ABSTRACT
Objective
This study aimed to investigate the relationship between tumor necrosis factor alpha (TNFα) polymorphisms and multiple myeloma (MM) risk.
Methods
Eligible studies were retrieved from PubMed, the Cochrane Library, Embase, CNKI and the Wanfang database. Polymorphisms of TNFα-308 G/A, TNFα-857 C/T, and TNFα-238 G/A were analyzed based on the allele, recessive, dominant, and additive-dominant models. The meta-analysis was conducted using R 3.12 software. Odds ratio (OR) and 95% confidence intervals (CI) were used as evaluation indicators. Heterogeneity among studies was detected. Publication bias was evaluated. Sensitivity and power analyses were also conducted.
Results
Significant associations existed between ‘TT vs. CC’ (OR = 2.3752, 95% CI = 1.1342–4.9740) and ‘TT vs. CC + TC’ (OR = 2.0802, 95% CI = 1.0250–4.2218) models of the TNFα-857 C/T gene and MM risk. There were no significant differences in other genetic models of TNFα-857 C/T or any genetic models of TNFα-308 G/A and TNFα-238 G/A. No significant publication bias existed among the studies. In addition, sensitivity analyses showed that meta-analysis results of all genetic models of the TNFα-238 G/A gene did not change after omitting one of these studies, but most models of TNFα-857 C/T and TNFα-308 G/A exhibited significant changes.
Conclusion
Our findings indicate that the ‘TT vs. CC’ and ‘TT vs. CC + TC’ of TNFα-857 C/T are correlated with MM risk. TNFα-857 C/T may be a risk factor for MM development. There is no association between TNFα-238/-308 polymorphisms and MM risk.
Introduction
Multiple myeloma (MM) is a type of hematologic tumor that features an abnormal growth of malignant plasma cells in the bone marrow [Citation1,Citation2] and accounts for approximately 10%–15% of all hematologic malignancies and 1% of total malignancies [Citation3,Citation4]. MM has a multifactorial etiology that is influenced by various environmental and genetic factors [Citation5–7]. Recent evidences have confirmed the association between genetic polymorphisms and MM risk [Citation8–10].
Tumor necrosis factor alpha (TNFα) is one of the most important cytokines and can function as a prominent mediator of immune regulation [Citation11]. An autocrine function for TNFα has been implicated in lymphoproliferative diseases, such as chronic lymphocytic leukemia and plasma cell myeloma [Citation12]. In MM, TNFα is a key regulator in the generation of malignant plasma cells [Citation13,Citation14], and an enhanced expression of TNFα is correlated with an increased aggressiveness of MM [Citation15].
Accumulating evidence has confirmed that TNFα polymorphisms are associated with increased MM risk. For example, TNFα-238 G/A is involved in MM development and prognosis following treatment [Citation14]. TNFα-857 T allele, or TT genotype, is correlated with a risk of developing MM among the Chinese population of Shangdong area [Citation16]. However, Basmaci et al. revealed that TNFα-308 GG polymorphism was lowly expressed in MM patients and might play a key role in MM pathogenesis, but TNFα-238/857 polymorphisms did not exhibit any difference between MM patients and healthy controls [Citation10]. Moreover, a previous meta-analysis has shown that TNFα-238/-308 polymorphisms cannot influence MM risk [Citation17]. These findings indicate that there remains a great controversy regarding the association between TNFα polymorphisms and MM risk. In addition, the previous meta-analysis did not analyze the link between TNFα-857 polymorphisms and MM risk. These findings prompted us to perform this updated meta-analysis. The findings of this study will provide new evidence for explaining the significance of TNFα polymorphisms in predicting MM risk.
Materials and methods
Search strategy
Relevant studies were searched for via Embase (http://www.embase.com), PubMed (http://www.ncbi.nlm.nih.gov/pubmed/), the Cochrane Library (http://www.cochranelibrary.com/), CNKI (http://www.cnki.net/) and the Wanfang database (http://med.wanfangdata.com.cn/).
Key words used for retrieving studies were ‘multiple myeloma’ OR ‘MM’ OR ‘Kahler’s disease’ OR ‘Huppert’s disease’ AND ‘tumor necrosis factor alpha’ OR ‘TNFα-308’ OR ‘TNFα-238’ OR ‘TNFα-857’ AND ‘polymorphi*’. The deadline for retrieval was 12 April 2018.
Study selection criteria
Inclusion criteria for eligible studies were (1) studies reported the distributions of TNFα-308 G/A, TNFα-238 G/A and TNFα-857 C/T polymorphisms in MM and non-MM patients; (2) accurate genotype or allele frequency data were available in the studies; (3) studies were case–control studies.
Exclusion criteria were (1) data were incomplete and could not be used for statistical analysis; (2) studies were reviews, reports, comments, letters, editorials, etc.; (3) repetitive publications and studies with data applied in several articles, except for one study with the latest research or the most complete information.
Data extraction
Useful data were independently extracted from retrieved articles by two investigators and included the first author’s name; publication year; geographical location; number and demographic data characteristics, including sex and age of participants in case and control groups; and the number of patients and controls with each gene polymorphism in each study. The Newcastle–Ottawa Scale (NOS) provided by the Agency for Healthcare Research and Quality was used for assessing the quality of the studies included [Citation18]. Divergences in data extraction and quality assessment were settled through discussion with a third investigator.
Statistical analysis
The Hardy–Weinberg equilibrium (HWE) was assessed in control participants using chi-squared (χ2) test [Citation19]. A meta-analysis was conducted using R 3.12 software. Odds ratio (OR) and 95% confidence intervals (CI) were applied as evaluation indicators to estimate data [Citation20]. Heterogeneity among studies was detected using chi-squared Q statistic [Citation21] and I2 value. If P < 0.05 or I2 > 50%, then heterogeneity was considered significant, and the random-effects model was thus applied to pool the effect sizes; otherwise, the fixed-effects model was chosen [Citation22]. Publication bias was evaluated using Egger’s method [Citation23]. Sensitivity analyses were conducted by omitting one study at a time. Power analyses were performed using Power Analysis and Sample Size (PASS) 2008 Statistical Software [Citation24].
Results
Characteristics of eligible studies
Based on the preliminary search strategy outlined above, study selection was conducted according to the flow chart shown in Figure . Briefly, 83 articles were initially recruited from PubMed [Citation20], Embase [Citation24], the Cochrane Library [Citation3], CNKI [Citation15] and the Wanfang database [Citation21]. Of these, 31 articles from repeated search results and 18 obviously irrelevant articles were removed. After reading the abstracts, three letters/editorials, three case series/reports, and four animal/cell studies were excluded. Upon further review of the full text of the remaining 24 articles, three reviews, five studies from which data could not be extracted, and three studies with duplicated populations were excluded. Finally, 13 eligible studies were included [Citation10,Citation14,Citation16,Citation25–34].
The eligible studies were published between 2000 and 2016. A total of 4110 participants, including 2421 MM patients and 1689 non-MM participants, were included from China, U.S.A., Turkey, U.K., Hungary, Italy, Russia, Sweden, and Germany. With regard to demographic characteristics, participants were middle-aged, and the distribution of men and women was evenly balanced, except for a study reported by Brown et al. [Citation31]. Single nucleotide polymorphisms (SNPs) were detected using PCR-RFLP and PCR-LDR. In addition, results of quality assessment showed that the NOS score was 3–8 points, indicating that the quality of most included studies was acceptable. Among the articles, 10 had an NOS score of ≥ 6 and the others had low quality. According to the results of the HWE test, all participants in the control group were in line with the HWE (Table ), indicating that the population representativeness was good.
Table 1. Characteristics of eligible studies.
Summary results of the meta-analysis
This meta-analysis investigated the relationship between MM risk and the allele model (TNFα-308, A vs. G; TNFα-238, A vs. G; TNFα-857, T vs. C), additive-dominant model (TNFα-308, AA vs. GG, AG vs. GG; TNFα-238, AA vs. GG, AG vs. GG; TNFα-857, TC vs. CC, TT vs. CC), recessive model (TNFα-308, AA vs. GG + AG; TNFα-238, AA vs. GG + AG; TNFα-857, TT vs. CC + TC), and dominant model (TNFα-308, AA + AG vs. GG; TNFα-238, AA + AG vs. GG; TNFα-857, TT + TC vs. CC), respectively.
A heterogeneity test was conducted to select a suitable effect model to pool the effect sizes. As shown in Table , significant heterogeneity existed in the ‘A vs. G’, ‘AG vs. GG’, and ‘AA + AG vs. GG’ models of the TNFα-308 G/A gene and the ‘T vs. C’ and ‘TT + TC vs. CC’ models of the TNFα-857 C/T gene; thus, the random-effects model was applied to combine the effect sizes of these models. Other models used the fixed-effects model.
Table 2. Summary results of meta-analysis.
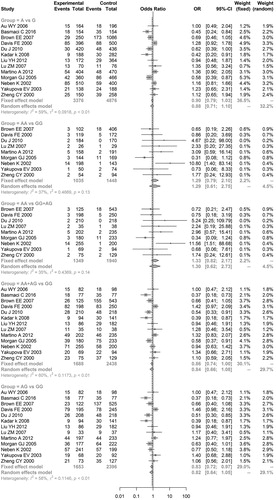
The results of the meta-analysis with regard to the relationship between TNFα polymorphisms and MM risk showed that significant associations existed between ‘TT vs. CC’ (OR = 2.3752, 95% CI = 1.1342–4.9740) and ‘TT vs. CC + TC’ (OR = 2.0802, 95% CI = 1.0250–4.2218) of TNFα-857 C/T and MM risk, indicating that TNFα-857 C/T might be a risk factor for MM development (Table and Figure ). However, no genetic models of TNFα-238 G/A and TNFα-308 G/A exhibited significant associations with MM risk (Table and and ), indicating that these two TNFα polymorphisms do not result in MM development.
Figure 2. Meta-analysis of associations between genetic models of tumor necrosis factor alpha-857 C/T polymorphisms and multiple myeloma risk.
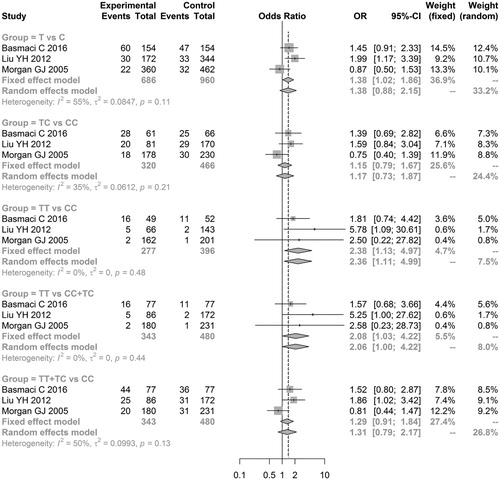
Figure 3. Meta-analysis of associations between genetic models of tumor necrosis factor alpha-238 G/A polymorphisms and multiple myeloma risk.
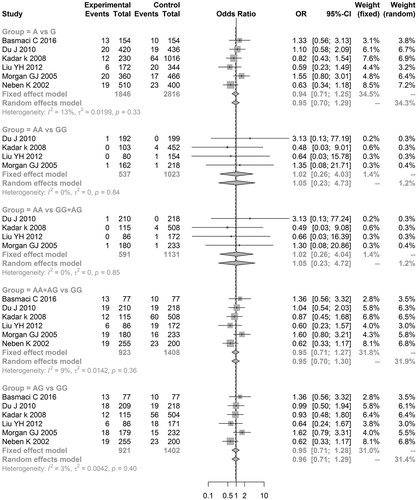
Figure 4. Meta-analysis of associations between genetic models of tumor necrosis factor alpha-308 G/A polymorphisms and multiple myeloma risk.
The results of Egger’s test did not display a significant publication bias among studies of the above three TNFα polymorphisms (Table ), confirming that our conclusions were more reliable.
In addition, sensitivity analysis results showed that the meta-analysis results for all genetic models of the TNFα-238 G/A gene did not markedly change after omitting one of these studies. However, meta-analysis results of the ‘AA vs. GG’, ‘AG vs. GG’, and ‘AA vs. GG + AG’ models of the TNFα-308 G/A gene and the ‘T vs. C’, ‘TT vs. CC’, ‘TT vs. CC + TC’, and ‘TT + TC vs. CC’ models of the TNFα-857 C/T gene exhibited significant changes (data not shown).
Further power analyses showed that all genetic models of the above three TNFα polymorphisms had a low statistical power (all power values <0.80), particularly TNFα-238 G/A. The power value of all genetic models of TNFα-238 G/A was <0.20.
Discussion
The present meta-analysis evaluated the relationship between TNFα polymorphisms and MM risk. The results showed that ‘TT vs. CC’ and ‘TT vs. CC + TC’ models of the TNFα-857 C/T gene had significant associations with MM risk. There were no significant differences in other genetic models of TNFα-857 C/T and any genetic models of TNFα-308 G/A and TNFα-238 G/A. These data suggest that TNFα-857 C/T polymorphisms are causative factors for MM.
Increasing evidences have reported that several proinflammatory cytokines, particularly TNFα, play crucial roles in the pathology of MM [Citation35,Citation36]. In addition, TNFα polymorphisms have been shown to influence TNFα production [Citation37,Citation38]. Consistent with the findings of Liu et al. that TNFα-857 T allele, or TT genotype, is associated with MM risk in the Chinese population of the Shangdong area [Citation16], our results showed that ‘TT vs. CC’ and ‘TT vs. CC + TC’ models of the TNFα-857 C/T gene have significant associations with MM risk. Although Basmaci et al. did not conclude any association of TNFα-857 polymorphism with MM risk, our analysis prompts us to speculate that TNFα-857 C/T is a risk factor for MM development. In addition, although previous studies have confirmed that TNFα-238 site variations are associated with a favorable overall response rate following thalidomide and dexamethasone therapy [Citation32] and that the TNFα-308 polymorphism might play a key role in MM pathogenesis [Citation10], our results were in line with the findings of a previous meta-analysis [Citation17] and we still cannot infer any association between TNFα-308/238 polymorphisms and MM risk.
Similar to other meta-analyses, significant heterogeneity existed among the articles included in the present meta-analysis in relation to the ‘A vs. G’, ‘AG vs. GG’, and ‘AA + AG vs. GG’ models of the TNFα-308 G/A gene and the ‘T vs. C’ and ‘TT + TC vs. CC’ models of the TNFα-857 C/T gene. For this meta-analysis, studies from different geographical regions, including China, U.S.A., Turkey, U.K., Hungary, Italy, Russia, Sweden and Germany, were included, which may explain the heterogeneity of genetic diversity caused by differences in regional living habits and living environments as well as disparities in economic development levels. Other confounding factors, such as sex and age, may also play a role in this heterogeneity.
Several limitations should be considered when explaining our results. First, because data from some studies were incomplete, particularly demographic data, the covariates were not calibrated in this study. These incomplete data may be potentially confounding factors affecting the results of this meta-analysis. Second, sensitivity analysis showed that meta-analysis results for the partial models of TNFα-308 G/A and TNFα-857 C/T exhibited significant changes after omitting one of these studies. Thus, more relevant studies are required to confirm our findings. Third, power analysis showed that all genetic models of the above three TNFα polymorphisms had a low statistical power, particularly in genetic models of TNFα-238 G/A, with all power values being <0.20. These data imply that further research is needed to support our results. Therefore, in future, more studies with bigger sample sizes should be conducted for further evaluation of any associations.
Taken together, the present meta-analysis reveals that the ‘TT vs. CC’ and ‘TT vs. CC + TC’ of TNFα-857 C/T are correlated with MM risk. TNFα-857 C/T may be a risk factor for MM development. There is no association between TNFα-238 or TNFα-308 polymorphisms and MM risk.
Acknowledgements
None.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Dhodapkar MV, Borrello I, Cohen AD, et al. Hematologic malignancies: plasma cell Disorders. Am Soc Clin Oncol Educ Book. 2017;37:561–568. doi: 10.14694/EDBK_175546
- Tang S, Cheng B, Zhe N, et al. Histone deacetylase inhibitor BG45-mediated HO-1 expression induces apoptosis of multiple myeloma cells by the JAK2/STAT3 pathway. Anticancer Drugs. 2018;29:61–74. doi: 10.1097/CAD.0000000000000568
- Rajkumar SV. Multiple myeloma: 2012 update on diagnosis, risk-stratification, and management. Am J Hematol. 2016;88:719–2734.
- Angtuaco EJC, Fassas ABT, Walker R, et al. Multiple myeloma: Clinical review and Diagnostic Imaging1. Radiology. 2004;231:11–23. doi: 10.1148/radiol.2311020452
- Stellrecht CM, Gandhi V. Myeloma antioxidant status: the good, the bad and the reactive. Leuk Lymphoma. 2009;50:691–693. doi: 10.1080/10428190902856832
- Becker N. Epidemiology of multiple myeloma. Berlin/Heidelberg: Springer; 2011; p. 1–15.
- Munshi NC, Avetloiseau H. Genomics in multiple myeloma. Clin Cancer Res. 2011;17:1234–1242. doi: 10.1158/1078-0432.CCR-10-1843
- Chakraborty B, Vishnoi G, Gowda SH, et al. Interleukin-6 gene-174 G/C promoter polymorphism and its association with clinical profile of patients with multiple myeloma. Asia-Pac J Clin Oncol. 2017;13:e402–e407. doi: 10.1111/ajco.12290
- Kasamatsu T, Kimoto M, Takahashi N, et al. IL17A and IL23R gene polymorphisms affect the clinical features and prognosis of patients with multiple myeloma. Hematol Oncol. 2018;36:196–201. doi: 10.1002/hon.2469
- Basmaci C, Pehlivan M, Tomatir AG, et al. Effects of TNFalpha, NOS3, MDR1 gene polymorphisms on clinical parameters, prognosis and survival of multiple myeloma cases. Asian Pac J Cancer Prev. 2016;17:1009–1014. doi: 10.7314/APJCP.2016.17.3.1009
- Chan KM, Siegel RM, Lenardo MJ. Signaling by the TNF receptor superfamily and T cell homeostasis. Immunity. 2000;13:419–422. doi: 10.1016/S1074-7613(00)00041-8
- Reiterer G, Toborek MB. Tumour necrosis factor gene polymorphisms in lymphoproliferative disease. Leuk Lymphoma. 2000;38:547–552. doi: 10.3109/10428190009059274
- Sawamura M, Murakami H, Tsuchiya J. Tumor necrosis factor-alpha and interleukin 4 in myeloma cell precursor differentiation. Leuk Lymphoma. 1996;21:31–36. doi: 10.3109/10428199609067576
- Neben K, Mytilineos J, Moehler TM, et al. Polymorphisms of the tumor necrosis factor-alpha gene promoter predict for outcome after thalidomide therapy in relapsed and refractory multiple myeloma. Blood. 2002;100:2263–2265.
- Jurišić V, Čolović M. Correlation of sera TNF-α with percentage of bone marrow plasma cells, LDH, β2-microglobulin, and clinical stage in multiple myeloma. Med Oncol. 2002;19:133–139. doi: 10.1385/MO:19:3:133
- Liu YH, Peng Z, Song WG, et al. Polymorphisms of tumor necrosis factor and susceptibility to multiple myeloma. Chin J Cancer Prev Treat. 2012;19:1696–1699.
- Li B, Wang XD, Sun X, et al. Lack of association between TNF-α promoter polymorphisms and multiple myeloma: a meta-analysis. Leuk Res. 2013;37:50–57. doi: 10.1016/j.leukres.2012.08.027
- Wells GA, Shea BJ, O’Connell D, et al. The Newcastle–Ottawa Scale (NOS) for assessing the quality of non-randomized studies in meta-analysis. Appl Eng Agric. 2014;18:727–734.
- Schaid DJ, Jacobsen SJ. Biased tests of association: comparisons of allele frequencies when departing from Hardy-Weinberg proportions. Am J Epidemiol. 1999;149:706–711. doi: 10.1093/oxfordjournals.aje.a009878
- Liu T, Xu QE, Zhang CH, et al. Occupational exposure to methylene chloride and risk of cancer: a meta-analysis. Cancer Causes Control. 2013;24:2037–2049. doi: 10.1007/s10552-013-0283-0
- Lau J, Ioannidis JP, Schmid CH. Quantitative synthesis in systematic reviews. Ann Intern Med. 1997;127:820–826. doi: 10.7326/0003-4819-127-9-199711010-00008
- Feng RN, Zhao C, Sun CH, et al. Meta-analysis of TNF 308 G/A polymorphism and type 2 diabetes mellitus. PLoS One. 2011;6:e18480. doi: 10.1371/journal.pone.0018480
- Minder C, Egger M, Smith GD, et al. Bias in meta-analysis detected by a simple, graphical test. Br Med J. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629
- Aykut V, Kirgiz A, Gül AB, et al. Comparison of pre-incision and single-stepped clear corneal incision in phacoemulsification surgery. Eur Rev Med Pharmacol Sci. 2014;18:1698–1703.
- Au WY, Fung A, Wong KF, et al. Tumor necrosis factor alpha promoter polymorphism and the risk of chronic lymphocytic leukemia and myeloma in the Chinese population. Leuk Lymphoma. 2006;47:2189–2193. doi: 10.1080/10428190600758645
- Davies FE, Rollinson SJ, Rawstron AC, et al. High-producer haplotypes of tumor necrosis factor alpha and lymphotoxin alpha are associated with an increased risk of myeloma and have an improved progression-free survival after treatment. J Clin Oncol. 2000;18:2843–2851. doi: 10.1200/JCO.2000.18.15.2843
- Martino A, Buda G, Maggini V, et al. Could age modify the effect of genetic variants in IL6 and TNF-α genes in multiple myeloma? Leuk Res. 2012;36:594–597. doi: 10.1016/j.leukres.2012.02.009
- Yakupova EV, Grinchuk OV, Kalimullina DK, et al. Molecular genetic analysis of the Interleukin 6 and tumor necrosis factor α gene polymorphisms in multiple myeloma. Mol Biol. 2003;37:358–361. doi: 10.1023/A:1024227008451
- Lu ZM, Zhang Q, Zu Y, et al. Polymorphisms of TNF-alpha and promoter genes in multiple myeloma. Chin J Intern Med. 2007;46:767–768.
- Morgan GJ, Adamson PJ, Mensah FK, et al. Haplotypes in the tumour necrosis factor region and myeloma. Br J Haematol. 2005;129:358–365. doi: 10.1111/j.1365-2141.2005.05467.x
- Brown EE, Lan Q, Zheng T, et al. Common variants in genes that mediate immunity and risk of multiple myeloma. Int J Cancer. 2007;120:2715–2722. doi: 10.1002/ijc.22618
- Du J, Yuan ZG, Zhang CY, et al. Role of the TNF-α promoter polymorphisms for development of multiple myeloma and clinical outcome in thalidomide plus dexamethasone. Leuk Res. 2010;34:1453–1458. doi: 10.1016/j.leukres.2010.01.011
- Kadar K, Kovacs M, Karadi I, et al. Polymorphisms of TNF-alpha and LT-alpha genes in multiple myeloma. Leuk Res. 2008;32:1499–1504. doi: 10.1016/j.leukres.2008.03.001
- Zheng CY, Huang DR, Bergenbrant S, et al. Interleukin 6, tumour necrosis factor α, interleukin 1β and interleukin 1 receptor antagonist promoter or coding gene polymorphisms in multiple myeloma. Br J Haematol. 2000;109:39–45. doi: 10.1046/j.1365-2141.2000.01963.x
- Sati HIA, Greaves M, Apperley JF, et al. Expression of interleukin-1β and tumour necrosis factor-α in plasma cells from patients with multiple myeloma. Br J Haematol. 2015;104:350–357. doi: 10.1046/j.1365-2141.1999.01193.x
- Trigo FMB, Luizon MR, Dutra HS, et al. Interaction between IL-6 and TNF-α genotypes associated with bacteremia in multiple myeloma patients submitted to autologous stem cell transplantation (ASCT). Leuk Res Rep. 2014;3:76–78.
- Wilson AG, Di GF, Blakemore AI, et al. Single base polymorphism in the human tumour necrosis factor alpha (TNF alpha) gene detectable by NcoI restriction of PCR product. Hum Mol Genet. 1992;1:353–353. doi: 10.1093/hmg/1.5.353
- Pociot F, D’Alfonso S, Compasso S, et al. Functional analysis of a New polymorphism in the Human TNF α gene Promoter. Scand J Immunol. 1995;42:501–504. doi: 10.1111/j.1365-3083.1995.tb03686.x

