ABSTRACT
Objectives: A total of 156 adult acute myeloid leukemia (AML) patients were enrolled in this study to explore the clinical characteristics and prognostic impact of ASXL1 mutations. Methods: Clinical characteristics, prognostic impact and the association between ASXL1 mutations and some other mutations were analyzed. Results: We found ASXL1 mutations were most frequently found in M5 subtype and intermediate risk karyotype and were correlated with TET2, DNMT3A and PHF6 mutations. A total of 145 patients were included in prognostic analysis; results showed ASXL1 mutations had no impact on OS and DFS. In normal karyotype-AML (CN-AML) and older (≥60 years) AML, ASXL1 mutations showed adverse impact on OS (P = 0.022; p = 0.019, respectively) and showed adverse prognostic tendency on DFS (p = 0.173; p = 0.108, respectively). ASXL1 mutations were also independent unfavourable prognostic factors for OS on CN-AML and older (≥60 years) AML patients and unfavourable factors for DFS on older (≥60 years) AML in multivariate analysis. Results also indicated that though ASXL1 mutations were associated with TET2, DNMT3A and PHF6 mutations, when coinciding with ASXL1 mutations, the prognosis of AML was not significantly impacted. Discussion: The reliability of our results need to be further confirmed by prospective randomized controlled studies covering a large numbers of AML patients. Conclusion: The results showed ASXL1 mutations may act as a poor prognostic index especially in elder AML and CN-AML patients.
Introduction
AML is a malignant clone disorder of the hematopoietic stem cell [Citation1, Citation2]. In clinical practice, karyotypic abnormality is the basis for classifying choice of treatment and assessing the prognosis of AML [Citation3]. Recently, along with the development of cytogenetics, molecular biology and gene sequencing, especially the rapid development of second generation sequencing, there is new understanding on pathogenetic, prognostic and stratification of treatment of AML, on which classifying choice of treatment and assessing the prognosis of new base and for which the treatment of AML is constantly optimized and the therapeutic outcomes are significantly enhanced. Recently, AML-related gene mutations have provided important evidence for diagnostic, prognostic stratification and individual therapy of AML [Citation4]. Many genetic mutations have been confirmed to associate with the prognosis of AML, such as Nucleophosmin 1(NPM1) and CCAAT/enhancer-binding protein (CEBPA) mutations are signs for good prognosis, and Fms-like tyrosine kinase-Internal Tandem Duplication(FLT3-ITD), Partial tandem duplication of the Mixed-Lineage Leukemia(MLL-PTD), Brain and acute leukemia cytoplasmic (BAALC) mutations are signs for bad prognosis. In addition, some genes like DNA-methyltransferase 3A(DNMT3A), Tet methylcytosine dioxygenase(TET2) and Isocitrate dehydrogenase(IDH), which relate to DNA or histone methylation, have been certified to be associated with the prognosis of AML [Citation4, Citation5]. The discovery of these molecular markers is important for the improvement of individual risk analysis and precise stratification treatment of AML. Thus, looking for AML-related molecular marker is an important issue in the current research on hematology.
Ever since Gelsi-Boyer detected Additional sex comb-like 1(ASXL1) gene mutations in patients with myelodysplastic syndrome (MDS), almost all hematologic malignancies have been reported to acquire ASXL1 mutations [Citation6]. The ASXL1 gene, located in chromosome band 20q11, involves in epigenetic regulation and encodes a nucleoprotein of 1541 amino acid that can act as a ligand-dependent co-activator for retinoic acid receptor and may associate with chromatin modifiers. ASXL1 mutations usually result in C-terminal truncation of the protein upstream of the PHD, causing gene function disability [Citation6–8]. Patients with ASXL1 mutations show different clinical characteristic in different diseases. Even in the same disease, different studies show different results. Most studies have shown that ASXL1 mutations are poor prognostic factors, especially in cytogenetically normal acute myeloid leukemia (CN-AML) patients [Citation9–12]. However, some studies suggest that ASXL1 mutations are not independent poor prognostic factors [Citation13]. Some studies indicated that ASXL1 mutations had no correlation with TET2 mutations; however, some others showed the reverse [Citation12, Citation14]. We carried out this clinical research, aiming to explore and assess whether ASXL1 mutations could be included in AML prognostic evaluation system, to improve AML prognostic evaluation system and to provide certain theoretical basis for individualized treatment.
Materials and methods
A total of 156 newly diagnosed non-M3 AML patients (150 cases de novo AML, 5 cases transformed from MDS, and 1 case transformed from MDS/MPN) between January 2013 and August 2015 were recruited in this research. All the patients recruited should meet the following criteria: 1, Newly diagnosed non-M3 AML, including AML transformed from MDS and MDS/MPN, but not from acute mixed leukemia (MAL) and chronic myelogenous leukemia (CML); 2, Detected the 11 common somatic mutations of AML; 3, age ≥ 14 years old. Finally, 156 patients (81 men and 75 women) with an average age of 47 years old (range 14–85) were included in the study. Complete follow-up data were available in 145 cases, with a mean follow-up interval of 29.9 months (range 1–38) and 97 cases of chromosome karyotype were analyzed.
All patients included were diagnosed by MICM classification and in line with the French-American-British (FAB) or the 2008 WHO diagnostic criteria of AML. SWOG was used for cytogenetic classification. According to FAB classification, the included patients were classified into M0 (2 cases, 1.3%), M1 (6 cases, 3.8%), M2 (144 cases, 28.2%), M4 (1 case, 0.6%), M5 (86 cases, 55.1%), M6 (10 cases, 6.4%) and 8 cases unclassified AML. Two patients refused chemotherapy and 1 patient died before chemotherapy; all the other patients received at least 2 cycles of chemotherapy. The majority of patients were treated by IA, DA or HA (I: idarubicin, D: daunomycin, H: Homoharringtonine, A: cytarabine) for induction therapy. Low-intensity CAG (C: Cytarabine, A: Adacinomycin, G: G-CSF) regiment or half-dose HA regiment or HA regiment was adopted for induction therapy in elderly AML patients. Most of the patients accepted high-dose cytarabine regiment for intensive treatment after complete remission. Patients with high white blood cell counts received hydroxycarbamide or leukapheresis as the pretreatment before induction therapy.
PCR and DNA sequencing were used to analyze the gene mutations. Diagnostic bone marrow samples were collected. Samples were stored at 4°C until DNA extraction was done with a TIANamp Blood DNA kit (Tiangen, Beijing, China). The concentration and purity of the DNA sample was determined using an ultraviolet Spectrophotometer (Biophotometer, Eppendorf, Hambury, Germany) by measuring the optical density at 260 vs. 280 nm absorbance, and the ratio was 1.8–2.0 for pure DNA. PCR was performed by life PRO PCR (Hangzhou, China). The NGS panel, targeting some common sequence of 11 gene, was applied using Life Technologies, Thermo Fisher, Ion Proton (Supplemental Table S1). A minimum cutoff of *250 bidirectional coverage and at least 5% allele frequencies were used for reporting.
We detected mutations of exon14 and exon15 in FLT3-ITD gene, mutations of exon12 in NPM1 gene, mutations of exon17 in C-kitD816, mutations in CEBPA, mutations of exon4in IDH1/IDH2, mutations in dupMLL, mutations of exon2, 4, 7, 8, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 22, 23 and the surrounding intron areas in DNMT3A gene, mutations of exon2–10 and the surrounding intron areas in 6 PHD finger protein (PHF6) gene, mutations of exon1–10 and the surrounding intron areas in TET2, mutations of exon12 in ASXL1 gene. The clinical data of patients when first diagnosed with AML between January 2013 and August 2015, including initial white blood cell counts in peripheral, proportion of immature cells in peripheral blood, platelet and hemoglobin counts, the serum levels of lactate of dehydrogenase, uric acid and α-HBAD, chromosome classification, the data of bone marrow examinations were collected. The chemotherapy regiments reduced time and recurrence time was also collected. The clinical outcome of enrolled patients was also traced.
The curative effect was assessed using Cheson’s criterion (2003) [Citation15]. Overall survival (OS) end points were measured from the date of first sample collection to the time of death from any cause or to the time at last follow-up (censored). Disease-free survival (DFS) end points were measured from the date of CR to the time of relapse or death from any cause or to the time at last follow-up (censored).
Data processing and statistical analysis were analyzed using SPSS19.0 statistical analysis package. The counting data were analyzed using the Chi-square test or Kruskal–Wallis single factor ANOVA analysis and the measurement data used independent sample of the Mann-Whitney U-nonparametric tests. The Kaplan-Meier method, log-rank test, and Cox proportional hazard models were used to estimate the distribution of OS and DFS and to compare differences between survival curves. A two-tailed P-value <0.05 was defined as statistically significant.
Results
Among the 156 AML cases, 18 cases (81.81%) were identified with ASXL1 missense mutations, 2 cases (9.09%) with deletion mutations, 1 case (4.54%) with frameshift mutations, and 1 case with nonsense mutations. We also detected some common somatic mutations of AML. Eighteen patients (11.54%) were detected with IDH1/2 gene mutations, 11 patients (7.05%) with NPM1 mutations, 28 patients (17.95%) with TET2 mutations, 27 patients (17.31%) with DNMT3A mutations, 22 patients (14.10%) with CEBPA mutations, 19 patients (12.18%) with FLT3-ITD gene mutations, 5 patients (3.2%) with C-kit mutations; 6 patients (3.8%) with PHF6 mutations,7 patients (4.5%) with dup-MLL mutations. Among the 22 patients with ASXL1 gene mutations, 12 cases were male, 10 cases were female, and the median age was 49 years old (20–75). Among the 134 AML patents without ASXL1 mutations, 69 were male, 65 were female, and the median age was 47 years old (14–85). Statistical analysis suggests that there was no significant difference for age and gender between ASXL-mutated and ASXL1 wild-type patients (P = 0.791 and P = 0.605, respectively). In addition, statistical analysis also showed that there was no significant difference for initial white blood cell, platelet and hemoglobin counts, proportion of immature cells in peripheral blood and bone marrow, the serum levels of lactate of dehydrogenase, uric acid, and α-HBAD (P > 0.05) (). A total of 149 patients were clearly identified into certain subtype of FAB classification. Statistical analysis showed that ASXL1 gene mutations were mostly occurred in M5; however, the distribution of ASXL1 gene mutations in each FAB subtype was not statistically significant (P = 0.416) ().
Table 1. Characteristics of AML patients with and without ASXL1 mutations.
Table 2. AML patients with and without ASXL1 mutations and FAB classification.
Of the 156 included AML patients, 97 had chromosome karyotype. Nine (9.3%) of them were grouped into the low-risk group, 78 (80.4%) of them were grouped into the moderate group, and 10 (10.3%) were grouped into the high-risk group. Statistical analysis showed that the distribution of ASXL1 mutations in each chromosome karyotype group had no statistical significance (P = 0.590). Of all the 97 patients, 57 were of normal karyotype and 40 were of abnormal karyotype. The distribution of ASXL1 mutations in normal karyotype group and abnormal karyotype group was not statistically significant (P = 0.642) ().
Table 3. AML patients with and without ASXL1 mutations and chromosome karyotype.
Analyses of the correlation between ASXL1 gene mutations and 10 other common somatic mutations of AML were done for the following: IDH1, IDH2, NPM1, TET2, DNMT3A, CEBPA, FLT3 – ITD, C – kitD816, PHF6, dupMLL mutation. Statistical analysis showed that the mutation rates of TET2, DNMT3A, PHF6 gene were statistically significant between the ASXL1 mutation group and wild-type group (P < 0.001, P = 0.001, and P = 0.037, respectively). The mutation rates of TET2 gene, DNMT3A gene, and PHF6 gene in the ASXL1 mutations group were higher than those without, which indicated that ASXL1 gene mutations may be associated with these three genetic mutations ().
Table 4. ASXL1 mutations and some other common AML prognosis-related mutations.
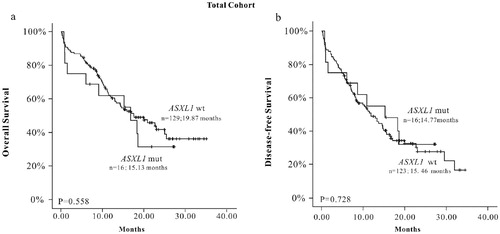
Among 145 patients available, the median follow-up interval was 15.3 (range from 9.97 to 20.30) months in the ASXL1 mutation group and 19.87 (range from 17.37 to 22.38) months in the wild-type group. There was no statistically significant difference for OS between the ASXL1 mutation and ASXL1 wild-type group (P = 0.558). Besides, when DFS data obtained from 139 cases were analyzed, the result showed that there was also no statistically significant difference between the ASXL1 mutation group and ASXL1 wild-type group (P = 0.728) ((a,b)).
Figure 1. The K-M survival curve of ASXL1 mutations in total AML patients. (a) OS (n = 145), (b) EFS (n = 139).
Meanwhile, we conducted a subtype analysis in CN-AML patients. The result showed that there was statistically significant difference OS between the ASXL1 mutation group and wild-type group in 55 normal karyotype cases (P = 0.016). The shorter OS in the ASXL1 mutation group indicated that ASXL1 mutation was a sign of poor prognosis in adult de novo CN-AML patients. Although no statistical significance was found on DFS (P = 0.165) between the two groups, the survival curve showed the ASXL1 mutation group seemed to have a worse prognosis ((a,b)). We also conducted a COX univariate analysis, including various related factors such as age, gender distribution, bone marrow blast percentage, α-HBDH, UA, initial WBC count, percentage of blasts in peripheral blood, HGB count, PLT lever, chromosome karyotype, ASXL1, TET2, CEBPA, DNMT3A, NPM1, FLT3-ITD mutations. The results of COX univariate analysis in CN-AML subgroup suggested that age was one of the adverse factors for OS and DFS and ASXL1 mutations and α-HBDH were adverse factors for OS (). In the COX multivariate regression model (), the results showed that age was still a negative factor for OS and DFS (HR: 2.338, 95% CI: 2.338–12.447; HR: 3.087, 95% CI: 1.454–6.554, respectively). ASXL1 gene mutation was an independent risk factor for OS (HR: 3.070, 95% CI: 1.095–8.604) in patients with CN-AML rather than DFS.
Table 5. Survival analysis (OS and DFS) in 55 CN-AML patients.
Table 6. Multivariate analysis of CN-AML patients on long-term survival.
Statistical analysis showed that there was statistically significance for OS between the ASXL1 mutation and ASXL1 wild-type group in older (≥60 years) AML patients (P = 0.011). No statistical significance was found for DFS (P = 0.096) between the two groups; however, the survival curve showed that ASXL1 mutations seemed to have a worse prognosis ( and (a,b)). The results of COX univariate analysis in the elder (≥60 years) AML subgroup () suggested that ASXL1 mutation was a poor prognostic factor for OS and α-HBDH was an adverse factor for OS and DFS. These two factors were included in COX multivariate analysis (), we found that ASXL1 mutation was still a poor prognosis factor for OS in the older AML subgroup (HR: 10.4, 95%CI: 2.594–41.977). In univariate analysis, we only found ASXL1 mutation had a tendency to have a poor prognosis for DFS in elder AML patients. However, in multivariate analysis, ASXL1 mutation was an independent poor prognostic factor for DFS in elder AML patients (HR: 6.336, 95%CI: 2.206–18.196).
Figure 2. The K-M survival curve of ASXL1 mutations in CN-AML patients, (a) OS (n = 55), (b) DFS (n = 55).
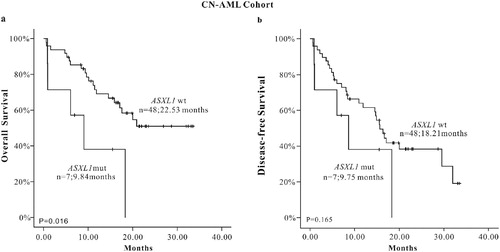
Figure 3. The K-M survival curve of ASXL1 mutations in AML (age > 0 years) patients, (a) OS (n = 36), (b) DFS (n = 33)
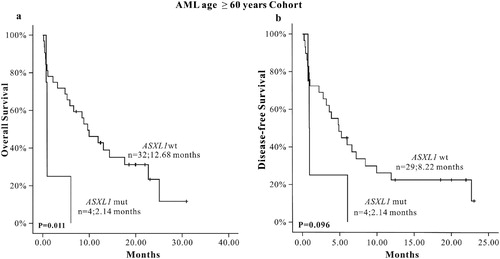
Table 7. Influencing factors analysis on survival (OS and DFS) of 36 elderly (≥60 years) AML patients.
Table 8. Multivariate analysis of elderly (≥60 years) AML patients on long-term survival.
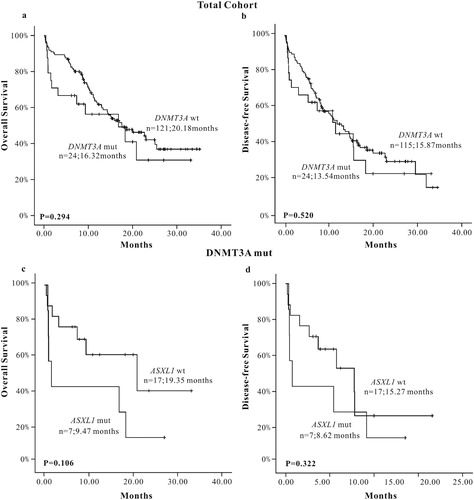
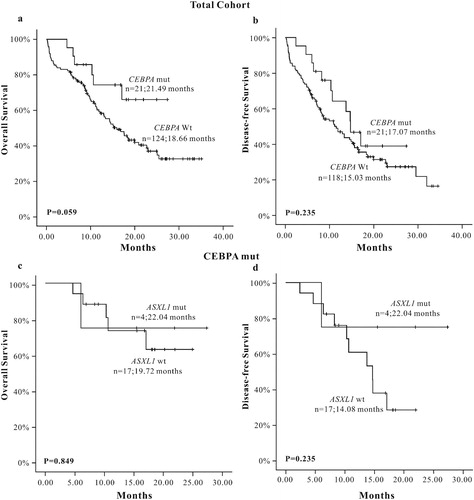
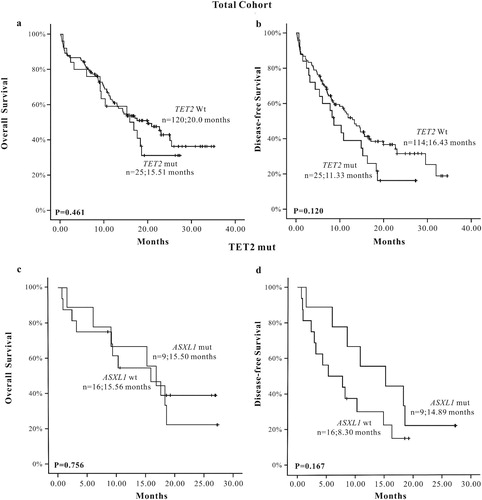
We chose three common somatic mutations of AML: TET2, CEBPA and DNMT3A genes, which had a higher mutation rate and lower loss follow-up rate to analyze the influence on the prognosis of AML patients when accompanied with ASXL1 mutations. The results showed that ASXL1 mutations did not affect the prognosis of AML patients in the TET2 mutation group (OS: P = 0.756, DFS: P = 0.167) ((a–d)), so did in the DNMT3A mutation group (OS: P = 0.106, DFS: P = 0.322) ((a–d)) and the CEBPA mutation group (OS: P = 0.849, DFS: P = 0.235) ((a–d)).
Figure 4. The K-M survival curve of TET2 mutations in total AML patients, (a) OS (n = 145), (b) DFS (n = 139); The K-M survival curve of TET2 mutations when accompanied with ASXL1 mutations in AML patients, (c) OS (n = 25), (d) DFS (n = 25).
Discussion
With the development of second generation sequencing, SNP probe sequences and comparative genomics, it is possible to detect complex genetic abnormality. More and more prognosis-related gene mutations have been found and detected. Epigenetic regulatory gene mutations, such as ASXL1, TET2, DNMT3A, EZH2 mutations, which play a role not only in the pathogenesis but also in the prognosis of AML, have been found. ASXL1 mutation was detected in MDS patients in 2009 and expressed in almost all myeloid malignancies [Citation6, Citation16]. Many studies suggested that ASXL1 mutation had an important effect on the pathogenesis and prognosis of AML, but the mechanism needed to be further elucidated.
In our studies, there was no statistically significant difference for gender distribution, initial WBC counts, HGB counts, PLT level, percentage of blasts in peripheral blood, LDH, UA, α-HBDH and bone marrow blast percentage(P > 0.05). These were different from many other previous studies [Citation9,Citation10, Citation12,Citation13, Citation17]. However, the association of ASXL1 mutations with the above factors is still controversial. Therefore, prospective randomized controlled studies, covering a large numbers and investigating the association of the clinical hematological characteristics with ASXL1 mutations, are needed.
We studied the distribution of FAB genotype in the ASXL1 mutations group and wild-type group and found that ASXL1 mutation was more common in AML-M5, but there was no statistically significant difference for the distribution of FAB genotype between the two groups. The exact relationship between these two factors needed further illustration.
We also analyzed the relationship between ASXL1 mutation and chromosome karyotype. Of the 97 cases, 57 of them were of normal karyotype and 40 were of abnormal karyotype. Also, no significant difference was found between the ASXL1 mutation group and ASXL1 wild-type group (P = 0.746). Our results were similar to those of Chou et al. study [Citation13]. However, S Schnittger et al.’s study showed that ASXL1 mutations were more common in abnormal karyotype [Citation12]. Although different centers reported different results, most studies suggested that ASXL1 mutations were more common in patients with CN-AML.
Some studies reported that ASXL1 mutations were not associated with TET2 and IDH1 mutations and showed a negative correlation with DNMT3A, FLT3-ITD, and NPM1 mutations and a negative correlative trend with IDH2 mutations [Citation9–13]. Some studies had shown that genes, such as ASXL1, TET2, IDH1/2, and DNMT3A, which belong to epigenetic genes, were not accompanied with each other [Citation18]. We analyzed the correction of ASXL1 mutation with some other somatic mutations of AML; results suggested that ASXL1 mutations were associated with TET2, DNMT3A, PHF6 mutations and showed a positive trend with IDH1 and Dup-MLL mutations. This was consistent with the results of PV Vlierberghe et al. [Citation19]. Some difference still existed between our study and previous studies. This may due to the missing of some data in some included studies, different analyses focus on different population and small study population. The difference needs further research activities and analyses.
Regarding prognosis analysis, the results suggested that there was no statistically significant for OS and DFS between the ASXL1 mutation group and wild-type group, but poor prognostic trend was observed in survival curves of OS. In the subgroup analysis of 55 cases of CN-AML patients, the results showed that the OS in the ASXL1 mutation group was shorter (P = 0.016) than that in the wild-type group, and poor prognostic trend was observed in survival curves of DFS in the ASXL1 mutation group. Meanwhile, we also carried prognostic analyses in elderly (≥60 years) newly diagnosed AML patients; the results showed that the OS in the ASXL1 mutation group was shorter (P = 0.011) than the wild-type group. ASXL1 mutation showed only a poor prognostic trend for DFS (P = 0.096). Most of the current research studies reported that ASXL1 mutation was a poor prognostic factor in AML patients. It may lead to shorter OS and DFS, especially in elderly CN-AML patients [Citation9–13]. Univariate survival analysis showed that ASXL1 mutation was a poor prognostic factor for OS in CN-AML and elderly AML patients. And the survival curve of DFS informed that ASXL1 mutation may be also a poor prognostic factor for DFS, which was consistent with most previous research results. The multivariate analysis in elderly AML patients showed that ASXL1 mutation was one of the factors that affected OS and DFS. Univariate and multivariate analyses showed that ASXL1 mutation was independent adverse prognostic factor in elderly AML patients. The possible reasons for the discrepancy between our results and some previous studies were as follows: (1) Different study population has different genetic background. (2) Our study covered a small number of patients. (3) Our study only detected the 12th exon of ASXL1 mutation and ignored other sites; this may be the cause of the slight difference between the ASXL1 mutation group and wild-type group. In addition, three other mutations, TET2, CEBPA, and DNMT3A, which had a high mutation rate and a lower lost follow-up rate were also screened from 156 included AML patients. We analyzed the prognostic role of those mutations and when they combined with ASXL1 mutations on AML patients, respectively. The results showed that the difference in OS and DFS between TET2, DNMT3A, CEBPA gene mutation group and wild-type group was not significant. The DFS survival curve inferred that AML patients with TET2 mutation showed poor prognosis than TET2 wild-type patients. Also, the OS and DFS survival curve showed that CEBPA mutations showed good prognosis for both OS and DFS. Clinical practices and previous studies have considered TET2, DNMT3A gene mutation as poor prognostic factors in AML patients [Citation9, Citation14, Citation20–25]. Also, it is generally believed that the double CEBPA mutation is a good prognostic factor [Citation26]. For covering a small number of patients and lack of some analytical data, some of the results of our study were not consistent with some previous studies. We also analyzed the effect of ASXL1 mutation on the prognosis of AML patients with TET2 or DNMT3A mutations. The results showed that the ASXL1 mutation was associated with TET2 mutation and DNMT3A mutation, but when combined with ASXL1 mutation, the prognosis was not significantly affected. Similarly, we analyzed the effect of ASXL1 mutation on the prognosis of AML patients with CEBPA mutations. The results also showed that when combined with ASXL1 mutations, the prognosis of patients with CEBPA mutation was not significantly affected. At present, few studies were conducted in genetic association study. However, the reliability of our results need to be further confirmed by prospective randomized controlled studies covering a large numbers of AML patients.
Conclusions
ASXL1 mutation is a common AML prognosis-related gene mutation, which is relatively common in AML M5 and CN-AML patients. ASXL1 mutation may be a molecular marker of poor prognosis in AML, especially in CN-AML and elderly AML patients. ASXL1 mutation is associated with TET2, DNMT3A, and PHF6 mutations, but when accompanied with ASXL1 mutations, the prognosis of AML was not significantly affected. Although our study has reported something useful, our analysis has its limitations. Firstly, the observational time of our study was limited; Secondly, the analysis covered a small number of AML patients; Beyond that, only the 12th exon of ASXL1 gene mutations was analyzed. Therefore, the reliability of the results needs to be confirmed by expanding the sample volume and observational time and analysis of all types of ASXL1 mutations and mutations in every location of ASXL1 gene.
Supplemental Material
Download PDF (76.9 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
Notes
*The P-value of the total distribution in FAB classification when compared ASXL1 mutation group with wild-type group.
a The P-value of the total distribution in karyotype when compared ASXL1 mutation group with wild-type group.
b The P-value of the normal karyotype and abnormal karyotype when compared ASXL1 mutation group with wild type-group.
a Take the median value as the cut-off point.
References
- Ahmad F, Mohota R, Sanap S, et al. Molecular evaluation of DNMT3A and IDH1/2 gene mutation: frequency, distribution pattern and associations with additional molecular markers in normal karyotype Indian acute myeloid leukemia patients. Asian Pac J Cancer Prev. 2014;15:1247–1253.
- Asif N, Hassan N. Acute myeloid leukemia amongst adults. J Islamabad Med Dental College. 2013;2:58–63.
- The Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–2074.
- Shih AH, Abdel-wallab O, Patel JP, et al. The role of mutations in epigenetic regulators in myeloid malignancies. Nat Rev Cancer. 2012;12:599–612.
- Tahiliani M, Koh KP, Shen Y, et al. Conversion of 5-methylcytosine to 5-Hydroxymethylcytosine in Mammalian DNA by MLL Partner TET1. Science. 2009;324:930–935.
- Gelsi-Boyer V, Brecqueville M, Devillier R, et al. Mutations in ASXL1 are associated with poor prognosis across the spectrum of malignant myeloid diseases. J Hematol Oncol. 2012;5:12.
- Cho YS, Kim EJ, Park UH, et al. Additional sex comb-like 1 (ASXL1) in cooperation with SRC-1 acts as a ligand-dependent coactivator for retinoic acid receptor. J Biol Chem. 2006;281(26):17588–17598.
- Lee SW, Cho YS, Na JM, et al. ASXL1 represses retinoic acid receptor-mediated transcription through associating with HP1 and LSD1. J Biol Chem. 2010;285:18–29.
- Pratcorona M, Abbas S, Sanders MA, et al. Acquired mutations in ASXL1 in acute myeloid leukemia: prevalence and prognostic value. Haematologica. 2012;97:388–392.
- Metzeler KH, Becker H, Maharry K, et al. ASXL1 mutations identify a high-risk subgroup of older patients with primary cytogenetically normal AML within the ELN Favorable genetic category. Blood. 2011;118:6920–6929.
- El-Sharkawi D, Ali A, Evans CM, et al. ASXL1 mutations are infrequent in young patients with primary acute myeloid leukemia and their detection has a limited role in therapeutic risk stratification. Leuk Lymph. 2014;55:1326–1331.
- Schnittger S, Eder C, Jeromin S, et al. ASXL1 exon 12 mutations are frequent in AML with intermediate risk karyotype and are independently associated with an adverse outcome. Leukemia. 2013;27:82–91.
- Chou WC, Huang HH, Hou HA, et al. Distinct clinical and biological features of de novo acute myeloid leukemia with additional sex comb-like 1 (ASXL1) mutations. Blood. 2010;116:4086–4094.
- Chou WC, Chou SC, Liu CY, et al. TET2 mutation is an unfavorable prognostic factor in acute myeloid leukemia patients with intermediate-risk cytogenetics. Blood. 2011;118:3803–3810.
- Cheson BD, Bennett JM, Kopecky KJ, et al. Revised recommendations of the international working group for diagnosis, standardization of response criteria, treatment outcomes, and reporting standards for therapeutic trials in acute myeloid leukemia. J Clin Oncol. 2003;21(24):4642–4649.
- Nolte F, Hofmann WK. Myelodysplastic syndromes: molecular pathogenesis and genomic changes. Ann Hematol. 2008;87:777–795.
- Shivarov V, Gueorguieva R, Ivanova M, et al. ASXL1 mutations define a subgroup of patients with acute myeloid leukemia with distinct gene expression profile and poor prognosis: a meta-analysis of 3311 adult patients with acute myeloid leukemia. Leuk Lymph. 2015;56:1881–1883.
- Wang F, Zhang Y, Chen X, et al. Mutaome analysis including 10 common mutated genes in newly diagnosed acute myeloid leukemia patients. J Leuk Lymph. 2015;24(3):161–164.
- Tie R, Zhang T, Fu H, et al. Association between DNMT3A mutations and prognosis of adults with de novo acute myeloid leukemia: a systematic review and meta-analysis. PLoS One. 2014;9:e93353.
- Ostronoff F, Othus M, Ho PA, et al. Mutations in the DNMT3A exon 23 independently predict poor outcome in older patients with acute myeloid leukemia: a SWOG report. Leukemia. 2013;27:238–241.
- Renneville A, Boissel N, Nibourel O, et al. Prognostic significance of DNA methyltransferase 3A mutations in cytogenetically normal acute myeloid leukemia: a study by the acute leukemia French association. Leukemia. 2012;26:1247–1254.
- Shivarov V, Gueorguieva R, Stoimenov A, et al. DNMT3A mutation is a poor prognosis biomarker in AML: results of a meta-analysis of 4500 AML patients. Leuk Res. 2013;37:1445–1450.
- Gaidzik V I, Paschka P, Spath D, et al. TET2 mutations in acute myeloid leukemia (AML): results from a Comprehensive genetic and clinical analysis of the AML study group. J Clin Oncol. 2012;30:1350–1357.
- Wei JF, Chen GH, Qiu HY, et al. The incidence of TET2 gene mutation and its clinical significance in acute myeloid leukemia patients. Zhonghua Xue Ye Xue Za Zhi. 2011;32:304–307.
- Liu WJ, Tan XH, Luo XP, et al. Prognostic significance of Tet methylcytosine dioxygenase 2 (TET2) gene mutations in adult patients with acute myeloid leukemia: a meta-analysis. Leuk Lymphoma. 2014;55(12):2691–2698.
- Green CL, Koo KK, Hills RK, et al. Prognostic significance of CEBPA mutations in a large cohort of younger adult patients With acute myeloid leukemia: impact of double CEBPA mutations and the Interaction With FLT3 and NPM1 mutations. J Clin Oncol. 2010;28:2739–2747.