, Ohtani M, Yamaguchi M, Toyooka K, Wakazaki M, Sato M, Kubo M, Nakano Y, Sano R, Hiwatashi Y, et al. Contribution of NAC transcription factors to plant adaptation to land. Science. 2014;343(6178):1505–1508. doi:10.1126/science.1248417.
- Rensing SA, Goffinet B, Meyberg R, Wu S-Z, Bezanilla M. The moss Physcomitrium (Physcomitrella) patens: a model organism for non-seed plants. Plant Cell. 2020;32(5):1361–1376. doi:10.1105/tpc.19.00828.
- Cove DJ, Perroud PF, Charron J, McDaniel SF, Khandelwal A, Quatrano RS. The moss Physcomitrella patens: a novel model system for plant development and genomic studies. Cold Spring Harb Protoc. 2009;2009(2):pdb.emo115. doi:10.1101/pdb.emo115.
- Whitewoods CD, Cammarata J, Nemec Venza Z, Sang S, Crook AD, Aoyama T, Wang XY, Waller M, Kamisugi Y, Cuming AC, et al. CLAVATA was a genetic novelty for the morphological innovation of 3D growth in land plants. Curr Biol. 2018;28(15):2365–2376.e5. doi:10.1016/j.cub.2018.05.068.
- Nemec-Venza Z, Madden C, Stewart A, Liu W, Novák O, Pěnčík A, Cuming AC, Kamisugi Y, Harrison CJ. CLAVATA modulates auxin homeostasis and transport to regulate stem cell identity and plant shape in a moss. New Phytol. 2022;234(1):149–163. doi:10.1111/nph.17969.
- Cammarata J, Morales Farfan C, Scanlon MJ, Roeder AHK. Cytokinin–CLAVATA cross-talk is an ancient mechanism regulating shoot meristem homeostasis in land plants. Proc Natl Acad Sci USA. 2022;119(14). doi:10.1073/pnas.2116860119.
- Nemec Venza Z, Greiff GRL, Harrison J. Mutant phenotypes and comprehensive expression analyses reveal roles for CLAVATA in moss vegetative 2 and reproductive development and fertility. bioRxiv. 2024; doi:10.1101/2024.04.05.585946.
- Demko V, Belova T, Messerer M, Hvidsten TR, Perroud P-F, Ako AE, Johansen W, Mayer KFX, Olsen O-A, Lang D, et al. Regulation of developmental gatekeeping and cell fate transition by the calpain protease DEK1 in Physcomitrium patens. Commun Biol. 2024;7(1). doi:10.1038/s42003-024-05933-z.
- Demko V, Ako E, Perroud PF, Quatrano R, Olsen O-A. The phenotype of the CRINKLY4 deletion mutant of Physcomitrella patens suggests a broad role in developmental regulation in early land plants. Planta. 2016;244(1):275–284. doi:10.1007/s00425-016-2526-2.
- Ashton N, Cove D. The isolation and preliminary characterisation of auxotrophic and analogue resistant mutants of the moss, Physcomitrella patens. Mol Gen Genet. 2004;154(1):87–95. doi:10.1007/BF00265581.
- Johansen W, Ako AE, Demko V, Perroud P-F, Rensing SA, Mekhlif AK, Olsen O-A. The DEK1 calpain linker functions in three-dimensional body patterning in Physcomitrella patens. Plant Physiol. 2016;172(2):1089–1104. doi:10.1104/pp.16.00925.
- Olsen OA, Perroud PF, Johansen W, Demko V. DEK1; missing piece in puzzle of plant development. Trends Plant Sci. 2015;20(2):70–71. doi:10.1016/j.tplants.2015.01.003.
- Barker EI, Ashton NW. Heteroblasty in the moss, Aphanoregma patens (Physcomitrella patens), results from progressive modulation of a single fundamental leaf developmental programme. J Bryol. 2013;35(3):185–196. doi:10.1179/1743282013Y.0000000058.
- Dennis RJ, Whitewoods CD, Harrison CJ. Quantitative methods in like-for-like comparative analyses of Aphanorrhegma (Physcomitrella) patens phyllid development. J Bryol. 2019;41(4):314–321. doi:10.1080/03736687.2019.1668109.
- Lin W, Wang Y, Coudert Y, Kierzkowski D. Leaf morphogenesis: insights from the moss Physcomitrium patens. Front Plant Sci. 2021;12:12. doi:10.3389/fpls.2021.736212.
- Nodine MD, Tax FE. Two receptor-like kinases required together for the establishment of Arabidopsis cotyledon primordia. Dev Biol. 2008;314(1):161–170. doi:10.1016/j.ydbio.2007.11.021.
- San-Bento R, Farcot E, Galletti R, Creff A, Ingram G. Epidermal identity is maintained by cell–cell communication via a universally active feedback loop in Arabidopsis thaliana. Plant J. 2014;77(1):46–58. doi:10.1111/tpj.12360.
