ABSTRACT
Objectives: DNA methylation is a well-known epigenetic modification, and it can be iteratively oxidized to 5-hydroxymethylcytosine (5-hmC), 5-formylcytosine (5-fC) and 5-carboxylcytosine (5-caC). Acute lymphoblastic leukemia (ALL) is a severe hematological disease, and it is essential to find out new biomarkers to better diagnose and cure ALL due to the development of chemo-resistance and low cure rate in adult ALL. This study aims to describe the role of global methylation and demethylation intermediates in ALL.
Methods: The levels of global methylation and its oxidation products in the peripheral blood (PB) of ALL patients and healthy controls were determined by Enzyme-Linked Immunosorbent Assay (ELISA).
Results: In this study, we described that global 5-mC, 5-hmC and 5-fC levels were dysregulated in ALL, and they were associated with clinical characteristics and genetic abnormalities of ALL patients. Interestingly, 5-mC and 5-hmC were closely related to inflammation, and the levels of 5-hmC were inversely correlated with C-Reactive protein (CRP) and ferritin. Finally, 5-mC and 5-hmC were associated with complete remission (CR), and 5-hmC was revealed as an independent prognostic indicator for ALL.
Conclusion: This study described a novel role for global methylation and demethylation intermediates in ALL detection and prognosis, and provided new clue to distinguish high-risk patients and improve the curative effect on ALL patients.
Introduction
Acute lymphoblastic leukemia (ALL), which is characterized by excessive proliferation of abnormal and undifferentiated lymphocytes, is one of the most common cancers in children, and also affects adults [Citation1]. A steady improvement in treatment outcome has been achieved due to optimal use of the antileukemic agents and a stringent application of prognostic factors for risk-directed therapy in the last several decades. The five-year survival rate in childhood ALL has been nearly 90%, while that in adults is still only 20–40% [Citation2]. The prognosis of some ALL patients can be particularly poor, especially among adults, because of the development of chemo-resistance and disease recurrence [Citation3, Citation4]. Therefore, it is essential to identify new biomarkers of ALL that can predict high-risk patients, and thus provide a hint for individualized therapy selection.
Epigenetic modifications, such as DNA methylation, have been described in ALL [Citation5]. DNA methylation is an epigenetic mark that can regulate the expression of specific genes, and therefore plays a critical role in numerous diseases, including ALL. It has been reported that DNA methylation was dysregulated in childhood ALL, and it was useful for subtype classification and prognosis evaluation [Citation6,Citation7]. In addition, molecular biology research showed that aberrant methylation in multiple genes inhibited their transcription, which was likely to play a key role in leukemogenesis [Citation7].
Despite its stability, DNA methylation can still be reversed to its unmodified state by DNA demethylation process. DNA demethylation is usually mediated by TET proteins, which oxidize 5-methylcytosine (5-mC) iteratively to 5-hydroxymethylcytosine (5-hmC), 5-formylcytosine (5-fC) and 5-carboxylcytosine (5-caC). 5-caC was further cleaved by thymine DNA glycosylase and restored to a normal cytosine by base-excision repair [Citation8]. TET proteins require iron and α-ketoglutarate (α-KG) as co-substrates, and catalyze oxidative decarboxylation of α-KG, thereby generating an enzyme-bound Fe(IV)-oxo intermediate that converts 5mC to 5hmC, 5-fC and 5-caC [Citation9]. The balance between DNA methylation and demethylation is frequently deregulated in cancer, leading to aberrant modification patterns. Mutations that disrupt the functions of TET genes cause changes in 5-mC and 5-hmC levels of hematopoietic stem cells and participate in the pathogenesis of hematopoietic malignancies [Citation10,Citation11]. However, the clinical implications of DNA methylation and its oxidation products have not been comprehensively evaluated in patients with ALL.
Herein, we aimed to evaluate the levels of global 5-mC, 5-hmC, 5-fC and 5-caC in ALL patients compared with those in healthy controls by a previously described Enzyme-Linked Immunosorbent Assay (ELISA). The relationships between these modifications and clinical characteristics and infection status of ALL patients were evaluated. Finally, the prognostic values of these modifications were assessed. Overall, we want to clarify the clinical significance of DNA methylation and demethylation intermediates in ALL.
Materials and methods
Study population
A total of 52 patients who had been diagnosed with ALL, including 37 B lineage and 15 T lineage ALL patients, in Peking University People’s Hospital from 2015 to 2018 were included in this study. The age range of patients was 11–68 years old, and 51.9% (27/52) was <30. In addition, 26 individuals without abnormality in the physical examination were used as healthy controls, and the age range was 12–65 years old, of which 53.8% (14/26) was <30. The clinical and laboratory characteristics of patients are summarized in . The study was carried out in accordance with the principles of the Declaration of Helsinki and approved by the Ethics Committee of Peking University People’s Hospital. The informed consents were obtained from the subjects before their participation. Peripheral blood (PB) samples from 52 patients and 26 healthy controls were collected separately.
Table 1. Clinical and laboratory characteristics of patients with acute lymphoblastic leukemia.
DNA extraction
Genomic DNA from PB was extracted using RelaxGene Blood DNA System Kit (Tiangen, Beijing, China) according to the manufacturer's instructions. The concentration and purity of DNA were measured by NanoDrop 2000 (Thermo Scientific, Wilmington, DE, U.S.A.). The ratio of 260/280 nm range was between 1.7 and 1.9.
Clinical indicator analysis
Hemoglobin (Hb) concentration, platelet (PLT) and white blood cell (WBC) counts were measured by Sysmex XE-2100 (TOA Medical Electronics, Kobe, Japan) with PB collected in EDTA Vacutainer tubes (Becton Dickinson, NJ, U.S.A.). For the detection of serum procalcitonin (PCT), C-Reactive protein (CRP) and ferritin, serum was separated from PB by centrifuging at 4000 × g for 5 min, and serum concentrations of PCT and CRP were detected by Roche Cobas e401 and Hitachi LABOSPECT008 separately. All tests were performed with original kits from the manufacturers according to the standard operation procedures, and calibration and quality control were performed sequentially before detection to ensure the measurement accuracy.
Global 5-mC, 5-hmc, 5-fC and 5-cac levels assessment by ELISA
An ELISA assay was applied to measure the global 5-mC, 5-hmC, 5-fC and 5-caC as previously described [Citation12]. Briefly, DNA samples were denatured and dispensed in high-affinity microplates, and the plates were incubated at 37°C for 90 min. Wash each well three times, block with bovine serum albumin (BSA) 1% at RT for 60 min. Next, wash each well three times, incubate with the diluted anti-5-mC, anti-5-hmC, anti-5-fC or anti-5-caC antibody at RT for 120 min separately. All anti-cytosine derivative antibodies or kits were purchased from Abcam (Cambridge, U.K.). After 6 washes, diluted horseradish peroxidase (HRP) -labelled goat anti-mouse or goat anti-rabbit (Zhongshan Jinqiao, Beijing, PR China) were added and the plate was incubated for 1 h at RT. The plates were then incubated with HRP Substrate Solution for several minutes until positive control wells turns medium blue. Finally, add the Stop Solution to each well to stop enzyme reaction and read the absorbance on a microplate reader (Bio-Rad, Hercules, CA, U.S.A.) at 450 nm within 15 min. The calibration curve was performed before the test, and non-specific background OD (well without DNA) was subtracted from the corresponding test sample.
Statistical analysis
All analyses were carried out with GraphPad Prime 5.01 or SPSS software. All data are expressed as mean ± standard deviation. Results between different groups were compared using Mann–Whitney U test. For correlation analysis, the Spearman correlation coefficients (r) were estimated between two variables. Multivariate logistic regression analyses were carried out using SPSS software to evaluate the factors influencing Complete Remission (CR). Analysis of the receiver operating characteristic (ROC) curves and the area under the ROC curves (AUC) were performed using SPSS software to evaluate the clinical values of the cytosine derivatives in ALL patients. All the statistical tests were 2-tailed. A p value <0.05 was considered to be statistically significant.
Results
Global 5-mC, 5-hmc and 5-fC levels in ALL patients are dysregulated in ALL patients
52 ALL patients and 26 healthy controls were enrolled in this study. ELISA assays were performed to detect global 5-mC, 5-hmC, 5-fC and 5-caC levels of PB in each individual. At first, calibration was performed to ensure the detection accuracy, and the calibration curves were plotted in . Global 5-mC, 5-hmC and 5-fC levels were measured and compared between ALL patients and healthy controls. As shown in (A–C), ALL patients possessed a higher level of 5-mC (p < 0.05), lower level of 5-hmC (p < 0.05) and a higher level of 5-fC (p < 0.001) globally than healthy controls. Unfortunately, we failed to detect 5-caC, and it might be due to low abundance of 5-caC in PB and low sensitivity of anti-5-caC antibody.
Figure 1. The calibration curve of the ELISA assay for detecting global 5-mC (A), 5-hmC (B) and 5-fC (C).
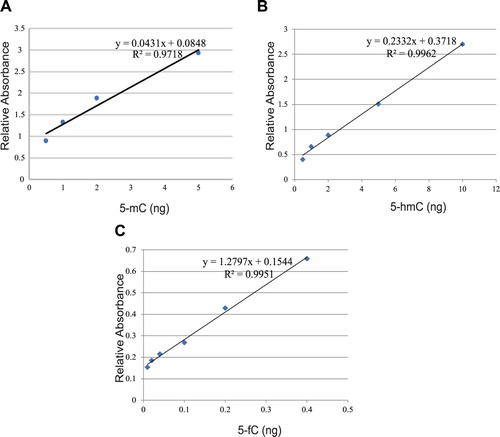
Figure 2. ALL patients possessed higher 5-mC, lower 5-hmC and higher 5-fC than Healthy controls. Global 5-mC (A), 5-hmC (B) and 5-fC (C) in ALL patients (n = 52) and Healthy controls (n = 26) was measured by ELISA separately. Results are presented as the mean value ± standard deviations. (D) The performance of 5-mC, 5-hmC, 5-fC and their combination to detect ALL. ROC curves were generated and each AUC was calculated separately. ROC, receiver operating characteristic; AUC, area under the curve.
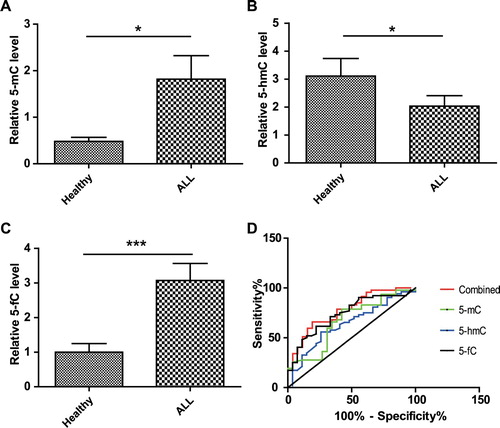
To investigate whether there is a correlation among 5-mC, 5-hmC and 5-fC, correlation analysis was conducted separately (). Interestingly, we found that the levels of 5-fC were positively correlated with 5-mC (r > 0), and the correlation was very significant (p < 0.001). However, the correlation between 5-hmC and 5-mC (p > 0.05), as well as that between 5-hmC and 5-fC (p > 0.05) was poor.
Figure 3. Correlation analysis among 5-mC, 5-hmC and 5-fC. (A) The correlation analysis was conducted between 5-hmC and 5-mC. (B) Correlation analysis between 5-fC and 5-hmC was conducted as described in A. (C) Correlation analysis between 5-mC and 5-fC was conducted as described in A. P values <0.05 are regarded as significant, ∗∗∗P < 0.001. Scatter plots with linear fit are shown and Spearman r and p values are listed.
Next, to clarify the performance of 5-mC, 5-hmC and 5-fC to detect ALL, a ROC analysis was performed and the AUC was calculated. As shown in (D), the AUC of 5-mC, 5-hmC and 5-fC were 0.67, 0.64 and 0.74, respectively, indicating the moderate performances of 5-mC, 5-hmC and 5-fC for detecting ALL. In addition, combined test of the three cytosine derivatives was conducted and the AUC was 0.78, suggesting a better performance than any single derivative. Generally, we found that the levels of global methylation and demethylation intermediates were dysregulated in ALL patients compared to healthy individuals.
Association of global 5-mC, 5-hmC and 5-fC levels with clinical characteristics and genetic abnormalities of ALL patients
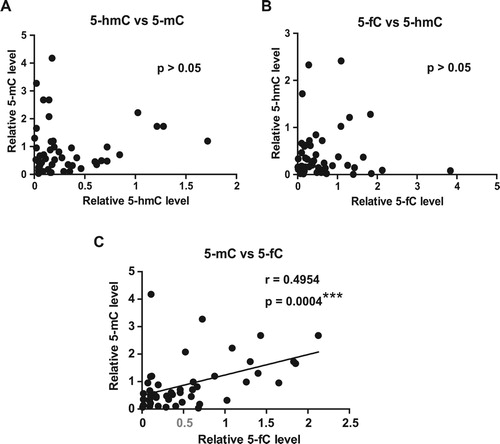
As the levels of global 5-mC, 5-hmC and 5-fC were dysregulated in ALL patients, we want to determine the association between these modifications and clinical characteristics of ALL patients. As shown in , the levels of global 5-mC, 5-hmC and 5-fC were associated with some clinical characteristics. Specifically, 5-mC was associated with risk stratification according to the NCCN guideline [Citation13]. It was illustrated that the levels of global 5-mC in high-risk group were significantly higher than low/intermediate-risk groups ((A)). In addition, both 5-mC and 5-fC were related to the lineage of ALL patients. We found that global 5-mC levels in common/pre-B lineage were significantly higher than pro-B and T lineage, while global 5-fC levels in pro-B, but not T lineage, were significantly lower than common/pre-B lineage ((B–C)). However, there is no significant relationship between 5-hmC and any clinical characteristic ().
Figure 4. The association of DNA methylation and demethylation intermediates with the clinical characteristics and genetic abnormalities of ALL patients. ALL patients were divided into different groups according to the clinical characteristics and genetic abnormalities, such as lineage, risk stratification, IKZF mutation and hyperdiploidy. Global 5-mC, 5-hmC and 5-fC levels in different groups were analyzed and compared, and results are presented as the mean value ± standard deviations. ns, not significant, ∗P < 0.05, ∗∗P < 0.01.
Table 2. Relationship between clinical characteristics and 5-mC, 5-hmC or 5-fC levels in ALL patients.
ALL patients possess many different genetic abnormalities, some of which are critical regulators of ALL progression [Citation14–16]. To determine the relationship between DNA methylation/demethylation intermediates and genetic abnormalities, we analyzed the association of 5-mC, 5-hmC and 5-fC with some common genetic alteration status, including BCR/ABL fusion, IgH rearrangement, hyperdiploidy and IKZF mutation, of ALL patients. As shown in (D–F) and , we found that the levels of global 5-mC in patients with IKZF mutation was significantly higher than those without IKZF mutation, while the levels of both 5-hmC and 5-fC in patients with hyperdiploidy was significantly lower than those without hyperdiploidy (p < 0.05). Collectively, these findings suggest that DNA methylation and its oxidation products were closely related to clinical characteristics and genetic abnormalities of ALL patients.
Global 5-mC and 5-hmC levels were associated with actual serum inflammation markers in ALL patients
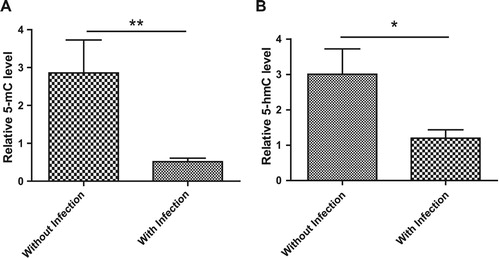
Infection is a major complication of acute leukemia, so we determined whether there is a possible relationship between infection and the levels of global 5-mC, 5-hmC and 5-fC in ALL patients. At first, the levels of global 5-mC, 5-hmC and 5-fC in ALL patients with and without infections were compared, and the results are shown in (A,B) and Figure S1. We found that global 5-mC and 5-hmC levels were significantly lower in patients with infection than patients without infection. However, global 5-fC levels in ALL patients with and without infections showed no difference.
Figure 5. The association of 5-mC and 5-hmC with infection status in ALL patients. (A,B) ALL patients were divided into two groups, patients with infection and patients without infection according to the diagnosis by physicians. The levels of global 5-mC (A) and 5-hmC (B) in 2 groups were compared with each other. Data points are shown with mean value ± standard deviations.
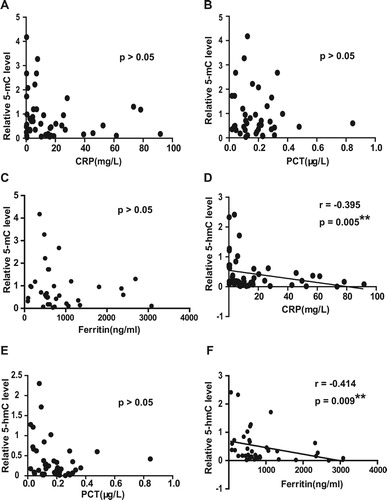
Infection can lead to inflammation in the body, so we measured levels of inflammation factors, including CRP, PCT and ferritin, in the serum of ALL patients. The correlation analysis was conducted between 5-mC / 5-hmC and inflammation factors, and the results were shown in . There is a slight correlation between global 5-hmC level and serum CRP concentration (r = −0.395, p = 0.005) ((D)), as well as serum ferritin level (r = −0.414, p = 0.009) ((F)). However, the correlations between 5-mC and CRP ((A)), 5-mC and PCT ((B)), 5-mC and ferritin ((C)) or 5-hmC and PCT ((E)) were poor. These data indicate that 5-mC and 5-hmC were associated with inflammation in ALL patients.
Figure 6. (A–C) Correlation analysis between 5-mC and serum CRP (A), PCT (B) or ferritin (C) in ALL patients. Serum CRP, PCT and ferritin levels of each ALL patients were measured. Correlation analysis was conducted between global 5-mC levels and these indicators among all the ALL patients. (D–F) Correlation analysis between 5-hmC and serum CRP (D), PCT (E) or ferritin (F) in ALL patients was conducted as described in A. Scatter plots with linear fit are shown and Spearman r and p values are listed, ∗P < 0.05, ∗∗P < 0.01.
Global 5-mC and 5-hmc levels are associated with complete remission of ALL patients
As IKZF mutation was associated with poor clinical outcomes and hyperdiploidy was identified as a favorable prognostic factor in ALL [Citation4], and it is hypothesized that 5-mC, 5-hmC and 5-fC is associated with complete remission (CR) of ALL patients. We compared global 5-mC, 5-hmC and 5-fC levels between patients with CR and those without CR. As shown in (A,B), patients achieving CR possessed lower 5-mC and 5-hmC levels than patients who did not achieve CR, indicating that global 5-mC and 5-hmC levels are risk factors for complete remission in ALL patients. However, the 5-fC levels in patients achieving CR and those who did not achieve CR showed no significant difference ((C)).
Figure 7. 5-mC and 5-hmC were potential prognostic indicator for ALL. ALL patients were divided into two groups, patients achieving CR and patients who did not achieve CR according to the diagnosis by physicians. The levels of global 5-mC (A), 5-hmC (B) and 5-fC (C) in two groups were compared with each other. Data points are shown with mean value ± standard deviations, ∗P < 0.05, ∗∗P < 0.01, ns, not significant. (D) ROC curves of 5-mC, 5-hmC, 5-fC and their combination as CR predictors of ALL patients. ROC, receiver operating characteristic; CR, complete remission; AUC, area under the curve.
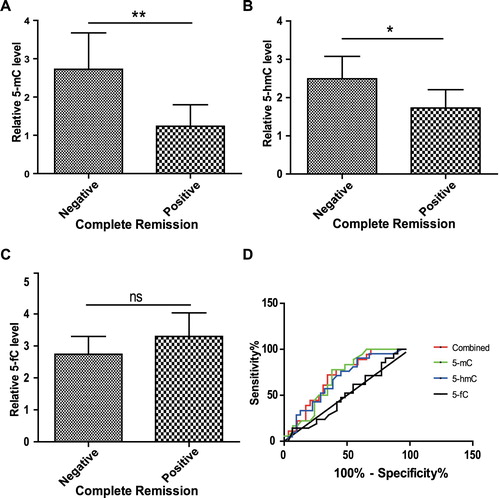
To further clarify the prognostic value of 5-mC, 5-hmC and 5-fC for CR of ALL patients, a ROC analysis was performed and the AUC was calculated. As shown in (D), the AUC of 5-mC, 5-hmC and 5-fC were 0.69, 0.67 and 0.50, respectively, indicating that 5-mC and 5-hmC, but not 5-fC, have moderate prognostic values. In addition, combined test of the three cytosine derivatives was performed and the AUC was 0.70, suggesting a better prognostic value than any single derivative.
To assess the independence of global methylation/demethylation intermediates and known risk factors for ALL prognosis, multivariate analysis was performed and 5-mC 5-hmC and 5-fC were entered into a logistic regression model as continuous variable in addition to other independent prognostic factors, including IKZF mutation, hyperdiploidy, IgH rearrangement and infection. As shown in , global 5-hmC level was independently associated with CR of ALL patients (p < 0.05), suggesting the independent prognostic significance of 5-hmC. Altogether, we found that global methylation / demethylation intermediates might be prognostic markers for ALL patients.
Table 3. Multivariate analysis for complete remission of ALL patients.
Discussion
In this study, DNA methylation and oxidation products were identified as potential tumor markers for ALL. Our data indicate that the levels of 5-mC, 5-hmC and 5-fC were dysregulated in ALL patients, and they were associated with some clinical characteristics and genetic abnormalities of ALL patients. Interestingly, the association of 5-mC and 5-hmC with inflammation was described, and global 5-hmC level was correlated with serum ferritin and CRP levels in ALL patients. Finally, 5-mC and 5-hmC were closely related to CR of ALL patients, and 5-hmC has the potential to be an independent predictor for complete remission in ALL patients.
It has been demonstrated that DNA methylation was closely associated with ALL. At first, abnormal hypermethylation at specific regions, such as the promoter region of some genes, such as p73, RASSF6 and RASSF10 [Citation17,Citation18], often exerted critical function in the pathogenesis and progression of ALL, and could be used as new biomarkers for prognostic evaluation of ALL patients. Besides the study of a single site, global DNA methylation was also determined, and the methylomes of control cells, ALL cells at diagnosis and relapse showed big differences, implying that aberrant global methylation is a signature of leukemic development and progression [Citation19]. As the tight relationship between DNA methylation and demethylation, it is hypothesized that DNA demethylation might also be critical for ALL progression and have implications for ALL diagnosis and prognosis. In this study, we indeed found that the aberrant levels of 5-mC, 5-hmC and 5-fC were associated with the clinic in ALL patients, confirming the crucial role of DNA methylation and demethylation in ALL.
Aberrant DNA demethylation is observed in various solid tumors because of the dysregulation of TET expression. For example, it was reported that diminished expression of TET family genes resulted in ‘loss of 5-hmC’ in human melanoma, and 5-hmC was a putative molecular marker for melanoma diagnosis and prognosis [Citation20]. Another study revealed that the levels of 5-hmC were dramatically reduced in several different human cancers, including breast, liver, lung, pancreatic and prostate cancer, and 5hmC decrease was also associated with a substantial reduction of TET gene expression in human tumors [Citation21]. For hamatopoietic malignancies, especially acute myelocytic leukemia, TET mutations were the main cause of dysregulated DNA demethylation. Many of these mutations compromise the enzymatic activity of TET and lead to 5-mC accumulation and 5-hmC reduction as solid tumors [Citation22–24]. Similarly, this study reports that 5-hmC was decreased and 5-fC was increased in ALL, further supporting the disruption of balance between DNA methylation and demethylation in cancers. However, the mutation rates of TET in ALL were not as high as AML [Citation25–27], and other factors that influence DNA methylation/demethylation process need to be further investigated.
Several methods currently exist for the detection and quantification of DNA methylation and demethylation intermediates. The most common method previously used for methylation analysis is based on bisulfite conversion, in which sodium bisulfite treatment of genomic DNA converts unmethylated cytosines to uracil, while methylated cytosine residues remain unaffected. It is usually combined with Polymerase Chain Reaction (PCR) or sequencing to resolve DNA methylation qualitatively and quantitatively [Citation28]. However, bisulfite conversion-based methods can't distinguish between 5-mC and 5-hmC in DNA, and novel approaches are required to detect different cytosine derivatives [Citation29]. Several other methods have been developed for the analysis of DNA methylation and demethylation intermediates, including high-performance liquid chromatography-tandem mass spectrometry (HPLC-MS/MS) and antibody-based immunochemical analysis. HPLC-MS/MS realizes high-resolution separation to obtain the total base composition of the genome after complete enzymatic hydrolysis of DNA [Citation30]. This method is highly sensitive for quantitative analysis, but it is very complicated to perform and requires specific expertise and equipment. Besides HPLC-MS/MS, antibody-based immunochemical analysis, such as ELISA, immunofluorescence and immunohistochemistry (IHC), has been proved as a powerful tool for studying global DNA methylation and demethylation intermediates [Citation31]. In this study, we used ELISA assay, which has been described before, to detect global DNA methylation and demethylation intermediates, and the detection accuracy was acceptable for the quantification of 5-mC, 5-hmC and 5-fC.
In summary, our data describe the clinical significance of DNA methylation / demethylation intermediates in ALL patients. This study indicates that 5-mC, 5-hmC and 5-fC might be potential biomarkers for ALL and provides a useful strategy with which to distinguish high-risk patients and improve the curative effect on ALL patients.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Harrison, C. J. Acute lymphoblastic leukemia. Clin Lab Med. 2011; 31(4): 631-647 , ix. doi: 10.1016/j.cll.2011.08.016
- Katz, A. J., Chia, V. M., Schoonen, W. M., et al. Acute lymphoblastic leukemia: an assessment of international incidence, survival, and disease burden. Cancer Causes Control. 2015; 26(11): 1627-1642. doi: 10.1007/s10552-015-0657-6
- Pui, C. H., and Evans, W. E. Treatment of acute lymphoblastic leukemia. N Engl J Med. 2006; 354(2): 166-178. doi: 10.1056/NEJMra052603
- Inaba, H., Greaves, M., and Mullighan, C. G. Acute lymphoblastic leukaemia. Lancet. 2013; 381(9881): 1943-1955. doi: 10.1016/S0140-6736(12)62187-4
- Gutierrez, M. I., Siraj, A. K., Bhargava, M., et al. Concurrent methylation of multiple genes in childhood ALL: correlation with phenotype and molecular subgroup. Leukemia. 2003; 17(9): 1845-1850. doi: 10.1038/sj.leu.2403060
- Milani, L., Lundmark, A., Kiialainen, A., et al. DNA methylation for subtype classification and prediction of treatment outcome in patients with childhood acute lymphoblastic leukemia. Blood. 2010; 115(6): 1214-1225. doi: 10.1182/blood-2009-04-214668
- Figueroa, M. E., Chen, S. C., Andersson, A. K., et al. Integrated genetic and epigenetic analysis of childhood acute lymphoblastic leukemia. J Clin Invest. 2013; 123(7): 3099-3111. doi: 10.1172/JCI66203
- Wu, X., and Zhang, Y. TET-mediated active DNA demethylation: mechanism, function and beyond. Nat Rev Genet. 2017; 18(9): 517-534. doi: 10.1038/nrg.2017.33
- Kohli, R. M., and Zhang, Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature. 2013; 502(7472): 472-479. doi: 10.1038/nature12750
- Moran-Crusio, K., Reavie, L., Shih, A., et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell. 2011; 20(1): 11-24. doi: 10.1016/j.ccr.2011.06.001
- Li, Z., Cai, X., Cai, C. L., et al. Deletion of Tet2 in mice leads to dysregulated hematopoietic stem cells and subsequent development of myeloid malignancies. Blood. 2011; 118(17): 4509-4518. doi: 10.1182/blood-2010-12-325241
- Thabet, Y., Le Dantec, C., Ghedira, I., et al. Epigenetic dysregulation in salivary glands from patients with primary Sjogren’s syndrome may be ascribed to infiltrating B cells. J Autoimmun. 2013; 41:175-181. doi: 10.1016/j.jaut.2013.02.002
- National Comprehensive Cancer Network. Clinical practice guidelines in oncology: acute lymphoblastic leukemia, 2. 2014
- Virely, C., Moulin, S., Cobaleda, C., et al. Haploinsufficiency of the IKZF1 (IKAROS) tumor suppressor gene cooperates with BCR-ABL in a transgenic model of acute lymphoblastic leukemia. Leukemia. 2010; 24(6): 1200-1204. doi: 10.1038/leu.2010.63
- Paganin, M., and Ferrando, A. Molecular pathogenesis and targeted therapies for NOTCH1-induced T-cell acute lymphoblastic leukemia. Blood Rev. 2011; 25(2): 83-90. doi: 10.1016/j.blre.2010.09.004
- Mullighan, C. G., Su, X., Zhang, J., et al. Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N Engl J Med. 2009; 360(5): 470-480. doi: 10.1056/NEJMoa0808253
- Corn, P. G., Kuerbitz, S. J., van Noesel, M. M., et al. Transcriptional silencing of the p73 gene in acute lymphoblastic leukemia and Burkitt’s lymphoma is associated with 5’ CpG island methylation. Cancer Res. 1999; 59(14): 3352-3356.
- Younesian, S., Shahkarami, S., Ghaffari, P., et al. DNA hypermethylation of tumor suppressor genes RASSF6 and RASSF10 as independent prognostic factors in adult acute lymphoblastic leukemia. Leuk Res. 2017; 61:33–38. doi: 10.1016/j.leukres.2017.08.016
- Nordlund, J., Backlin, C. L., Wahlberg, P., et al. Genome-wide signatures of differential DNA methylation in pediatric acute lymphoblastic leukemia. Genome Biol. 2013; 14(9): r105. doi: 10.1186/gb-2013-14-9-r105
- Lian, C. G., Xu, Y., Ceol, C., et al. Loss of 5-hydroxymethylcytosine is an epigenetic hallmark of melanoma. Cell. 2012; 150(6): 1135-1146. doi: 10.1016/j.cell.2012.07.033
- Yang, H., Liu, Y., Bai, F., et al. Tumor development is associated with decrease of TET gene expression and 5-methylcytosine hydroxylation. Oncogene. 2013; 32(5): 663-669. doi: 10.1038/onc.2012.67
- Ko, M., Huang, Y., Jankowska, A. M., et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010; 468(7325): 839-843. doi: 10.1038/nature09586
- Abdel-Wahab, O., Mullally, A., Hedvat, C., et al. Genetic characterization of TET1, TET2, and TET3 alterations in myeloid malignancies. Blood. 2009; 114(1): 144-147. doi: 10.1182/blood-2009-03-210039
- Figueroa, M. E., Abdel-Wahab, O., Lu, C., et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010; 18(6): 553-567. doi: 10.1016/j.ccr.2010.11.015
- Ntziachristos, P., Abdel-Wahab, O., and Aifantis, I. Emerging concepts of epigenetic dysregulation in hematological malignancies. Nat Immunol. 2016; 17(9): 1016-1024. doi: 10.1038/ni.3517
- McKenney, A. S., and Levine, R. L. Isocitrate dehydrogenase mutations in leukemia. J Clin Invest. 2013; 123(9): 3672-3677. doi: 10.1172/JCI67266
- Scourzic, L., Mouly, E., and Bernard, O. A. TET proteins and the control of cytosine demethylation in cancer. Genome Med. 2015; 7(1): 9. doi: 10.1186/s13073-015-0134-6
- Yokoi K., Yamashita K., Watanabe M. Analysis of DNA methylation status in bodily fluids for early detection of cancer. Int J Mol Sci. 2017;18:735. doi: 10.3390/ijms18040735
- Nestor, C., Ruzov, A., Meehan, R., et al. Enzymatic approaches and bisulfite sequencing cannot distinguish between 5-methylcytosine and 5-hydroxymethylcytosine in DNA. Biotechniques. 2010; 48(4): 317-319. doi: 10.2144/000113403
- Tang, Y., Zheng, S. J., Qi, C. B., et al. Sensitive and simultaneous determination of 5-methylcytosine and its oxidation products in genomic DNA by chemical derivatization coupled with liquid chromatography-tandem mass spectrometry analysis. Anal Chem. 2015; 87(6): 3445-3452. doi: 10.1021/ac504786r
- Chowdhury, B., Cho, I. H., and Irudayaraj, J. Technical advances in global DNA methylation analysis in human cancers. J Biol Eng. 2017; 11:10. doi: 10.1186/s13036-017-0052-9
