ABSTRACT
Objectives
Acute lymphoblastic leukemia (ALL) is one of the most common malignancies in children. Our aim was to identify a novel miRNA that can predict prognosis of childhood ALL patients and explore its potential mechanism.
Methods
The miRNA expression profiles of childhood ALL were analyzed using GEO database and HiSeq instruments. The expression of miR-155 was examined by RT–PCR in 42 ALL patients. To investigate the role of miR-155 in ALL, four ALL cell lines (CEM-C1, Jurkat, MOLT-3 and MOLT-4) were transfected with miR-155 mimics, miR-155 inhibitors or corresponding controls. Dual-luciferase reporter system was applied to confirm the miR-155 target ZNF238. Moreover, proliferation and apoptosis were evaluated by MTT and flow cytometry.
Results
Dataset GSE56489 and GSE23024 demonstrated that miR-155 was up-regulated and ZNF238 was down-regulated at diagnosis status of ALL. High miR-155 expression was associated with poor outcome. Overexpressed miR-155 promoted ALL cell proliferation and inhibited apoptosis. Dual-luciferase reporter result showed that miR-155 directly regulated ZNF238. Silencing ZNF238 promoted cell proliferation in ALL cells.
Discussion
Our research indicating that miR-155 might possess potential value as a biomarker for predicting the prognosis of individuals. However, the role of ZNF238 in childhood ALL remain unknown. In the present study, we found the possible role of ZNF238 as a new tumor suppressor in ALL, which might be necessary for the antiproliferative functions of normal cells to counteract ALL formation.
Conclusion
Our results propose that miR-155 is in association with poor prognosis of childhood ALL. Furthermore, miR-155 could promote cell proliferation targeting ZNF238.
Abbreviations: ALL: Acute Lymphoblastic Leukemia; miRNAs: MicroRNAs; 3’-UTR: 3’untranslated region; WBC: white blood cell; CNSL: central nervous system leukemia; SR: standard risk; IR: intermediate risk; EFS: event-free survival; OS: overall survival.
1. Introduction
Acute lymphoblastic leukemia (ALL) is a hematological malignancy characterized by the proliferation of early lymphoid precursors that replace normal hematopoietic cells in the bone marrow [Citation1]. It is the most common childhood malignant disease [Citation2]. ALL accounts for about 25% of cancer diagnosis in children under 15 years old. Though 5-year event-free survival rate for children with ALL has been improved to approximately 80% [Citation3], some patients still experience poor prognosis such as high recurrence rate. Therefore, it’s crucial to identify the molecular mechanisms of ALL for early diagnosis and treatment.
MicroRNAs (miRNAs) are a class of non-coding small RNAs. By linking the 3’ untranslated region (3’-UTR) of the downstream target gene, the miRNA can inhibit gene and protein expression at the post-transcriptional level. Leukemia is one of the earliest malignancies, which is associated with abnormal expression of miRNAs [Citation4]. It has been reported that a number of abnormally expressed miRNAs are in association with diagnosis and relapse in acute myeloid leukemia [Citation5]. The aberrant expression of miRNAs can be involved in many biological processes of leukemia such as cell proliferation [Citation6], apoptosis [Citation7], and hematopoietic differentiation [Citation8,Citation9]. Furthermore, some miRNAs have been identified to be as biomarkers for relapse and poor prognosis in acute myeloid leukemia and to be associated with treatment outcomes [Citation10,Citation11]. Among them, miR-155 has been addressed to be involved in the tumorigenesis of acute myeloid leukemia [Citation12,Citation13]. But whether it is an independent prognostic risk factor for children ALL still remains unknown.
In this research, we analyzed the miRNA expression profiles of childhood ALL and confirmed that up-regulated miR-155 was associated with poor prognosis of childhood ALL. A target gene ZNF238 of miR-155 was predicted and further confirmed that miR-155 directly bound to the 3’ UTR region of ZNF238 and inhibited ZNF238 expression at the protein levels. Furthermore, miR-155 and ZNF238 affected proliferation of ALL cell lines. The data suggest that miR-155 expression may partly responsible for improving prognostic stratification of childhood ALL. Moreover, it may participate in cell proliferation by targeting ZNF238 in ALL cells.
2. Materials and methods
2.1. Clinical specimens
Forty-two childhood ALL patients from the hospital were enrolled in this study. The detail information is presented in . All bone marrow samples were gathered at initial diagnosis and the first relapse. Our research was approved by the Ethics Committee of the hospital. All participants signed an informed consent form.
Table 1. Characteristics of patients.
2.2. MiRNA expression profiles
Data for miRNA expression profiles in childhood ALL compared with healthy controls were collected from the GEO database (www.ncbi.nlm.nih.gov/geo), two datasets meet standards (GEO record GSE56489 [Citation14], and GSE23024). MiRNA expression analysis of datasets GSE56489 was performed by array while dataset GSE23024 was performed by RT-qPCR. MiRNA-array analysis of GSE56489 was performed on bone marrow samples, at diagnosis, including 43 childhood ALL patients and 14 age-matched healthy controls. MiRNA expression analysis of datasets GSE23024 was performed on bone marrow samples, including 81 childhood ALL patients and 7 age-matched healthy controls. In addition, in order to obtain a novel miRNA that can predict poor prognosis of childhood ALL, we collected 6 bone marrow samples, at diagnosis, among them, 3 patients experienced relapse. MiRNA expression analysis was achieved by Gene Denovo Biotechnology Co. (Guangzhou, China) using an Illumina HiSeq™ 2500 instrument [Citation15].
2.3. RNA isolation and quantitative real-time PCR (qRT-PCR)
Trizol (Invitrogen, Carlsbad, CA) was used for RNA isolation from bone marrow samples in line with the manufacturer’s instruction. The qRT-PCR was achieved to investigate miRNA expression. cDNA was synthesized using stem-loop reverse transcription primer and PCR amplification primers (Taqman probe, Invitrogen). U6 was used as an internal reference. The miRNA expression was quantified by 2-ΔΔCT formula. The primers of miRNAs or mRNAs are as follows: miR-155; miR-199b; ZNF238; U6.
2.4. Cell culture
CEM-C1, Jurkat, MOLT-3 and MOLT-4 cells were cultured in RPMI 1640 medium (Gibco) supplemented with 10% fetal bovine serum (fetal bovine serum, Australia) at 37°C and 5% CO2 in a humidified atmosphere.
2.5. MiRNA mimics/inhibitor transfection
Transfections were performed with 100 nM of the miR-155 mimics, miR-155 inhibitors or corresponding controls (GenePharma, Shanghai, China) using the Neon Transfection System (Invitrogen). The electroporation conditions were 1230 V, 30 ms width and one pulse for CEM-C1, Jurkat, MOLT-3 and MOLT-4 cells [Citation16]. Cells were placed in 12-well plates (1 × 106 cells/well) in 1 ml of medium supplemented with 10% fetal bovine serum. After transfection, miRNA expression was evaluated by qRT-PCR.
2.6. MTT assay
CEM-C1, Jurkat, MOLT-3 and MOLT-4 cells were respectively plated in 96-well plates (3 × 104/well). The cells were transfected with 100 nM of the miR-155 mimics, miR-155 inhibitors or corresponding controls. After transfection at 24, 48 and 72 h, the cells were hatched with Dye Solution (5 g/L) for 4 h until purple precipitate was clear. Then, the cells were left in the dark for 2 h at room temperature after adding 100 uL Stop Mix. Finally, the absorbance was recorded (570 nm).
2.7. Apoptosis assay
CEM-C1, Jurkat, MOLT-3 and MOLT-4 cells were transfected with miR-155 mimics, miR-155 inhibitors or corresponding control (100 nM) using the Neon Transfection System. The cells were gathered at 96 h after transfection. Then, the cells were marked with Annexin V-FITC (BD Pharmingen, San Diego, CA). After that, the cells were examined by flow cytometry.
2.8. Bioinformatics analysis
Target genes of miR-155 were predicted using starBase database [Citation17], which contains five prediction programs: [TargetScan [Citation18], PicTar [Citation19], miRanda [Citation20], PITA [Citation21] and RNA22 [Citation22]] for animal miRNAs. We analyzed the predicted target genes of miR-155 in these five prediction programs, respectively. For the purpose of reducing false-positive predictions, targets were accepted if they were predicted by at least four programs. Then, we performed Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of these targets for miR-155 using KOBAS 3.0 tool [Citation23], and only p-value <0.05 corrected by Fisher’s exact test was set as statistical significance.
2.9. Western blot assay
Protein isolation was relied on RIPA buffer (Pierce, Rockford, IL, USA) with protease and phosphatase inhibitors (Roche, Beijing, China). First, the protein was segregated in a 10% SDS-PAGE gel and migrated to a methanol-activated PVDF membrane (Milliport, Beijing, China). Next, the membrane was blocked for 2 h in Tris-buffered saline Tween-20 (TBST) containing 5% milk, followed by incubation with rabbit anti-ZNF238 (Abcam, UK, ab118471, 1:500) and anti-GAPDH (Cell Signaling Technology, USA), overnight at 4°C. After incubation with secondary antibody (Sigma-Aldrich) for 1 h, the membrane was then blotted using ECL Substrate (Pierce) following manufacturer’s instructions.
2.10. Dual luciferase reporter assay
Wild-type (WT) or mutant (Mut) 3’ UTR segments of ZNF238 containing miR-155 binding site were synthesized and cloned into HEK-293T cells using luciferase reporter vectors (pGL3- ZNF238-WT and pGL3-ZNF238-Mut). The WT or mutant luciferase reporter vector was co-transfected with the miR-155 mimics, and miR-155 control using Lipofectamine 2000 (Invitrogen, NY, USA). At 48 h after transfection, the relative luciferase activities were examined using a dual-luciferase reporter assay system (Promega, USA).
2.11. Statistical analysis
SPSS 22.0 software and GraphPad Prism software 7.0 were used to achieve statistical analysis. Statistically significance was approved when P < 0.05. Data were displayed as mean ± standard deviation. The Mann–Whitney U test was used to analyze the difference between two samples. Two-tailed test was used for data analysis.
3. Results
3.1. MiR-155 is highly expressed in childhood ALL at diagnosis and relapse status, and high miR-155 expression is associated with poor prognosis
To analyze differentially expressed miRNAs in childhood ALL patients compared to healthy controls, two datasets GSE56489 (including 3 children with ALL and 14 age-matched healthy controls) and GSE23024 (including 81 pediatric leukemia cases and 17 normal hematopoietic control cases) from GEO database were selected. A total of 40 miRNAs were aberrantly expressed in childhood ALL in dataset GSE56489 (FC ≥ 1.5 or ≤ −1.5, P < 0.05) (Figure S-B), in addition, the most significant miRNAs are showed in (A). 88 miRNAs were aberrantly expressed in childhood ALL in dataset GSE23024 (FC ≥ 1.5 or ≤ −1.5, P < 0.05) (Figure S–C), and the most significantly aberrantly expressed miRNAs in childhood ALL in dataset GSE23024 are showed in (B). Moreover, the dataset GSE56489 mentioned that the expression of miR-146a, miR-155, miR-181a and miR-195 were significantly decreased in ALL patients after a six-month treatment period [Citation14]. To further select a functional miRNA associated with the clinical outcome of childhood ALL, we collected 6 bone marrow samples of childhood ALL patients at diagnosis, among them, 3 patients suffered from relapse. MiRNA expression analysis was performed by an Illumina HiSeq™ 2500 instrument. 1200 miRNAs were detected, including 151 novel miRNAs. Differentially expressed miRNAs were analyzed using hierarchical clustering analysis, as shown in (C), ALL samples were divided into relapse and CR patients. By comparing above three miRNA expression profiles, miR-199b and miR-155 were overlapped ((D)). The qRT-PCR was used to detect the expression of the two miRNAs on 42 bone marrow samples of childhood ALL patients at diagnosis (including 10 patients with relapse). The results showed that only miR-155 was significantly higher in relapse group compared to no relapse group (P=0.0004) ((A)). However, there was no significant change of miR-199b expression between relapse group compared to no relapse group ((B)). Therefore, miR-155 was identified for further analysis. To investigate miR-155 expression in different clinical and molecular characteristics groups (including white blood cell numbers, immuno-type, central nervous system leukemia (CNSL), risk status, disease stages) of childhood ALL, we analyzed the expression of miR-155 on 42 newly diagnosed samples (). MiR-155 expression was higher in CNSL patients ((E)) (p=0.0261). What’s more, miR-155 expression was higher in patients at relapse compared to whom at diagnosis ((H)) (p=0.0157). However, there was no significant change of miR-155 expression in white blood cell numbers, immuno-type and risk status ((C, D, E, and G). We conducted a multivariate logistic regression analysis and found that miR-155, immunophenotyping, and risk status are independent risk factors for recurrence. Next, we divided the patients into high and low miR-155 expression groups based on the median expression of miR-155. We found that patients with high miR-155 expression had shorter OS (P = 0.0043; (I)) and EFS (P = 0.0017; (J)). Additionally, we observed no significant difference on OS and EFS between standard risk status and intermediate risk status patients (P>0.05) ((K,L)). As a consequence, high miR-155 expression is associated with poorer outcome of childhood ALL cases.
Figure 1. MiRNA expression profiles in childhood acute lymphoblastic leukemia. Clustering analysis of the most significantly differentially expressed miRNAs in childhood acute lymphoblastic leukemia compared with healthy controls (FC ≥ −1.5 or ≤ 1.5, P < 0.05) in GEO datasets GSE56489 (A) and GSE23024 (B). (C) Hierarchical clustering of differentially expressed miRNAs in 6 bone marrow samples, at diagnosis, three of them relapse latter (P < 0.05). (D) Venn diagram showing the intersection of the abnormally expressed miRNAs on the above three datasets.
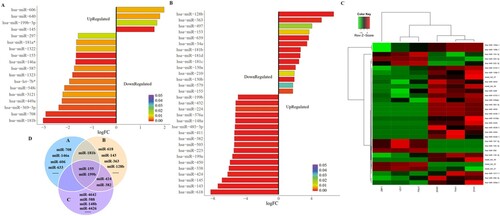
Figure 2. Up-regulated miR-155 is associated with specific risk groups and survival rate of childhood ALL patients. (A, B) MiRNA expression was evaluated by qRT-PCR in 42 newly diagnosed bone marrow samples, ten of whom relapsed latter. U6 was used as an internal reference. (C, D) MiR-155 expression levels in white blood cells (WBCs) and immunophenotype at diagnosis (P>0.05). (E) MiR-155 expression was significantly higher in CNSL patients compared to control (P < 0.05). (F) MiR-155 expression between standard risk status and intermediate risk status patients (P > 0.05). (G) The newly diagnosed bone marrow samples were divided into diagnosis (have no-relapse lately, n=32) and relapse (who relapse lately, n = 10) groups (P > 0.05). (H) Four paired diagnosis-relapse samples were detected. (I, J) High miR-155 expression was associated with poorer OS and EFS rate (P < 0.01). (K, L) The OS and EFS of patients between intermediate risk status and standard risk status (P > 0.05).
In addition, multivariate regression analysis of recurrence factors for pediatric ALL showed that miR-155 expression is an independent prognostic factor (Table S1).
3.2. MiR-155 promotes cell proliferation and inhibits apoptosis in ALL
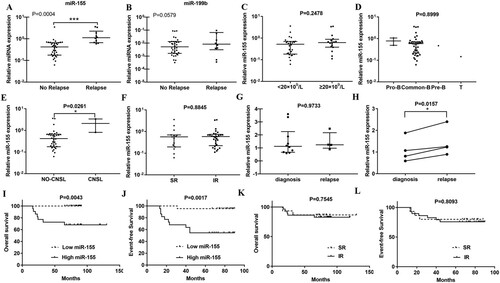
To further elucidate the role of miR-155 in childhood ALL, proliferation of four ALL cell lines was analyzed when miR-155 was over-expressed or inhibited. We found that cell proliferation in miR-155 over-expressing groups was significantly higher than controls ((A)). Furthermore, the proliferation of four human ALL cell lines transfected with miR-155 inhibitors was significantly reduced compared to controls. Furthermore, Figure S-A proves high transfection efficiency after 36 h. These results proposed that miR-155 could promote cell proliferation of four human ALL cell lines. After that, we further assessed the apoptotic effect of miR-155 in ALL cells. As supposed, (B) presents that the apoptosis rate in four human ALL cell lines was higher when the expression of miR-155 was inhibited. (C) shows that no significant difference of the apoptosis rate was captured between miR-155 groups and their corresponding NCs in four human ALL cell lines. Consequently, above results indicated that miR-155 may be closely associated with proliferation and apoptosis of ALL.
Figure 3. MiR-155 promotes proliferation and inhibits apoptosis of CEM-C1, Jurkat, MOLT-3 and MOLT-4 cells. (A) MTT assay shows that miR-155 obviously promotes cell proliferation. (B) Inhibitor-miR-155 promotes cell apoptosis in the presence of 0.1uM dexamethasone. (C) MiR-155 inhibits the apoptosis in the presence of 1uM dexamethasone. Apoptotic cells were detected at 96 h after transfection.
3.3. MiR-155 could promote cell proliferation by directly targeting ZNF238 in ALL
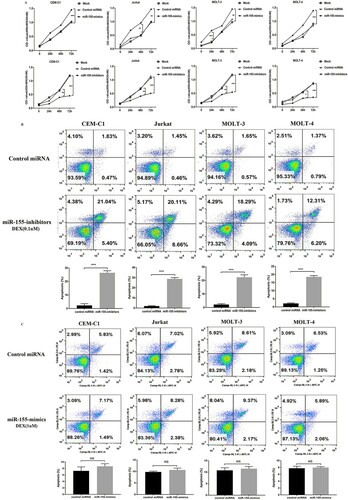
Computational prediction of human miRNA targets from StarBase suggested that miR-155 can modulate more than 1000 genes ((A)). To further explore the underlying mechanism of miR-155 regulatory network, we performed KEGG analysis of these targets of miR-155. The results proposed that miR-155 might participate in cellular mechanisms associated with ALL dysregulation (eg, PI3K-Akt, mTOR, and AMPK) ((B)). To further identify a functional miR-155 target, we chosen the genes that were predicted by at least four databases simultaneously. 75 of the putative miR-155 targets met the requirements. Then, we performed correlation analysis of miRNA and target genes in leukemia (p>0.05). Sixteen of the target non-oncogenes in leukemia were significantly correlated with miR-155 (). Among them, JARID2, PHF17, RREB1, SDCBP and BACH1 had been already demonstrated as the targets of miR-155 in leukemia. Previous studies have proposed that ZNF238 is the novel tumor suppressor in human brain cancer [Citation24], which can drastically decrease proliferation and promote cell death in medulloblastoma and glioblastoma multiforme cells [Citation25]. However, the role of ZNF238 in childhood ALL is unknown. (C) shows the predicted binding sites between miR-155 and ZNF238. Dual luciferase reporter assay was performed in HEK-293 T cells ((D)). Furthermore, the expression of ZNF238 was inhibited at the protein and mRNA levels in CEM-C1 and MOLT-4 cells transfected with miR-155 mimics, miR-155 inhibitors, and corresponding controls ((E,G)). To analyze ZNF238 expression between childhood ALL patients and healthy controls, we chose Coustan-Smith Leukemia dataset in ONCOMINE database, which contains 46 children with T-ALL, 238 children with B- ALL and 4 healthy controls. We found that ZNF238 was significantly reduced in children ALL ((H)).
Figure 4. MiR-155 could promote ALL cell proliferation by directly targeting ZNF238. (A) Venn diagram showing the predicted targets of miR-155 in five prediction programs. (B) KEGG pathway enrichment analysis of miR-155 targets. (C) The site sequence of predicted target ZNF238 for miR-155. (D) Luciferase reporter showing luciferase activity in wild-type ZNF238, mutant ZNF238 and controls. (E-G) The protein and mRNA expression of ZNF238 in CEM-C1 and MOLT-4 cell transfected with miR-155 mimics, miR-155 inhibitors or control duplex. (H) ZNF238 expression in childhood ALL. (I, J) MTT assay shows that cell proliferation was significantly inhibited in CEM-C1 and MOLT-4 cells transfected with si-ZNF238.
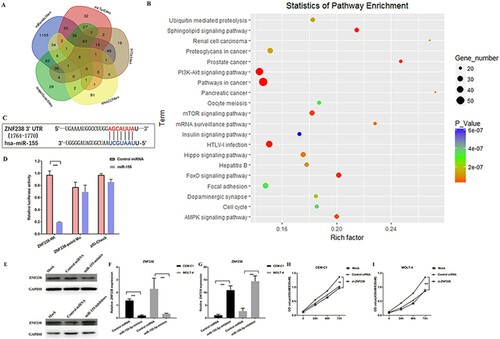
Table 2. The non-oncogene targets which are significantly correlated to miR-155 in leukemia.
To further investigate whether miR-155 regulates cell proliferation by targeting ZNF238 in ALL cells, we analyzed the cell proliferation after silencing the expression of ZNF238. MTT assay result shows that inhibiting ZNF238 promoted ALL cell proliferation ((I,J)). Combing above results, up-regulated miR-155 could promote ALL cell proliferation by directly targeting ZNF238.
4. Discussion
MiRNA dysregulation is a common event in leukemia [Citation26]. Recent study indicated that unusual expressions of relevant miRNAs may promote tumors [Citation27]. Their abnormal expressions are closely involved in the incidence, development, treatment response and prognosis of leukemia [Citation28,Citation29]. In our study, we analyzed miRNA expression profiles of childhood ALL. The results showed that miR-155 expression was obviously up-regulated in patients with ALL and significantly higher in relapsed patients. In addition, miR-155 expression can be a prognostic risk factor in pediatric ALL, indicating that miR-155 might possess potential value as a biomarker for predicting the prognosis of individuals. Furthermore, miR-155 has been identified in monoclonal B-cell lymphocytosis [Citation30] and AML [Citation31]. Additionally, MTT and flow cytometry assay proved that up-regulated miR-155 could promote cell proliferation and inhibit apoptosis in ALL cells. MiR-155 has been addressed to be involved in many human cancers. It has been shown to have both oncogenic and tumor suppressor functions [Citation32]. For example, high miR-155 expression is associated with therapy resistance in chronic lymphocytic leukemia [Citation33] and acts as an anti-leukemic role in human FLT3-wildtype AML [Citation34]. Our results suggest that miR-155 may serve as an oncogene in childhood ALL. What’s more, it has been demonstrated that combining anti-miR-155 with chemotherapy can successfully resensitize tumors to chemotherapy in vivo for lung cancer. In AML, therapeutic inhibition of miRNA-21 and miRNA-196b lead to leukemia-free survival in a murine model [Citation35], suggesting the therapeutic value of microRNA antagonists in leukemia [Citation35]. Furthermore, anti-miR-155-DOPC can be considered non-toxic in vivo [Citation36]. Therefore, miR-155 could be considered as a potential therapeutic target in childhood ALL, however, more research needs to be performed.
However, the perceive signals that participate in ALL cell proliferation and cease apoptosis are still under exploring. Bioinformatics analyses predicted that miR-155 can target more than 1000 regulatory genes. Here in, we validated ZNF238 as target of miR155 in ALL cells. ZNF238 is a member of the BTB/POZ-ZF protein family, which contains several proteins involved in development and/or cancer formation. It is critical in the developing cerebral cortex [Citation37–41]. Potential mechanisms include repressing Ngn2 transcription to regulate the Ngn2-Rnd2 pathway [Citation37], negatively regulating all four Id genes (Id1–Id4) [Citation40]. ZNF238 also displays tumor suppressor activity. Previous study verified that reinstating ZNF238 expression in medulloblastoma and glioblastoma multiforme cells drastically decreases cell proliferation and promotes cell death [Citation25]. ZNF238 antagonizes medulloblastoma and glioblastoma multiforme tumor growth in vivo [Citation25]. All the data suggest an anti-proliferative function of ZNF238 in medulloblastoma and glioblastoma multiforme tumor. However, the role of ZNF238 in childhood ALL remain unknown. In the present study, we found that ZNF238 could play a part in regulating cell proliferation in human ALL cells. Dual luciferase reporter assay results showed that miR-155 could directly regulate ZNF238 expression. These results indicated that miR-155 could promote proliferation by mediating down-regulated ZNF238 in ALL cells. In addition, the possible role of ZNF238 as a new tumor suppressor in ALL might be necessary for the antiproliferative functions of normal cells to counteract ALL formation. The precise mechanisms that how ZNF238 orchestrates ALL cell proliferation, in particular transcriptional regulation that it controls, remain to be refined in future studies.
5. Conclusion
Our findings showed that miR-155 is up-regulated in ALL and associated with poor prognosis. Furthermore, we found that miR-155 could promote ALL cell proliferation by targeting ZNF238.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Ultimo S, Martelli AM, Zauli G, et al. Roles and clinical implications of microRNAs in acute lymphoblastic leukemia. J Cell Physiol. 2018 Aug;233:5642–5654.
- Scheijen B, Boer JM, Marke R, et al. Tumor suppressors BTG1 and IKZF1 cooperate during mouse leukemia development and increase relapse risk in B-cell precursor acute lymphoblastic leukemia patients. Haematologica. 2017 Mar;102(3):541–551.
- Tuckuviene R, Ranta S, Albertsen BK, et al. Prospective study of thromboembolism in 1038 children with acute lymphoblastic leukemia: a Nordic Society of Pediatric Hematology and Oncology (NOPHO) study. J Thromb Haemost. 2016 Mar;14(3):485–494.
- Fernandes Q. MicroRNA: Defining a new niche in leukemia. Blood Rev. 2017 May;31(3):129–138.
- Hackl H, Astanina K, Wieser R. Molecular and genetic alterations associated with therapy resistance and relapse of acute myeloid leukemia. J Hematol Oncol. 2017 Feb 20;10(1):51.
- Zenz T, Habe S, Denzel T, et al. Detailed analysis of p53 pathway defects in fludarabine-refractory chronic lymphocytic leukemia (CLL): dissecting the contribution of 17p deletion, TP53 mutation, p53-p21 dysfunction, and miR34a in a prospective clinical trial. Blood. 2009 Sep 24;114(13):2589–2597.
- Zauli G, Voltan R, di Iasio MG, et al. miR-34a induces the downregulation of both E2F1 and B-Myb oncogenes in leukemic cells. Clin Cancer Res. 2011 May 1;17(9):2712–2724.
- De Luca L, Trino S, Laurenzana I, et al. Knockdown of miR-128a induces Lin28a expression and reverts myeloid differentiation blockage in acute myeloid leukemia. Cell Death Dis. 2017 Jun 1;8(6):e2849.
- Jian P, Li ZW, Fang TY, et al. Retinoic acid induces HL-60 cell differentiation via the upregulation of miR-663. J Hematol Oncol. 2011 Apr 25;4:20.
- de Leeuw DC, Verhagen HJ, Denkers F, et al. MicroRNA-551b is highly expressed in hematopoietic stem cells and a biomarker for relapse and poor prognosis in acute myeloid leukemia. Leukemia. 2016 Mar;30(3):742–746.
- Piatopoulou D, Avgeris M, Marmarinos A, et al. miR-125b predicts childhood acute lymphoblastic leukaemia poor response to BFM chemotherapy treatment. Br J Cancer. 2017 Sep 5;117(6):801–812.
- Faraoni I, Laterza S, Ardiri D, et al. MiR-424 and miR-155 deregulated expression in cytogenetically normal acute myeloid leukaemia: correlation with NPM1 and FLT3 mutation status. J Hematol Oncol. 2012 Jun 8;5:26.
- Marcucci G, Maharry KS, Metzeler KH, et al. Clinical role of microRNAs in cytogenetically normal acute myeloid leukemia: miR-155 upregulation independently identifies high-risk patients. J Clin Oncol. 2013 Jun 10;31(17):2086–2093.
- Duyu M, Durmaz B, Gunduz C, et al. Prospective evaluation of whole genome microRNA expression profiling in childhood acute lymphoblastic leukemia. Biomed Res Int. 2014;2014:1–7.
- Meng Y, Tian H, Hu Q, et al. MicroRNA repertoire and comparative analysis of Andrias davidianus infected with ranavirus using deep sequencing. Dev Comp Immunol. 2018 Aug;85:108–114.
- Liang YN, Tang YL, Ke ZY, et al. MiR-124 contributes to glucocorticoid resistance in acute lymphoblastic leukemia by promoting proliferation, inhibiting apoptosis and targeting the glucocorticoid receptor. J Steroid Biochem Mol Biol. 2017 Sep;172:62–68.
- Yang JH, Li JH, Shao P, et al. Starbase: a database for exploring microRNA-mRNA interaction maps from Argonaute CLIP-Seq and Degradome-Seq data. Nucleic Acids Res. 2011 Jan;39(Database issue):D202–D209.
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005 Jan 14;120(1):15–20.
- Krek A, Grun D, Poy MN, et al. Combinatorial microRNA target predictions. Nat Genet. 2005 May;37(5):495–500.
- John B, Enright AJ, Aravin A, et al. Human MicroRNA targets. PLoS Biol. 2004 Nov;2(11):e363.
- Kertesz M, Iovino N, Unnerstall U, et al. The role of site accessibility in microRNA target recognition. Nat Genet. 2007 Oct;39(10):1278–1284.
- Miranda KC, Huynh T, Tay Y, et al. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006 Sep 22;126(6):1203–1217.
- Xie C, Mao X, Huang J, et al. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011 Jul;39(Web Server issue):W316–W322.
- Fedele V, Dai F, Masilamani AP, et al. Epigenetic regulation of ZBTB18 promotes glioblastoma Progression. Mol Cancer Res. 2017 Aug;15(8):998–1011.
- Tatard VM, Xiang C, Biegel JA, et al. ZNF238 is expressed in postmitotic brain cells and inhibits brain tumor growth. Cancer Res. 2010 Feb 1;70(3):1236–1246.
- Dzikiewicz-Krawczyk A, Macieja A, Maly E, et al. Polymorphisms in microRNA target sites modulate risk of lymphoblastic and myeloid leukemias and affect microRNA binding. J Hematol Oncol. 2014 Jun 2;7:43.
- Lin S, Gregory RI. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer. 2015 Jun;15(6):321–333.
- Fernando TR, Rodriguez-Malave NI, Rao DS. MicroRNAs in B cell development and malignancy. J Hematol Oncol. 2012;5:7.
- Yeh CH, Moles R, Nicot C. Clinical significance of microRNAs in chronic and acute human leukemia. Mol Cancer. 2016 May 14;15(1):37.
- Ferrajoli A, Shanafelt TD, Ivan C, et al. Prognostic value of miR-155 in individuals with monoclonal B-cell lymphocytosis and patients with B chronic lymphocytic leukemia. Blood. 2013 Sep 12;122(11):1891–1899.
- Diaz-Beya M, Brunet S, Nomdedeu J, et al. MicroRNA expression at diagnosis adds relevant prognostic information to molecular categorization in patients with intermediate-risk cytogenetic acute myeloid leukemia. Leukemia. 2014 Apr;28(4):804–812.
- Narayan N, Morenos L, Phipson B, et al. Functionally distinct roles for different miR-155 expression levels through contrasting effects on gene expression, in acute myeloid leukaemia. Leukemia. 2017 Apr;31(4):808–820.
- Cui B, Chen L, Zhang S, et al. MicroRNA-155 influences B-cell receptor signaling and associates with aggressive disease in chronic lymphocytic leukemia. Blood. 2014 Jul 24;124(4):546–554.
- Palma CA, Al Sheikha D, Lim TK, et al. MicroRNA-155 as an inducer of apoptosis and cell differentiation in acute myeloid Leukaemia. Mol Cancer. 2014 Apr 5;13:79.
- Velu CS, Chaubey A, Phelan JD, et al. Therapeutic antagonists of microRNAs deplete leukemia-initiating cell activity. J Clin Invest. 2014 Jan;124(1):222–236.
- Van Roosbroeck K, Fanini F, Setoyama T, et al. Combining anti-Mir-155 with chemotherapy for the treatment of lung cancers. Clin Cancer Res. 2017 Jun 1;23(11):2891–2904.
- Ohtaka-Maruyama C, Hirai S, Miwa A, et al. RP58 regulates the multipolar-bipolar transition of newborn neurons in the developing cerebral cortex. Cell Rep. 2013 Feb 21;3(2):458–471.
- Baubet V, Xiang C, Molczan A, et al. Rp58 is essential for the growth and patterning of the cerebellum and for glutamatergic and GABAergic neuron development. Development. 2012 Jun;139(11):1903–1909.
- Xiang C, Baubet V, Pal S, et al. RP58/ZNF238 directly modulates proneurogenic gene levels and is required for neuronal differentiation and brain expansion. Cell Death Differ. 2012 Apr;19(4):692–702.
- Hirai S, Miwa A, Ohtaka-Maruyama C, et al. RP58 controls neuron and astrocyte differentiation by downregulating the expression of Id1-4 genes in the developing cortex. EMBO J. 2012 Mar 7;31(5):1190–1202.
- Heng JI, Qu Z, Ohtaka-Maruyama C, et al. The zinc finger transcription factor RP58 negatively regulates Rnd2 for the control of neuronal migration during cerebral cortical development. Cereb Cortex. 2015 Mar;25(3):806–816.
- Nakagawa R, Leyland R, Meyer-Hermann M, et al. MicroRNA-155 controls affinity-based selection by protecting c-MYC+ B cells from apoptosis. J Clin Invest. 2016 Jan;126(1):377–388.
- Skalsky RL, Samols MA, Plaisance KB, et al. Kaposi's sarcoma-associated herpesvirus encodes an ortholog of miR-155. J Virol. 2007 Dec;81(23):12836–12845.
- Muylkens B, Coupeau D, Dambrine G, et al. Marek's disease virus microRNA designated Mdv1-pre-miR-M4 targets both cellular and viral genes. Arch Virol. 2010 Nov;155(11):1823–1837.
- Sahmatova L, Tankov S, Prans E, et al. MicroRNA-155 is Dysregulated in the Skin of patients with Vitiligo and inhibits Melanogenesis-associated genes in Melanocytes and Keratinocytes. Acta Derm Venereol. 2016 Aug 23;96(6):742–747.
- Elton TS, Selemon H, Elton SM, et al. Regulation of the MIR155 host gene in physiological and pathological processes. Gene. 2013 Dec 10;532(1):1–12.
- Pottier N, Maurin T, Chevalier B, et al. Identification of keratinocyte growth factor as a target of microRNA-155 in lung fibroblasts: implication in epithelial-mesenchymal interactions. PloS one. 2009 Aug 24;4(8):e6718.
- Xie Q, Chen X, Lu F, et al. Aberrant expression of microRNA 155 may accelerate cell proliferation by targeting sex-determining region Y box 6 in hepatocellular carcinoma. Cancer. 2012 May 1;118(9):2431–2442.
- Yin Q, Wang X, Fewell C, et al. MicroRNA miR-155 inhibits bone morphogenetic protein (BMP) signaling and BMP-mediated Epstein-Barr virus reactivation. J Virol. 2010 Jul;84(13):6318–6327.
- Chen C, Luo F, Yang Q, et al. NF-kappaB-regulated miR-155, via repression of QKI, contributes to the acquisition of CSC-like phenotype during the neoplastic transformation of hepatic cells induced by arsenite. Mol Carcinog. 2018 Apr;57(4):483–493.
- He B, Gao SQ, Huang LD, et al. MicroRNA-155 promotes the proliferation and invasion abilities of colon cancer cells by targeting quaking. Mol Med Rep. 2015 Mar;11(3):2355–2359.
