ABSTRACT
Objectives: Although DNA (cytosine-5)-methyltransferase 3 alpha (DNMT3A) gene mutations have been widely reported in myelodysplastic syndromes (MDS), the prognostic significance of DNMT3A mutations is still controversial. In this study, we conducted a meta-analysis to determine the prognostic effect of DNMT3A mutations in patients with MDS.
Methods: Eligible studies from PubMed, Embase, Web of Science, Clinical Trials and the Cochrane Library were searched. Hazard ratios (HRs) and their 95% confidence intervals (CIs) for overall survival (OS) and leukemia-free survival (LFS) were pooled to assess the effect of DNMT3A mutations on the prognosis in MDS patients.
Results: A total of 12 studies with 2236 patients were included in this meta-analysis. The pooled HRs for OS and LFS revealed that MDS patients with DNMT3A mutations had a significantly poor prognosis as compared with those without mutations (OS: HR = 1.654, 95% CI = 1.387–1.973, p < 0.001; LFS: HR = 4.624, 95% CI = 3.121–6.851, p < 0.001).
Discussion and Conclusion: This meta-analysis showed an adverse prognostic effect of DNMT3A mutations in patients with MDS, which will contribute to risk stratification and prognostic assessment in the disease.
Introduction
Myelodysplastic syndromes (MDS) are a group of heterogeneous myeloid clonal diseases originated from hematopoietic stem cells, which are characterized by cytopenia, ineffective hematopoiesis, hematopoietic failure and high-risk transformation to acute myeloid leukemia (AML) [Citation1]. With the development of gene sequencing technology, an increasing number of somatic mutant genes have been detected in MDS. DNMT3A is one of the most common mutated genes found in MDS patients [Citation2,Citation3], belonging to DNA methyltransferase (DNMT) family. There are mainly three functional DNA methyltransferases in mammals: DNMT1, DNMT3A, DNMT3B [Citation4]. DNMT1 is a maintenance methylase [Citation5], by the way, DNMT2 is not a DNA methyltransferase. DNMT3A gene is encoded by 23 exons on human chromosome 2p23, and encodes a 908-amino acid protein that, along with DNMT3B, is responsible for de novo CpG methylation independent of replication [Citation6,Citation7].
DNA methylation is one of the common epigenetic alterations in cancer [Citation8]. In MDS, aberrant DNA methylation is a dominant epigenetic change, which has been deeply studied and plays an important role in the pathogenesis and transformation to leukemia [Citation9,Citation10]. Due to the close relationship between DNA methylation and cell development, DNA methyltransferases have become important research hotspots in various diseases. So far, epigenetic regulation mechanism in MDS has been revealed well. DNMT3A mutations are detected frequently in patients with MDS, and often associated with prognosis [Citation11,Citation12].
Recently, somatic mutations in DNMT3A gene have been found in acute myeloid leukemia (AML) with a frequency of 22.1% [Citation13], and DNMT3A mutations were widely believed to be independently associated with inferior prognosis in AML patients [Citation14]. Also, some studies have identified DNMT3A mutations in patients with MDS [Citation15,Citation16]. Although DNMT3A mutations have been reported a lot in MDS, the prognostic significance still remains controversial. Many studies have reported the clinical impact of DNMT3A mutations in MDS, most of which indicated that the DNMT3A mutations were associated with poor prognosis [Citation17–22], while other studies showed they had no significant effect on overall survival [Citation23–29]. Thus, to further illustrate the prognostic effect of DNMT3A mutations in patients with MDS, we performed this meta-analysis.
Materials and methods
Literature search
Relevant literatures were systematically searched from electronic databases including PubMed, Embase, Web of Science, Clinical Trials, and the Cochrane Library. Articles (published before October 2018) were obtained by using the following search terms: ‘MDS OR “myelodysplastic syndrome” OR “myelodysplasia” OR “preleukemia” OR CMML OR “chronic myelomonocytic leukemia”’ and ‘DNMT3A OR “DNA methyltransferase 3A” OR “DNA (cytosine-5)-methyltransferase 3 alpha” OR “DNA (cytosine-5)-methyltransferase 3A”’.
Study selection
Studies were included when they fulfilled all criteria as follows: Published in English before October 2018; original articles as cohort studies; focused on prognostic effect of DNMT3A mutations in MDS patients; and provided data on overall survival (OS) or leukemia-free survival (LFS). The following studies were excluded from the analysis: meta-analysis, letters, comments, case reports and reviews; unavailable full-text. And literatures were managed and removed by EndNote X7.
Data extraction
Two researchers independently went over all the articles that met the inclusion criteria and extracted relevant information. Information included first author, year of publication, study region, the total number of patients and the number of patients with DNMT3A mutations, sex distribution, median age, the French–American–British (FAB) or WHO subtype, cytogenetic features and International Prognostic Scoring System (IPSS) or the Revised International Prognostic Scoring System (IPSS-R) classification. Furthermore, the corresponding hazard ratios (HRs) with 95% confidence intervals (95% CIs) for OS and LFS were extracted from COX multivariable models, or from analysis of original data in supplemental information via COX models. If HRs and 95% CIs were not available, we calculated them according to the reported Kaplan–Meier curves or p-value or other statistical parameters given in the text using the method [Citation30].
Quality assessment
Newcastle–Ottawa Scale (NOS) was used to evaluate the methodological quality of included literatures [Citation31]. The NOS consists of three dimensions (Selection, Comparability, and Outcome), which assigns 4, 2, and 3 stars for the three dimensions, respectively, with a total maximum of 9 scores. In the Selection and Outcome categories, each item within each category is entitled to a maximum of 1 star, whereas in the Comparability category, high-quality research could be entitled to a maximum of 2 stars. According to the NOS, these studies were classified into three types: high-quality studies (7–9 scores), moderate-quality studies (4–6 scores), and low-quality studies (0–3 scores).
Statistical analysis
All analyses were performed with the use of Stata 14.0 (College Station, TX, USA). Prognostic roles of DNMT3A mutations on OS and LFS were assessed by the pooled HRs and 95% CIs. Heterogeneity among these studies was evaluated by using the Q test (p < 0.10 was considered significant) and I2 statistic (I2 = 0–25%, no heterogeneity; I2 = 25–50%, moderate heterogeneity; I2 = 50–75%, large heterogeneity; I2 = 75–100% extreme heterogeneity). When there was no statistically significant heterogeneity (p-value > 0.1 and I2 < 50%), the pooled HRs were evaluated using a fixed-effect model. Otherwise, a random-effect model was applied. Subgroup analysis and meta-regression analysis were implemented to probe the potential sources of heterogeneity. Publication bias was assessed by Begg’s and Egger’s tests. A two-sided p value less than 0.05 was considered to be statistically significant.
Results
Study selection
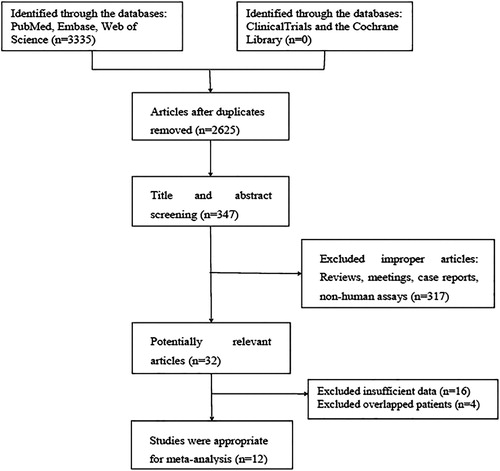
The total number of identified references from the databases was 3325, leaving 347 articles after removing duplications and screening the rest records. Of these, 32 references were considered potentially relevant based on the titles and abstracts according to the inclusion and exclusion criteria. Then, another 20 articles were excluded after full-text review for insufficient or overlapping data. Finally, 12 studies met the inclusion criteria and were available for the meta-analysis ().
Study characteristics
Twelve studies containing a total of 2236 patients were included for this meta-analysis. The characteristics are listed in . There are 232 patients with mutant DNMT3A and 2004 patients with wild-type DNMT3A. These studies were published between 2011 and 2018, two of which were originated from Europe, five from USA, and five from Asia. Most studies were based on the WHO classification except for four based on the FAB classification. The sample size ranged from 65 to 469 and the frequency of DNMT3A mutations ranged from 2.73% to 18.39%. The total mutation frequency was 11.58%.
Table 1. Summary of the data extracted from the 12 studies included.
Quality assessment
Quality assessment was evaluated based on nine items of NOS. Six studies (50%) scored 8 stars, three studies (25%) scored 7 stars, and three studies (25%) scored 6 stars. Relevant details are presented in .
Table 2. Quality assessment of individual study.
Prognostic impact of DNMT3A mutations in patients with MDS
OS and LFS were the main outcomes we analyzed to determine the prognostic impact of DNMT3A mutations in patients with MDS.
1. Prognostic impact of DNMT3A mutations on overall survival (OS) in patients with MDS
(1) Prognostic power of DNMT3A mutations in patients with MDS
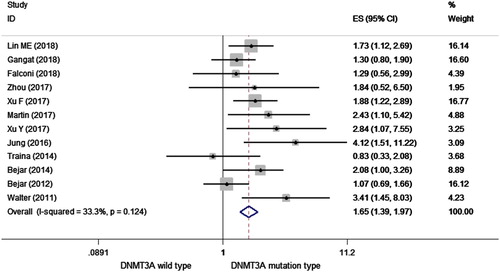
HRs were pooled for OS from 12 studies with 2236 patients totally. As shown in , the overall HR for the OS was 1.654 (95% CI = 1.387–1.973, p < 0.001, I2 =33.4%) in MDS patients with DNMT3A mutations, which indicated that DNMT3A mutations can predict a poorer OS in patients with MDS.
Figure 2. Forest plot of the HR and 95% CI for OS in MDS patients with DNMT3A mutations by a fixed-effect model.
(2) Prognostic power of DNMT3A mutations stratified by age (<60 and ≥60)
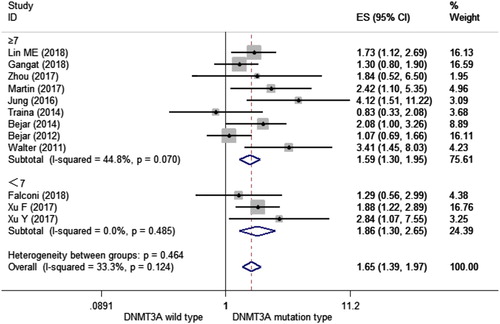
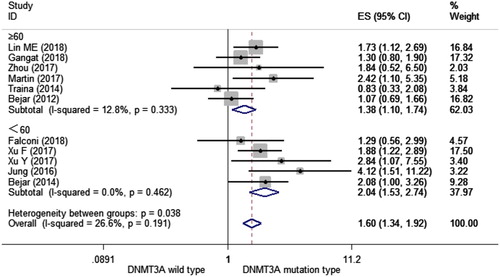
Age is a major prognostic parameter for overall survival in MDS [Citation32]. To further confirm the prognostic ability of DNMT3A mutations, we performed a subgroup analysis stratified by age (<60 and ≥60). We observed a significant shorter OS in MDS patients with mutant DNMT3A than those with wild-type DNMT3A in both subgroups of age < 60 (HR = 2.043, 95% CI = 1.526–2.736, p < 0.001, I2 = 0.0%) and age ≥ 60 (HR = 1.381, 95% CI = 1.099–1.736, p = 0.006, I2 = 12.8%) (). This result demonstrated that DNMT3A mutations could predict a shorter OS in MDS patients regardless of patients’ age.
Figure 3. Forest plot of the HR and 95% CI for OS in MDS patients with DNMT3A mutations stratified by age (<60 and ≥60) by a fixed-effect model.
2. Prognostic impact of DNMT3A mutations on leukemia-free survival (LFS) in patients with MDS
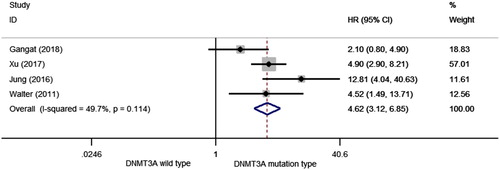
HR was pooled for LFS extracted from four studies with a total of 877 patients. As shown in , the overall HR for LFS was 4.624 (95% CI: 3.121–6.851, p < 0.001, I2 = 49.7%) in MDS patients with mutant DNMT3A compared with those with wild-type DNMT3A, suggesting that DNMT3A mutations may predicted a higher probability of transformation to leukemia.
Subgroup analysis
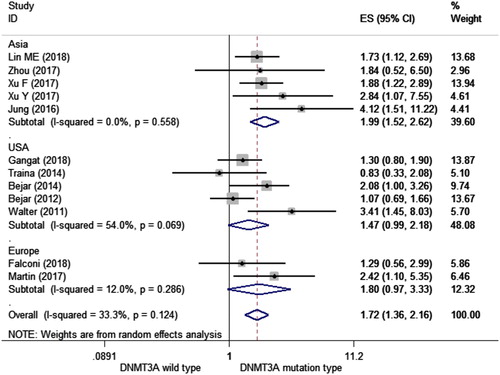
Different subgroup analyses were performed. We conducted subgroup analyses of several factors that may alter the association between DNMT3A mutations and OS, including the year of publication, NOS, region, and median age of each study (). First, we found that articles published in 2018 and 2017 showed an association between DNMT3A mutations and OS (HR = 1.472, 95%CI = 1.102–1.966, I2 = 0.0%; HR = 2.068, 95%CI = 1.472–2.906, I2 = 0.0%, respectively). As for the quality of each study, both levels showed an association DNMT3A mutations and OS (HR = 1.594, 95%CI = 1.301–1.952, I2 = 44.8%; HR = 1.858, 95%CI = 1.300–2.654, I2 = 0.0%, respectively) (). Then according to the study region, we found an association between DNMT3A mutations and OS in Asia (HR = 1.994, 95%CI = 1.515–2.624, I2 = 0.0%) and Europe (HR = 1.802, 95%CI = 1.013–3.208, I2 = 12.0%), but not in the United States (HR = 1.469, 95%CI = 0.990–2.180, I2 = 54.0%) ().
Table 3. Results of subgroup analyses.
Sensitivity analysis, meta-regression analysis and publication bias
Sensitivity analysis showed no individual study changed the pooled HR significantly (), indicating that the results were reliable and stable. The Begg’s funnel plot was roughly symmetric (). And the Begg’s (p = 0.244) and Egger’s (p = 0.172) tests revealed there was no evidence for significant publication bias in this meta-analysis. Then, meta-regression analysis was analyzed to further explore the resources of heterogeneity, and the results showed us that none of the following covariates affected the prognostic value of DNMT3A mutations on OS in patients with MDS, except for the gender distribution (coefficient = −0.5297291, p = 0.021) (). The covariates included: year of publication (coefficient = 0.007595, p = 0.889), NOS (coefficient=−0.0560025, p = 0.713), n/N (the ratio of the number of patients with DNMT3A mutations to all patients) (coefficient = −2.678812, p = 0.385), median age (coefficient = −0.0246394, p = 0.166), median follow-up (coefficient = 0.1884168, p = 0.168), median white blood cell count (coefficient = 0.0038781, p = 0.344), median hemoglobin (coefficient = 0.0035804, p = 0.363), and median platelet count (coefficient = 0.0005092, p = 0.835).
Figure 7. Sensitivity analysis for OS with a fixed-effect model for all included studies. The middle vertical axis represents the pooled HR and the 2 vertical axes indicate the corresponding 95% CI. Each hollow circle represents the pooled HR when the left study was omitted in this meta-analysis, and the 2 ends of every broken line indicate the 95% CI.
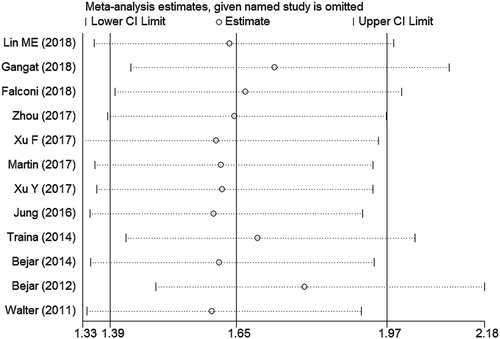
Figure 8. Begg’s funnel plot for publication bias analysis. Each point represents an individual study and horizontal line represents the mean effect size. The distribution is roughly symmetrical.
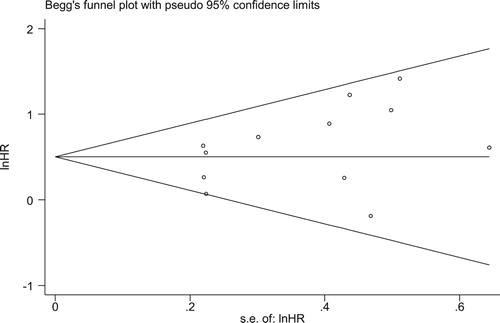
Table 4. Fixed-effects meta-regression analysis for OS in all available studies.
Discussion
The main purpose we conducted this meta-analysis was to explore whether mutant DNMT3A is a poor prognostic factor in patients with MDS. This meta-analysis included a total of 12 articles with 2236 MDS patients, covering 232 patients with DNMT3A mutations and 2004 patients without DNMT3A mutations. The pooled HR value (HR = 1.654, 95% CI = 1.387–1.973, p < 0.001, I2 = 33.3%) showed that MDS patients with DNMT3A mutations had a significant shorter OS than those without mutations. Age is one of the most important prognostic factors in MDS patients, and usually adults over 60 have a shorter OS than younger adults [Citation32]. Next, we assessed the prognosis power of DNMT3A mutations on overall survival stratified by age (<60 and ≥60) and found that OS was shorter in MDS patients with DNMT3A mutations than those without DNMT3A mutations in the two subgroups of age <60 and age ≥ 60. The result demonstrated that DNMT3A mutations may act as a poor prognostic indicator in MDS patients, which is independent of age. Likewise, for LFS, data from four studies covering only 877 MDS patients were limited, indicating that the patients without DNMT3A mutations were less likely to transform from MDS to AML.
It’s of note that hypomethylating agents (HMAs) were mentioned in several studies. To date, there was no clear clinical evidence linking HMAs to DNMT3A mutations [Citation33,Citation34]. But one meta-analysis came to the conclusion that no gene mutation in DNA methylation pathway is confirmed to have prognostic value to HMAs on OS, whereas mutant DNMT3A is a favorable biomarker on overall response rate in MDS patients by HMAs treatment [Citation35]. In a word, MDS patients with DNMT3A mutations are encouraged to have HMAs as major therapy.
The heterogeneity was moderate (I2 =33.3%) and acceptable, and the sensitivity analysis, containing all studies by excluding one study at a time, suggested that the results of the meta-analysis were stable and reliable. As for publication bias, both the Begg’s test and Egger’ test showed no significant bias. Then we performed subgroup analyses, including the year of publications, study region, the qualities of studies, and median age of each study. However, there is no difference in heterogeneity among these factors. The regression analysis showed that only gender distribution might be the resource of heterogeneity, whereas other factors did not have any effect.
This meta-analysis has a few limitations. First, not all the included studies directly provided the HRs and 95% CIs. HR with 95% CI for OS of one article could only be calculated from Kaplan–Meier curves by manual work, which might lead to less accurate results. Second, the effects of some relatively small sample size studies may be overestimated compared with other large size studies. Third, only studies published in English were included in this analysis so that language bias may have affected the results. In addition, there were numerous clinical factors affected the outcomes, such as the various treatment regimens, age and gender distribution of patients, MDS subtype, cytogenetic and molecular abnormalities, time of follow-up, and the methods of gene sequencing. Therefore, heterogeneity and publication bias could not be avoided.
MDS are clinically heterogeneous diseases, which led to develop a prognostic scoring system to assess overall survival and leukemia-free survival, and to promote clinical and therapeutic decisions [Citation36]. The prognosis factors include age, the percentage of bone marrow blast cells, peripheral blood anemia, presence of thrombocytopenia and leukocytosis, previous treatment and so on. Nowadays, more and more studies focus on the value of genetic and molecular profiling of target genes for subclassification and prognostication in MDS patients [Citation37–39], which is of great importance.
In conclusion, our meta-analysis results indicated that mutant DNMT3A is an independent factor to predict poor outcomes in MDS patients, which were consistent with those previous studies showing DNMT3A mutations can be a negative prognostic marker in MDS [Citation15–18,Citation29,Citation36]. Therefore, it can be helpful to assess prognosis and guide treatment in MDS patients. Importantly, further prospective studies or randomized trials covering a large number of patients to confirm the prognostic impact of DNMT3A mutations are needed.
Acknowledgement
The authors are thankful to all the patients and clinical investigators who were involved in the studies selected for this meta-analysis.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Greenberg PL, Attar E, Battiwalla M, et al. Myelodysplastic syndromes. J Natl Compr Canc Netw. 2008;6(9):902–926.
- Papaemmanuil E, Gerstung M, Malcovati L, et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood. 2013;122(22):3616–3627. doi:10.1182/blood-2013-08-518886.
- Xu Y, Li Y, Xu Q, et al. Implications of mutational spectrum in myelodysplastic syndromes based on targeted next-generation sequencing. Oncotarget. 2017;8(47):82475–82490. doi:10.18632/oncotarget.19628.
- Okano M, Xie S, Li E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat Genet. 1998;19(3):219–220. doi:10.1038/890.
- Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3(6):415–428. doi:10.1038/nrg816.
- Simpson DJ, Hibberts NA, McNicol AM, et al. Loss of pRb expression in pituitary adenomas is associated with methylation of the RB1 CpG island. Cancer Res. 2000;60(5):1211–1216.
- Auclair G, Guibert S, Bender A, et al. Ontogeny of CpG island methylation and specificity of DNMT3 methyltransferases during embryonic development in the mouse. Genome Biol. 2014;15(12):545, doi:10.1186/s13059-014-0545-5.
- Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128(4):683–692. doi:10.1016/j.cell.2007.01.029.
- Khan H, Vale C, Bhagat T, et al. Role of DNA methylation in the pathogenesis and treatment of myelodysplastic syndromes. Semin Hematol. 2013;50(1):16–37. doi:10.1053/j.seminhematol.2013.01.001.
- Jiang Y, Dunbar A, Gondek LP, et al. Aberrant DNA methylation is a dominant mechanism in MDS progression to AML. Blood. 2009;113(6):1315–1325. doi:10.1182/blood-2008-06-163246.
- Haferlach T, Nagata Y, Grossmann V, et al. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia. 2014;28(2):241–247. doi:10.1038/leu.2013.336.
- Ghannam D E, Taalab MM, Ghazy HF, et al. DNMT3A r882 mutations in patients with cytogenetically normal acute myeloid leukemia and myelodysplastic syndrome. Blood Cells Mol Dis. 2014;53(1-2):61–66. doi:10.1016/j.bcmd.2014.01.004.
- Ley TJ, Ding L, Walter MJ, et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010;363(25):2424–2433. doi:10.1056/NEJMoa1005143.
- Yuan XQ, Peng L, Zeng WJ, et al. DNMT3A r882 mutations predict a poor prognosis in AML: a meta-analysis from 4474 patients. Medicine (Baltimore). 2016;95(18):e3519. doi:10.1097/MD.0000000000003519.
- Thol F, Winschel C, Lüdeking A, et al. Rare occurrence of DNMT3A mutations in myelodysplastic syndromes. Haematologica. 2011;96(12):1870–1873. doi:10.3324/haematol.2011.045559.
- Brecqueville M, Cervera N, Gelsi-Boyer V, et al. Rare mutations in DNMT3A in myeloproliferative neoplasms and myelodysplastic syndromes. Blood Cancer J. 2011;1(5):e18. doi:10.1038/bcj.2011.15.
- Lin ME, Hou HA, Tsai CH, et al. Dynamics of DNMT3A mutation and prognostic relevance in patients with primary myelodysplastic syndrome. Clin Epigenetics. 2018;10:42. doi:10.1186/s13148-018-0476-1.
- Walter MJ, Ding L, Shen D, et al. Recurrent DNMT3A mutations in patients with myelodysplastic syndromes. Leukemia. 2011;25(7):1153–1158. doi:10.1038/leu.2011.44.
- Xu F, Wu LY, He Q, et al. Exploration of the role of gene mutations in myelodysplastic syndromes through a sequencing design involving a small number of target genes. Sci Rep. 2017;7:43113, doi:10.1038/srep43113.
- Martín I, Such E, Navarro B, et al. Negative impact on clinical outcome of the mutational co-occurrence of SF3B1 and DNMT3A in refractory anemia with ring sideroblasts (RARS). Leuk Lymphoma. 2017;58(7):1686–1693. doi:10.1080/10428194.2016.1246725.
- Jung SH, Kim YJ, Yim SH, et al. Somatic mutations predict outcomes of hypomethylating therapy in patients with myelodysplastic syndrome. Oncotarget. 2016;7(34):55264–55275. doi:10.18632/oncotarget.10526.
- Bejar R, Stevenson KE, Caughey B, et al. Somatic mutations predict poor outcome in patients with myelodysplastic syndrome after hematopoietic stem-cell transplantation. J Clin Oncol. 2014;32(25):2691–2698. doi:10.1200/JCO.2013.52.3381.
- Zhang YY, Zhou JD, Yang DQ, et al. Intragenic hypomethylation of DNMT3A in patients with myelodysplastic syndrome. Clin Chem Lab Med. 2018;56(3):485–491. doi:10.1515/cclm-2016-0142.
- Lin CC, Hou HA, Chou WC, et al. IDH mutations are closely associated with mutations of DNMT3A, ASXL1 and SRSF2 in patients with myelodysplastic syndromes and are stable during disease evolution. Am J Hematol. 2014;89(2):137–144. doi:10.1002/ajh.23596.
- Gangat N, Mudireddy M. Mutations and prognosis in myelodysplastic syndromes: karyotype-adjusted analysis of targeted sequencing in 300 consecutive cases and development of a genetic risk model. Am J Hematol. 2018;93(5):691–697. doi:10.1002/ajh.25064.
- Zhou JD, Lin J, Zhang TJ, et al. GPX3 methylation in bone marrow predicts adverse prognosis and leukemia transformation in myelodysplastic syndrome. Cancer Med. 2017;6(1):267–274. doi:10.1002/cam4.984.
- Falconi G, Fabiani E, Piciocchi A, et al. Somatic mutations as markers of outcome after azacitidine and allogeneic stem cell transplantation in higher-risk myelodysplastic syndromes. Leukemia. 2019;33(3):785–790. doi:10.1038/s41375-018-0284-9.
- Traina F, Visconte V, Elson P, et al. Impact of molecular mutations on treatment response to DNMT inhibitors in myelodysplasia and related neoplasms. Leukemia. 2014;28(1):78–87. doi:10.1038/leu.2013.269.
- Bejar R, Stevenson KE, Caughey BA, et al. Validation of a prognostic model and the impact of mutations in patients with lower-risk myelodysplastic syndromes. J Clin Oncol. 2012;30(27):3376–3382. doi:10.1200/JCO.2011.40.7379.
- Tierney JF, Stewart LA, Ghersi D, et al. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16. doi:10.1186/1745-6215-8-16.
- Wells GA, Shea B, O’Connell D, et al. The Newcastle-Ottawa Scale (NOS) for assessing the quality if nonrandomized studies in meta-analyses. Available from: http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp.
- Greenberg PL, Tuechler H, Schanz J, et al. Revised International prognostic Scoring System for myelodysplastic syndromes. Blood. 2012;120(12):2454–2465. doi:10.1182/blood-2012-03-420489.
- Cedena MT, Rapado I, Santos-Lozano A, et al. Mutations in the DNA methylation pathway and number of driver mutations predict response to azacitidine in myelodysplastic syndromes. Oncotarget. 2018;9(56):30936. doi:10.18632/oncotarget.25856.
- Cabezón M, Bargay J, Xicoy B, et al. Impact of mutational studies on the diagnosis and the outcome of high-risk myelodysplastic syndromes and secondary acute myeloid leukemia patients treated with 5-azacytidine. Oncotarget. 2018;9(27):19342–19355. doi:10.18632/oncotarget.25046.
- Du M, Zhou F, Jin R, et al. Mutations in the DNA methylation pathway predict clinical efficacy to hypomethylating agents in myelodysplastic syndromes: a meta-analysis. Leuk Res. 2019;80:11–18. doi:10.1016/j.leukres.2019.03.001.
- Zini G. Diagnostics and prognostication of myelodysplastic syndromes. Ann Lab Med. 2017;37(6):465–474. doi:10.3343/alm.2017.37.6.465.
- Bartels S, Schipper E, Hasemeier B, et al. Routine clinical mutation profiling using next generation sequencing and a customized gene panel improves diagnostic precision in myeloid neoplasms. Oncotarget. 2016;7(21):30084–30093. doi:10.18632/oncotarget.8310.
- Nazha A, Bejar R. Molecular data and the IPSS-R: How Mutational Burden Can Affect prognostication in MDS. Curr Hematol Malig Rep. 2017;12(5):461–467. doi:10.1007/s11899-017-0407-9.
- Lindsley RC, Saber W, Mar BG, et al. Prognostic mutations in myelodysplastic syndrome after stem-cell transplantation. N Engl J Med. 2017;376(6):536–547. doi:10.1056/NEJMoa1611604.