ABSTRACT
Objective: This study aims to investigate the gene mutation characteristics of chronic neutrophilic leukaemia (CNL).
Method: This study retrospectively analyses the molecular biological characteristics, laboratory characteristics and clinical data of four patients with CNL that were admitted in the second Hospital of Shanxi Medical University from May 2014 to October 2016. On the basis of the molecular biological data of 22 patients with CNL and 4 patients with CNL, we further analysed the characteristics of CNL molecular mutation.
Results: Two out of the four patients with CNL were carriers of colony-stimulating factor 3 receptor (CSF3R) mutation, among which two were carriers of CSF3R T618I mutation combined with ASXL1 mutation and SETBP1 mutation, and two were only carriers of JAK2 V617F mutation. According to the molecular biological data of 22 patients with CNL, 20 patients were positive for CSF3R mutation. Two patients were positive for JAK2 V617F mutation. A total of 10 patients were positive for SETBP1 mutation which was correlated with the CSF3R T618I gene mutation (P = 0.03). A total of 13 patients were positive for ASXL1 mutation. No patients carried mutations in ASXL2 and MPL genes.
Conclusion and Discussion: CSF3R mutation is the main tumorigenic mutation in CNL, in which CSF3R T618I mutation is the main mutation, and an extremely small number of CNL patients may be caused by JAK2 V617F mutation. SETBP1 and ASXL1 are the most common concomitant mutations in CNL with CSF3R mutation, and SETBP1 and CSF3R T618Imutations may have a certain correlation.
Introduction
Chronic neutrophilic leukaemia (CNL) is a potentially aggressive myeloprolife-rative neoplasm with a low incidence which is characterized mainly by a continuous increase in mature neutrophils in peripheral blood [Citation1]. Colony-stimulating factor 3 receptor (CSF3R) gene mutation is a highly specific and sensitive molecular marker of CNL [Citation2,Citation3]. Other mutated genes, such as JAK2 V617F and CALR, can also be detected in some patientswith CNL [Citation4–8], thereby suggesting that these mutated genes may also be involved in CNL pathogenesis and development. The discovery of all mutated molecules suggested the heterogeneity of molecular pathogenesis in CNL. In this study, the molecular mutation characteristics of CNL are analysed based on the gene mutation in 4 patients with CNL in our hospital and in 18 patients in the literature to improve the understanding of the molecular mechanism of CNL.
Materials and methods
Reagents and instruments
Genomic DNA was extracted from the bone marrow of patients with CNL by OMAGE extraction. The concentration and purity of the samples were determined by an ultraviolet spectrophotometer. The samplesthatmet the requirements (A260/A280 > 1.7) were stored at −20°Cfor standby. TaqDNA polymerase reaction system (B300097-0500) and DL-1000 Marker (SD4210) were obtainedfrom TaKaRa. The primer sequence designed by the Premier 5.0 software () were synthesized by Beijing Orco Dingsheng Biotechnology. Agarose was purchased from Sigma USA, and the amplification reaction was carried out in a T100TM Thermal Cycler instrument (BIO-RAD). UVP gel imager (FlourChem HD2) was purchased from UVP USA.
Table 1. Primer sequences, amplification conditions and product length of PCR amplification of various genes.
Cases data
The four CNL cases were selected from May 2014 to July 2016 in our hospital, and all patients were females, with a median age of 70.5 years (53–8). The diagnostic criteria were based on the WHO (2016) classification criteria ().
Table 2. CNL Diagnostic Criteria (2016).
CSF3R, JAK2 V617F, CALR, MPL, SETBP1, ASXL1 and ASXL2 gene mutation detection
DNA extraction kit was used to extract genomic DNA from the bone marrow or peripheral blood of patients. The primer sequences of CSF3R exon14-17, CALR exon 9, MPL exon 10, ASXL1, ASXL2 and JAK2 V617F areshown in below. The SETBP1primer design was referred to in the literature [Citation2,Citation3]. Gene mutation was detected by PCR amplification combined with Sanger sequencing, as follows: PCR reaction system(25 µl): 2.5 µl of 10×buffer, 1.5 µl of 25 mmol/lMgCl2, 0.5 µl of 10×dNTPs, 1 µl of upstream and downstream primers, 1.5 µl of Taq DNA polymerase(1.5 U), 3 µl of DNA and 25 µl of ddH2O. The PCR thermal cycling conditions are as follows: predenaturation at 94°C for 3 min, denaturation at 95°C for 30 s, annealing at 60°C (CALR,56°C)for 30 s, extension at 72°C for 45 s, cycle of 35 times, total extension at 72°C for 10 min. The amplified products were sent to Beijing Huada Gene Technology Service Co., Ltd., and the sequencing results were compared with the National Biotechnology Information Centre (NCBI) gene bank sequence.
JAK2 V617F mutation was detected byAS-PCR, as follows: PCR reaction system (25 µl): 2.5 µl of 10×buffer; 1.5 µl of 25 mmol/l MgCl2;0.5 µl of 10 mmol/l dNTPs; 0.8, 0.6 and 0.5 µl of 20 µmol/l V617F primers F, R1 and R2, respectively; 1.5 µl (1.5 U) of Taq DNA polymerase; 100 ng DNA and 25 µl of ddH2O. The PCR thermal cycling conditions are as follows: predenaturation at 94°C for 3 min, denaturation at 95°C for 30 s, annealing at 60°C for 30 s, extension at 72°C for 45 s, cycle for 35 times and total extension at 72°C for 10 min. Positive, negative and blank controls were set up for each reaction and analysed by the UVP gel image analysis system, where in 451 and 295 bp fragments were JAK2 V617F mutant-type, and only 451 bp was JAK2 V617F wild-type.
Patient data and case collection
In this study, the laboratory data of fourpatients were collected from inpatient or outpatient medical records, including peripheral blood cell count, neutrophilic alkaline phosphatase activity (NAP) and spleen size. The clinical data of the four patients in this study were collected according to the follow-up records. The follow-up time was 39.5 (3–59) months until October 2016. The other cases were collected through PubMed and CNKI databases. The data of CNL cases reported from 2013 to 2016 were collected, and the general data of patients (including sex and age, gene mutation and survival time) were recorded.
Results
Gene mutation analyses of CSF3R, JAK2 V617F, CALR, MPL, ASXL1, ASXL2 and SETBP1
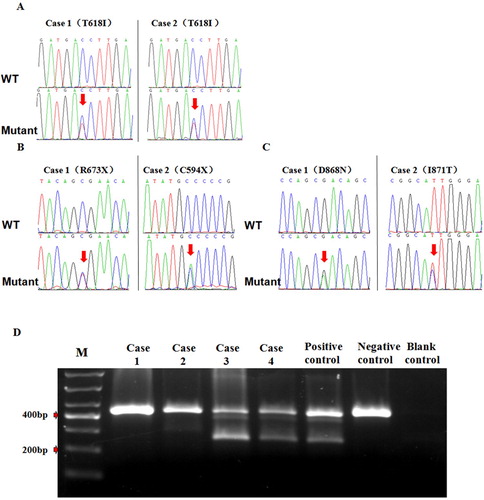
Two cases with CSF3R mutation were detected in four patients with CNL, all of which exhibited CSF3R T618I mutation combined with ASXL1(1c.2509C > T, 2c.2214C > A) and SETBP1mutations (D868N and I871 T). The two other cases were patients with JAK2 V617F mutation. All four patients with CALR, MPL and ASXL2 gene mutations were negative ().
Figure 1. Mutation test results in 4 cases of patients with CNL gene. A: Sequencing results of CSF3R gene mutation in 2 patients with CNL. B: Sequencing results of ASXL1 gene mutation in 2 patients with CNL. C: Sequencing results of SETBP1 gene mutation in 2 patients with CNL. D: Analysis JAK2 V617F mutation using allele specific polymerase chain reaction. Patients (case3.and case4) strongly exhibit positive.
Clinical and laboratory characteristics
The granulocyte of the peripheral blood cells in four patients with CNL increased, whilst those of erythroid, megakaryocyte, lymphoid, mononuclear, acidophilic and alkalophilic were normal or slightly decreased. The NAP+ rate and integral value in four patients were significantly higher than those in the normal range. The two other patients with JAK2 V617F mutation showed severe splenomegaly, and the two patients with CSF3R T618I mutation showed moderate or insignificant splenomegaly. The two patients with JAK2 V617F mutation had persistent neutropenia for more than 3 months with splenomegaly and excluded potential causes of reactive neutropenia such as infection, inflammation, tumors and no evidence of other myeloid tumors. All four patients survived at the end of follow-up, with the survival times of 28, 3, 49 and 59 months, respectively, and no progress was made in the direction of myelofibrosis and acute myeloid leukaemia (AML) ().
Table 3. Analysis of laboratory and clinical features of 4 patients with CNL.
Characteristics of molecular mutation in 22 patients with CNL in this study and literature
Among 22 patients with CNL, 9 were male and 13 were female, with a median age of 68.5 (6–80). CSF3R, JAK2 V617F, SETBP1, ASXL1 and CALR gene mutations were detected in all patients. A total of 20 patients were positive for CSF3R mutation, including 17 patients with CSF3RT618I mutation, 2 patients with CSF3RM696 T mutation and 1 patient with CSF3RI598I mutation. Two patients were in the position for JAK2V617F mutation. Ten patients with SETBP1 mutation were positive and coexisted with the CSF3R T618I mutation. The results showed a correlation between the occurrence of SETBP1 gene and CSF3R T618Imutations in CNL (P = 0.03), and the correlation coefficient was ϕ = 0.495. Out of the 13 patients with ASXL1 mutation, 8 patients with ASXL1,CSF3R T618I and SETBP1 gene mutations; 3 patients with CSF3R T618I mutation; 2 patients with CSF3R M696 T mutation; 2 patients with CSF3R T618IandSETBP1 gene mutations; 4 patients with CSF3R T618I and 2 patients with JAK2 V 617F mutation positive were ASXL1-negative. The frequency of ASXL1 mutation distribution among the above mutation groups was statistically analysed. The results showed that the difference in the frequency of ASXL1 mutation among the groups was insignificant (P > 0.05). Only two patients were positive for CALR mutation, one patient with CSF3R T618I and one patient with CSF3R T618I, SETBP1 and ASXL1 mutations ().
Table 4. Mutations of CSF3R, JAK2 V617F, SETBP1, ASXL1 and CALR gene in 22 patients with CNL
Discussion
Chronic neutrophilic leukaemia is a rare aggressive myeloproliferative neoplasm [Citation1]. Since Touhy was first reported in 1920, <200 cases have been reported in the literature locally and worldwide [Citation9]. In 2013, Maxson et al. [Citation2] found that the oncogenic mutation of the CSF3R gene is a molecular marker specific to CNL. Reports on CSF3R mutations in patients with CNL have been available since then [Citation4,Citation5]. Approximately 80%–95% of patients with CNL have CSF3R mutations, in which CSF3R T618I which is a membrane proximal mutation is the main mutation type. Some truncated mutations are present in the intracellular region (D771fs,S783fs,Y752X and W791X) [Citation2,Citation3]. Out of the four patients with CNL, two had CSF3R mutations, all of which were CSF3R T618I mutations. JAK2 V617F, CALR and MPLmutations are the main molecular aetiology of polycythaemia vera, essential thrombocythemia and primary myelofibrosis [Citation10]. However, JAK2 V617F and CALR mutations are also present in some patients with CNL, and MPL mutation has not been reported. The mutation rate of SETBP1 in CNL is 33% [Citation3]. The gene encodes SET binding protein and is the binding partner of the multifunctional SET protein. SETBP1 is mainly involved in cell apoptosis, transcription and nucleosome assembly [Citation11]. ASXL1 and ASXL2 genes which belong to the ASXL gene family are the genes related to chromosome modification and regulation. These genes play a dual regulatory function of transcription inhibition and activation [Citation12–14]. Elliott et al. [Citation8] found that ASXL1 gene mutation is as high as 57% in CSF3R mutation-positive patients with CNL.. ASXL2 mutations have been found in some patients with AML [Citation15], but the ASXL2 gene mutation in patients with CNL is unclear. Therefore, in the present study, in addition to CSF3R mutation, JAK2 V617F, CALR, MPL, SETBP1, ASXL1 and ASXL2 gene mutations were analyzed in four patients with CNL. JAK2 V617F mutation was independent of CSF3R mutation in two patients. Two patients with CSF3R T618I mutation were accompanied by SETBP1 and ASXL1 mutations. CALR, MPL and ASXL2 mutations were undetected in this group.
To understand the characteristics and possible role of the gene mutations in CNL further, we analysed the mutation data of 22 patients with CNL in the literature. Firstly, JAK2 V617F is a mutagenic gene independent of CSF3R mutation which suggested that JAK2 V617F may be another molecular cause of CNL development except CSF3R mutation. However, the frequency of JAK2 V617F mutation was significantly lower than that of CSF3R mutation. Secondly, SETBP1 and ASXL1 gene mutations possessed high frequencies in patients with CNL, but they did not exist independently in CNL. These mutations were always accompanied in CSF3R mutation-positive patients either alone or in combination. Hence, these mutations are not involved in driving CNL but may play a role in the CNL disease progression. The occurrence of SETBP1 mutation was also related to the mutation type of CSF3R T618I which suggested that the occurrence of SETBP1 mutation is not a random event but has a certain relationship with CSF3R T618I. The mechanism underlying SETBP1 mutation still needs to be explored. Thirdly, the CALR mutation rate in CNL was low, and mutation coexisted with CSF3R mutation. The case of JAK2 V617F combined with BCR-ABL1 is characterized by the transformation of the phenotypes of MPN and chronic myeloid leukaemia [Citation16,Citation17] which suggested that the patient had these two diseases at the same time. Therefore, CALR mutation is a driving gene that can cause MPN [Citation18]. Therefore, we preliminarily speculated that the CALR here may not be a mutated molecule in CNL pathogenesis but is a case similar to that of JAK2 V617F combined with BCR-ABL1. The CALR and CSF3R mutations were independent of different clone cells, and the patients were complicated with CNL and another MPN disease at the same time. On the basis of the gene mutation distribution in 22 patients, we preliminarily considered that CSF3R, JAK2 V617F, SETBP1 and ASXL1 are the main mutation genes associated with CNL. CSF3R and JAK2 V617F gene mutations are two phenotypic-driven mutant genes which can independently cause CNL, whilst SETBP1 and ASXL1 mutations are nonphenotypic-driven mutant genes which are associated with CSF3R mutations and may affect the progression of CNL. CSF3R mutation has been included in the diagnostic criteria of CNL. JAK2 V617Fmutation can be considered as the preferred examination method in CSF3R mutation-negative patients.
The median survival time of patients with CNL is 2 years [Citation19]. The question on how does the mutation gene affect the prognosis of patients with CNL should be addressed. Pardanani et al. [Citation3]conducted a small sample analysis and found that SETBP1 mutation is associated with high leukocyte and poor prognosis in patients with CNL, and patients with CNL with this mutation have short survival time and are independent risk factors for prognosis. Elliott et al. [Citation9] conducted a multivariate analysis of 14 CSF3R mutation-positive patients with CNL and found that ASXL1 mutation is an independent prognostic factor for short survival period. In this study, we also followed up four patients with CNL, all of whom survived during the follow-up period and did not progress to the malignant direction of bone marrow fibrosis and acute leukaemia. With the exception of one patient with a short follow-up time (3 months), the survival times of the three other patients were longer than that of the median 2-year survival period. One patient hadCSF3R T618I mutation combined with SETBP1 and ASXL1 mutation, and two patients possessed JAK2 V617F mutation.
The current treatment for CNL is based on oral hydroxyurea or interferon α- injection [Citation19]. The discovery of CNL-related molecular mutations may provide a therapeutic target for CNL. JAK and SRC kinase are the downstream genes of CSF3R [Citation20,Citation21]. CSF3R mutations near the transmembrane region, such asCSF3RT618Imutation, mainly activate the downstream JAK kinasesignalling molecules, and CSF3R-truncated mutations in intracellular region mainly activate the SRC kinase signalling downstream molecules of CSF3R. The near transmembrane of CSF3R mutation may be sensitive to JAK inhibitors such as Lucertini, whilst the intracellular CSF3R mutation may be sensitive to SRC kinase inhibitors such as Dasatinib [Citation2,Citation20,Citation21]. For patients with JAK2 V617F-positive CNL, Lukotinib can be used as the target drug. Hence, with the development of technology, targeted therapy will become the main treatment of CNL in the future, and targeted drug selection needs to be based on the accurate molecular CNL diagnosis.
Acknowledgements
This work was supported by National Natural Science Foundation of China (NO.81670126; NO.81500104), The Shanxi Natural Science Foundation of China (NO.201801D111003;NO.201801D221409).
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bohm J, Kock S, Schaefer HE, et al. Evidence of clonality in chronic neutrophilic leukaemia. J Clin Pathol. 2003;56(4):292–295. doi: 10.1136/jcp.56.4.292
- Maxson JE, Gotlib J, Pollyea DA, et al. Oncogenic CSF3R mutations in chronic neutrophilic leukemia and atypical CML. N Engl J Med. 2013;368(19):1781–1790. doi: 10.1056/NEJMoa1214514
- Pardanani A, Lasho TL, Laborde RR, et al. CSF3R t618i is a highly prevalent and specific mutation in chronic neutrophilic leukemia. Leukemia. 2013;27(9):1870–1873. doi: 10.1038/leu.2013.122
- Cui Y, Li B, Jiang Q, et al. [CSF3R, ASXL1,SETBP1, JAK2 V617F and CALR mutations in chronic neutrophilic leukemia]. Zhonghua xue ye xue za zhi. 2014;35(12):1069–1073.
- Lasho TL, Elliott MA, Pardanani A, et al. . CALR mutation studies in chronic neutrophilic leukemia. Am J Hematol. 2014;89(4):450. doi: 10.1002/ajh.23665
- Gajendra S, Gupta R, Chandgothia M, et al. Chronic neutrophilic Leukemia with V617F JAK2 mutation. Indian J Hematol Blo. 2014;30(2):139–142. doi: 10.1007/s12288-012-0203-6
- Imashuku S, Kudo N, Kubo K, et al. Rituximab for managing acquired hemophilia A in a case of chronic neutrophilic leukemia with the JAK2 kinase V617F mutation. J Blood Med; 2012;3:157–161. doi: 10.2147/JBM.S37631
- Elliott MA, Pardanani A, Hanson CA, et al. ASXL1 mutations are frequent and prognostically detrimental in CSF3R-mutated chronic neutrophilic leukemia. Am J Hematol. 2015;90(7):653–656. doi: 10.1002/ajh.24031
- Gotlib J, Maxson JE, George TI, et al. The new genetics of chronic neutrophilic leukemia and atypical CML: implications for diagnosis and treatment. Blood. 2013;122(10):1707–1711. doi: 10.1182/blood-2013-05-500959
- Vainchenker W, Kralovics R. Genetic basis and molecular pathophysiology of classical myeloproliferative neoplasms. Blood. 2017;129(6):667–679. doi: 10.1182/blood-2016-10-695940
- Minakuchi M, Kakazu N, Gorrin-Rivas MJ, et al. Identification and characterization of SEB, a novel protein that binds to the acute undifferentiated leukemia-associated protein SET. Eur J Biochem. 2001;268(5):1340–1351. doi: 10.1046/j.1432-1327.2001.02000.x
- Katoh M. Functional and cancer genomics of ASXL family members. Br J Cancer. 2013;109(2):299–306. doi: 10.1038/bjc.2013.281
- Fisher CL, Randazzo F, Humphries RK, et al. Characterization of Asxl1, a murine homolog of Additional sex combs, and analysis of the Asx-like gene family. Gene. 2006;369:109–118. doi: 10.1016/j.gene.2005.10.033
- Lai HL, Wang QT. Additional sex combs-like 2 is required for polycomb repressive complex 2 binding at select targets. PloS one. 2013;8(9):e73983. doi: 10.1371/journal.pone.0073983
- Micol JB, Duployez N, Boissel N, et al. Frequent ASXL2 mutations in acute myeloid leukemia patients with t(8;21)/RUNX1-RUNX1T1 chromosomal translocations. Blood. 2014;124(9):1445–1449. doi: 10.1182/blood-2014-04-571018
- Bornhauser M, Mohr B, Oelschlaegel U, et al. Concurrent JAK2(V617F) mutation and BCR-ABL translocation within committed myeloid progenitors in myelofibrosis. Leukemia. 2007;21(8):1824–1826. doi: 10.1038/sj.leu.2404730
- Hussein K, Bock O, Theophile K, et al. Chronic myeloproliferative diseases with concurrent BCR-ABL junction and JAK2V617F mutation. Leukemia. 2008;22(5):1059–1062. doi: 10.1038/sj.leu.2404993
- Klampfl T, Gisslinger H, Harutyunyan AS, et al. Somatic mutations of calreticulin in myeloproliferative neoplasms. N Engl J Med. 2013;369(25):2379–2390. doi: 10.1056/NEJMoa1311347
- Elliott MA, Hanson CA, Dewald GW, et al. WHO-defined chronic neutrophilic leukemia: a long-term analysis of 12 cases and a critical review of the literature. Leukemia. 2005;19(2):313–317. doi: 10.1038/sj.leu.2403562
- Lasho TL, Mims A, Elliott MA, et al. Chronic neutrophilic leukemia with concurrent CSF3R and SETBP1 mutations: single colony clonality studies, in vitro sensitivity to JAK inhibitors and lack of treatment response to ruxolitinib. Leukemia. 2014;28(6):1363–1365. doi: 10.1038/leu.2014.39
- Dao KH, Solti MB, Maxson JE, et al. Significant clinical response to JAK1/2 inhibition in a patient with CSF3R-T618I-positive atypical chronic myeloid leukemia. Leuk Res Rep. 2014;3(2):67–69.
