ABSTRACT
Objectives
To investigate the role of Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) in acute myeloid leukemia (AML) and analyze the potential regulatory network of MALAT1/miR-146a/ CXCR4.
Methods
The expressions of MALAT1, miR-146a and CXCR4 were performed by qRT-PCR and Western Blot. We conducted trans-well assay, CCK-8 assay and flow cytometry to evaluate the migration, proliferation and apoptosis of AML cells. Also by using luciferase reporter assay, we investigated the interaction between miR-146a and MALAT1 or CXCR4.
Results
Firstly, MALAT1 and CXCR4 were upregulated while miR-146a was downregulated in AML patients compared with healthy controls. We observed a negative correlation between miR-146a and MALAT1 or CXCR4, but a positive correlation between MALAT1 and CXCR4 in AML patients. MALAT1 knockdown inhibited migration and proliferation but induced apoptosis of HL-60 cells. MALAT1 restrained miR-146a expression by acting as a ceRNA. miR-146a regulated HL-60 cells migration, proliferation and apoptosis by directly targeting CXCR4 expression. Finally, we found that CXCR4 expression was downregulated by MALAT1 knockdown and partially restored by miR-146a abrogation.
Conclusions
Our results showed that MALAT1 regulates migration, proliferation and apoptosis by sponging miR-146a to regulate CXCR4 expression in AML cells, providing novel insights into the role of MALAT1 as a therapeutic target in AML.
Introduction
Acute myeloid leukemia (AML) is a heterogeneous hematological malignancy characterized by the clonal expansion of myeloid blasts in peripheral blood, bone marrow (BM), and/or other tissues [Citation1]. The outcomes of AML patients improved due to intensive chemotherapy and stem cell transplantation (HSCT). In the last decades, however, most patients experienced relapse and drug resistance [Citation2–4]. To improve the outcomes of AML, new insights into mechanisms for relapse and drug resistance are required. There is an urgent need to find more new biomarkers that could identify high risk patients and be therapeutic targets.
In recent years, many scientists have focused on the roles of non-coding RNAs in AML development and therapy, including microRNAs (miRNAs) and long non-coding RNAs (lncRNAs) [Citation5]. MiRNAs are a class of single-stranded small non-coding RNAs with 19–22 nucleotides in length, which modulate gene expression at the post-transcriptional level via binding to the 3′-untranslated regions of target genes [Citation6]. Some studies found that microRNA-14a (miR-146a) was downregulated in some AML patients [Citation7–9], which suggested miR-146a played a critical pathogenic role in AML. Our previous studies demonstrated that miR-146a and its target gene CXCR4 as novel biomarkers in AML patients. However, the potential molecular mechanisms remain to be further investigated.
LncRNAs are a family of non-protein-coding transcripts with a length of over 200 nucleotides. Various evidence demonstrated that lncRNAs played an vital role in biological functions, such as epigenetic regulation and gene transcription [Citation10–12]. In recent years, it has been reported that some lncRNAs are involved in AML progression by regulating cell processes, including proliferation, apoptosis, migration and drug resistance [Citation13–15]. Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), also known as nuclear enriched abundant transcripts (NEAT2), is an 8.7 kb oncogenic lncRNA located on chromosome 11. MALAT1 is overexpressed in several solid tumors and hematopoietic malignancies, including multiple myeloma (MM), chronic lymphocytic leukemia (CLL), chronic myelogenous leukemia (CML) and acute lymphoblastic leukemia (ALL) [Citation16–20]. But till now, the biological function of MALAT1 in AML remains unknown.
LncRNAs generally act as miRNA sponges through a mechanism of competing for endogenous RNAs (ceRNAs) that regulates expression of the targeting miRNAs [Citation19]. Some reports confirmed that MALAT1 promoted inflammatory response through holding miR-146a by acting as a ceRNA [Citation21,Citation22]. Tan et al suggested that MALAT1 was involved in promoting tumor cell migration, invasion, and metastasis in various tumors by regulating CXCR4 expression [Citation23]. Our previous studies demonstrated that CXCR4 mediates AML cell migration, adhesion and drug resistance held by miR-146a. Thus, we hypothesized that MALAT1 might act as a ceRNA for miR-146a to regulate CXCR4 in AML. This study aimed to investigate the effect of MALAT1 on AML cells migration, proliferation, and apoptosis in vitro and analyzed the potential ceRNA regulatory network of MALAT1/miR-146a/CXCR4.
Materials and methods
Patients
A total of 20 eligible patients with de novo AML(Clinical data of AML cells are shown in ) and 20 volunteers (Hematopoietic stem cell donors) were enrolled in the present study. AML patients were diagnosed and classified according to the World Health Organization (WHO) criteria at our Hospital in the period from January 2016 to December 2018. The peripheral blood mononuclear cells (PBMCs) were isolated by using Ficol-Histopaque gradient centrifugation method. All participants have signed the informed consent, and this study was approved by the Research Ethics Committee of Hospital.
Table 1. The clinical characteristics of patients.
Cell culture and treatment
The human AML cell line (HL-60) was a gift from the Institute of Hematology at the China Academy of Chinese Medical Sciences (Beijing, China). As described previously, HL-60 cells were cultured in Iscove's modified Dulbecco's medium (IMDM; Gibco Invitrogen, Grand Island, NY, USA) supplemented with 20% fetal bovine serum (FBS; Gibco, Grand Island, NY, USA) and 100 U/ml penicillin–streptomycin at 37˚C in a humidified incubator with 5% CO2.
Small interfering RNA (siRNA) against MALAT1 (si-MALAT1), siRNA negative control (si-NC), miR-146a mimic, miR-146a inhibitor (anti-miR-146a) and miR-146a negative control (mi-NC) were synthesized by Genepharma (Shanghai, China). All transfections were performed in HL-60 by using Lipofectamine 2000 (Invitrogen) following the manufacturer's instructions.
Quantitative real-time polymerase chain reaction (qRT-PCR) analysis
Total RNA was extracted from HL-60 cells or AML cells using TRIzol reagent (Vazyme Biotech Co., Ltd., Nanjing, China) and reverse transcribed using HiScript II Q RT SuperMix (Vazyme Biotech) according to the manufacturer's protocols. qRT-PCR was performed using ChamQ SYBR Color qPCR Master Mix (Vazyme Biotech) on a Bio-Rad iQ5 PCR thermal cycler (Bio-Rad Laboratories, Inc., Hercules, CA, USA). Each gene was tested and the respective melting curves were detected. All samples were measured in triplicate. The relative levels of RNAs were measured with U6 small RNA or GAPDH as an internal control using the 2−△△t method. Primers are shown in .
Table 2. Primer sequences.
Western Blot
HL-60 cells were lysed using RIPA buffer (Beyotime Institute of Biotechnology, Shanghai, China) containing 50 mM Tris (pH 7.4), 0.1% sodium dodecyl sulfate (SDS) and 1 mM PMSF protease inhibitor. Lysates were maintained on ice for 30 min following centrifugation at 14,000 x g for 3 min at 4˚C. The supernatant's protein concentration was determined using a Bicinchoninic Acid Protein Assay kit (Beyotime Institute of Biotechnology), according to the manufacturer's protocol. Total protein (30 µg) from each sample was separated by 10% SDS-polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membranes (Millipore, Billerica, MA, USA). Membranes were blocked with 5% non-fat milk in Tris-buffered saline containing Tween-20 (TBST; Sigma-Aldrich) at room temperature for 30 min, after which the membranes were incubated overnight at 4˚C with anti-CXCR4 (1:250) and β-tubulin (1:3,000) primary antibodies. Following rinsing three times with 20 ml TBST for 5 min, the membranes were incubated with HRP-conjugated goat anti-rabbit immunoglobulin G secondary antibody (1:1,500) for 1 h at room temperature. Bands were detected by Western lightning enhanced chemiluminescence reagent (PerkinElmer, Inc., Waltham, MA, USA). Results were analyzed on a Tanon 5500 Chemiluminescence.
Dual luciferase reporter assay
The 3′-untranslated region (UTR) sequence of MALAT1 or CXCR4 containing wild-type (WT) or mutant (MUT) binding sites of miR-146a were amplified by PCR and cloned into the pGL3 vector (Promega, Madison, WI, USA) to generate the recombinant luciferase reporter vectors (MALAT1-WT or MALAT1-MUT, CXCR4-WT or CXCR4-MUT). HL-60 cells were transfected with 0.2 μg MALAT1 or CXCR4 reporter constructs, 10 ng control vector and 50 nM miR-146a or miR-NC using Lipofectamine 2000 according to the manufacturer’ protocols. After 48 h post-transfection, luciferase activities were measured using the luciferase assay kit (Promega).
Migration assay
Migration assays were performed using Transwell assays with 8.0 µm pore polycarbonate membrane inserts (Sigma-Aldrich). As we reported previously, HL-60 cells were starved for 24 h before performing the migration assays, and the FBS was replaced with 0.5% BSA. The lower chamber was briefly supplemented with chemoattractant (600 µl IMDM medium containing 100 ng/ml SDF-1α). Assay medium (IMDM with 0.5% BSA) without SDF-1α was used as baseline control. Subsequently, 5 × 105 cells in 100 µl medium were added to the upper chamber and incubated for 4 h under 37˚C and 5% CO2 culture conditions. Following this, cells that had migrated into the lower chamber were collected and counted by flow cytometry (BD Pharmingen) at high flow for 20 sec. Results are shown migration index,represents the ratio of the number of cells migrated in the presence of chemoattractant to the number of cells migrated without chemoattractant.
Cell viability assays
Cell viability was assessed using a Cell Counting kit-8 (CCK-8) assay (Dojindo Molecular Technologies, Inc., Kumamoto, Japan). Briefly, 100 µl HL-60 cell suspension (1 × 104 cells/ml) per well was seeded into a 96-well plate, and 10 µl CCK-8 solution was added to each well, followed by incubation under culture conditions for 4 h. The absorbance was determined at OD 450 nm using a spectrophotometer.
Flow cytometry assay
Apoptosis was analyzed by flow cytometry using Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) apoptosis detection kit (Yeasen, Shanghai, China) according to the manufacturer's protocol. After the culture for 48 h, HL-60 cells were washed with cold PBS and resuspended in binding buffer, followed by staining with 5 µl Annexin V-FITC and 10 µl PI for 10 min in the dark at room temperature. The stained cells stained were evaluated by flow cytometry as soon as possible.
Statistical analysis
Data are presented as the mean ± standard deviation (SD) of at least three independent experiments. Comparisons were performed by one-way analysis of variance (ANOVA) or two-tailed, unpaired Student's t-test using SPSS 21.0 software (SPSS Inc., Chicago, USA). P < 0.05 was regarded as statistically significant.
Results
The expression of MALAT1, mir-146a and CXCR4 in AML
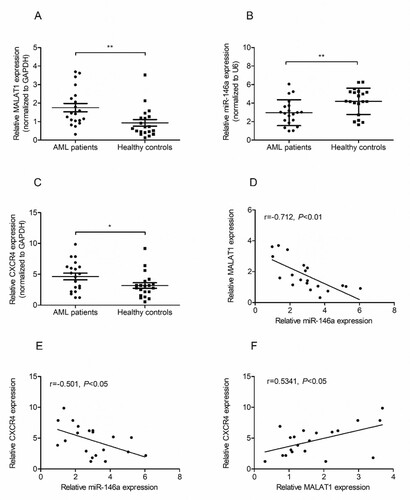
Firstly, we identified the expression status of MALAT1, miR-146a and CXCR4 in PBMCs samples from 20 AML patients and 20 healthy donors by qRT-PCR. As shown in Figures 1A and 1B, mRNA level of MALAT1 was significantly increased and miR-146a was decreased in AML patients compared with that in healthy donors. Also, we observed a significant upregulation of CXCR4 mRNA expression in AML patients than that in healthy controls (C). Interestingly, negative correlations between miR-146a and MALAT1, CXCR4 and miR-146a were observed in AML patients (D and 1E). But we observed a positive correlation between MALAT1 and CXCR4 expression (F).
Figure 1. MALAT1 and CXCR4 are upregulated while miR-146a is downregulated in AML patients. The expression of MALAT1 (A), miR-146a (B) and CXCR4 (C) were measured in PBMSCs cells from AML patients (n = 20) or normal controls (n = 20) by qRT-PCR. The Spearman's correlation analyses were performed to analyze the correlation between MALAT1 and miR-146a (D), the correlation between CXCR4 and miR-146a (E), the correlation between MALAT1 and CXCR4 (F). *P < 0.05, **P < 0.01.
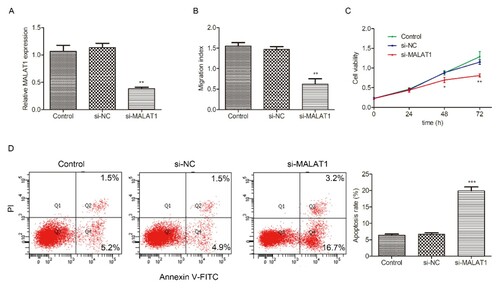
Knockdown MALAT1 suppresses migration, proliferation and induced apoptosis of HL-60 cells
To investigate the biological functions of MALAT1 in AML progression, we transfected HL-60 cells with si-MALAT1 or si-NC. As shown in A, MALAT1 expression was downregulated in HL-60 cells after transection with si-MALAT1 when compared with control. Then we performed CCK-8 assay, trans-well assay and flow cytometry assay to explore the effects of MALAT1 on migration, proliferation and apoptosis in HL-60 cells, respectively. Results showed that MALAT1 silencing significantly suppressed cell migration (B) and proliferation (C), but induced apoptosis (D) in HL-60 cells compared with the control group.
Figure 2. Effects of MALAT1 knockdown on migration, proliferation and apoptosis of HL-60 cells. HL-60 cells were transfected with si-NC or si-MALAT1 for 48h, then the transfection efficiency was evaluated by qRT-PCR (A). (B) Tran-swell assay was implemented to analyze cell migration at 48h in H-60 cells transfected with si-NC or si-MALAT1. (C) CCK-8 assay was performed to measure cell proliferation at 0, 24, 48 and 72h in H-60 cells transfected with si-NC or si-MALAT1. (D) Flow cytometry was conducted to determine cell apoptosis at 48h in HL-60 cells transfected with si-NC or si-MALAT1. Control: non-transfected group. *P < 0.05, **P < 0.01, *** P < 0.001.
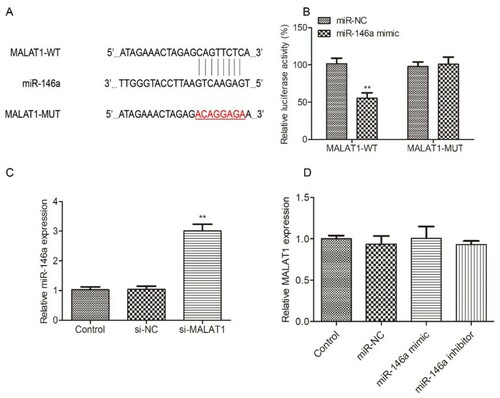
MALAT1 acts as a ceRNA for miR-146a
As we all know, lncRNAs usually act as ceRNA or miRNA sponge, the potential miRNAs bound to MALAT1 were explored by StarBase v2.0 (http://starbase.sysu.edu.cn/) to verify the interaction between MALAT1 and miR-146a. As shown in A, miR-146a formed the complementary bases pairing with MALAT1, indicating that MALAT1 might be a ceRNA for miR-146a. Firstly, we used luciferase assay to explore the interaction between MALAT1 and miR-146a. As shown in B, overexpression of miR-146a reduced the luciferase activity of HL-60 cells transfected with MALAT1-WT, while there was no significant difference in MALTA1-MUT group. Moreover, the expression of miR-146a was greatly increased by the interference of MALAT1 in HL-60 (C). However, miR-146a overexpression/knockdown did not affect MALAT1 mRNA expression (D). These results indicated that miR-146a is a target of MALAT1.
Figure 3. The interaction between MALAT1 and miR-146a in HL-60 cells. (A) The potential binding sites of MALAT1 and miR-146a were predicted by StarBase website. (B) Luciferase activity was measured by luciferase reporter assay 48h after HL-60 cells were transfected with MALAT1-WT or MALAT1-MUT and miR-146a mimic or mi-NC. (C) The expression of miR-146a was detected in HL-60 cells transfected with si-NC or si-MALAT1 for 48h by qRT-PCR. Control: non-transfected group. (D) The expression of MALAT1 was measured in HL-60 cells transfected with mi-NC, miR-146a mimic or miR-146a inhibitor for 48h by qRT-PCR. Control: non-transfected group. *P < 0.05, **P < 0.01.
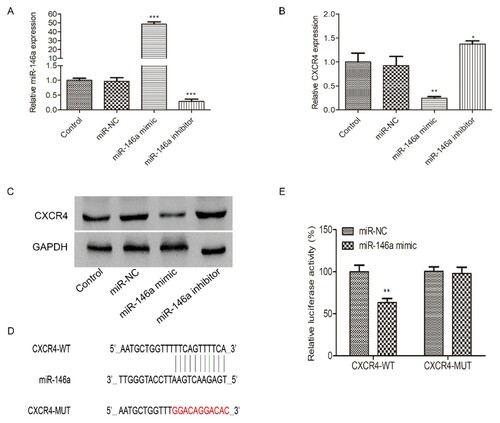
miR-146a regulates HL-60 cells migration, proliferation and apoptosis by targeting CXCR4 expression
As we previously reported that CXCR4 might be a target of miR-146a. Firstly, we constructed miR-146a overexpression and inhibition cell lines by miR-146a mimics or inhibitors. The infection efficiency was confirmed by qRT-PCR (A). We used qRT-PCR and Western Blot assay to verify that overexpression of miR-146a markedly suppressed the CXCR4 mRNA and protein expression, while miR-146a inhibition significantly increased CXCR4 mRNA and protein expression (B and 4C). We also used luciferase assay to validate the interaction between CXCR4 and miR-146a. As reported by Labbaye, miR-146a has three putative binding sites with CXCR4 [Citation24]. We constructed the wild or mutant type CXCR4 3′UTR luciferase reporter vector (4D). Results showed that overexpression of miR-146a caused an obvious reduction of luciferase activity in HL-60 cells transfected with CXCR4-WT, but it failed in CXCR4-MUT group (E).
Figure 4. The interaction between miR-146a and CXCR4 in HL-60 cells. mi-NC, miR-146a mimic or miR-146a inhibitor was transfected into HL-60 cells to achieve miR-146a overexpression or inhibition. The transfection efficiency was measured by qRT-PCR (A). The expression of CXCR4 mRNA (B) and protein (C) were evaluated in HL-60 cells transfected with mi-NC, miR-146a mimic or miR-146a inhibitor at 48h by qRT-PCR and Western Blot assays. Control: non-transfected group. (D) The potential binding sites of CXCR4 and miR-146a. (E) Luciferase activity was measured by luciferase reporter assay 48h after HL-60 cells were transfected with CXCR4-WT or CXCR4-MUT and miR-146a mimic or mi-NC. *P < 0.05, **P < 0.01, *** P < 0.001.
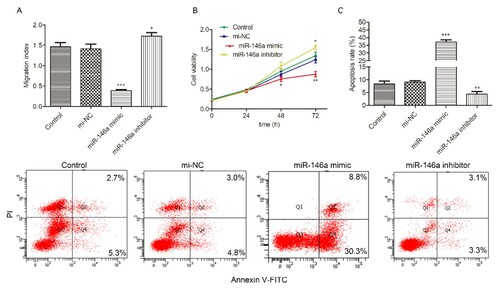
Then in functional assays, we further identified that miR-146a regulated HL-60 cells’ biological behaviors through targeting CXCR4. As shown in A, overexpression of miR-146a resulted in significant inhibition of HL-60 cell migration, while miR-146a inhibition improved HL-60 cell migration. Similarly, we observed that proliferation was depressed by miR-146a overexpression while promoted by miR-146a downregulation (5B). However, upregulation of miR-146a induced apoptosis and downregulation of miR-146a reduced apoptosis (C). All these results revealed that miR-146a regulated HL-60 cells migration, proliferation and apoptosis by targeting CXCR4 expression.
Figure 5. miR-146a regulates migration, proliferation and apoptosis in HL-60 cells. mi-NC, miR-146a mimic or miR-146a inhibitor was transfected into HL-60 cells to achieve miR-146a overexpression or inhibition. (A) Tran-swell assay was implemented to analyze cell migration at 48h in H-60 cells transfected with mi-NC, miR-146a mimic or miR-146a inhibitor. (B) CCK-8 assay was performed to measure cell proliferation at 0, 24, 48 and 72h in H-60 cells transfected with mi-NC, miR-146a mimic or miR-146a inhibitor. (C) Flow cytometry was conducted to determine cell apoptosis at 48h in HL-60 cells transfected with mi-NC, miR-146a mimic or miR-146a inhibitor. Control: non-transfected group. *P < 0.05, **P < 0.01, *** P < 0.001.
MALAT1 regulates CXCR4 expression by sponging to mir-146a in HL-60 cells
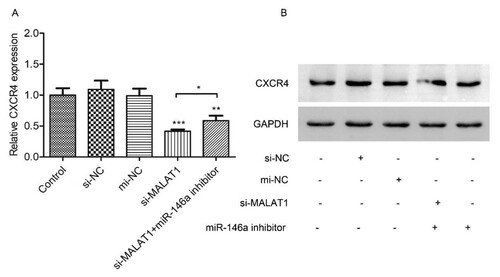
As we have proved that MALAT1 sponges to miR-146a and that miR-146a regulates CXCR4 expression by targeting, we next further elucidated that whether MALAT1 regulated CXCR4 expression through miR-146a. HL-60 cells were transfected with si-MALAT1, si-NC and anti-miR-146a or mi-NC. As results exhibited that knockdown of MALAT1 significantly inhibited CXCR4 mRNA (A), CXCR4 protein (B). Moreover, miR-146a inhibitor partially abolished the effect of MALAT1 ( A and B). Taken together, these results revealed that MALAT1 knockdown suppressed the downstream targets CXCR4 via sponging miR-146a.
Figure 6. MALAT1 regulates CXCR4 expression through miR-146a. HL-60 cells were co-transfected with si-NC/si-MALAT1 and mi-NC/mi-R146a inhibitor for 48h. Then CXCR4 mRNA (A) and protein (B) were measured by qRT-PCR and Western Blot assay, respectively. Control: non-transfected group. *P < 0.05, **P < 0.01, *** P < 0.001.
Discussion
With fast progress in intensive chemotherapy, stem cell transplantation, cell therapy and molecular target therapy in AML, up to 50% of young patients achieve a clinical cure, but 90% of older patients face the risk of failure in the course of treatment [Citation25]. Therefore, a further understanding of the underlying mechanism of AML pathogenesis was very urgent, which pushed us to find effective therapeutic targets for AML. In recent years, a number of reports of the lncRNAs have emerged in the study of AML development and progression, indicating that lncRNAs may be novel therapeutic targets [Citation26–28]. In the present study, we focused on the role of MALAT1 and miR-146a in AML. We proved that MALAT1 was upregulated and miR-146a was downregulated in AML patients compared with normal. Functional and mechanistic studies revealed that MALAT1 regulated AML cell migration, proliferation and apoptosis by sponging miR-146a to mediate CXCR4 expression.
MALAT1 has been reported to be a highly conservative lncRNA and overexpressed in many human cancers, such as lung cancer, hepatocellular carcinoma, breast cancer, colorectal and MM [Citation29–32]. These studies reported that MALAT1 is a cancer-promoting and metastasis-promoting lncRNA through regulating cancer cell proliferation, migration and invasion by different mechanisms [Citation33]. So far, we are the first to demonstrate that MALAT1 is upregulated in AML cells compared with PBCs from healthy donors. Similarly, knockdown of MALAT1 by siRNA suppressed HL-60 cells migration and proliferation, promoted cell apoptosis. These results suggested that MALAT1 may act as a metastasis and progress promoter in AML.
LncRNAs are non-protein-coding RNAs and function through binding to other RNA, genomic DNA, or protein. MALAT1 is a long lncRNA that contains many potential binding sites of miRNAs. Many studies found that MALAT1 served as a ceRNA through sponging miRNAs, including miR-145, miR-202, miR-328 and so on [Citation19,Citation34,Citation35]. Our previous study reported that high miR-146a expression levels in AML patients predicted better clinical outcomes, which suggested that miR-146a may be a therapeutic target. We hypothesized that MALAT1 might serve as a miRNA sponge to interact with miR-146a, which was proved by StarBase. Interestingly, we found a negative correlation between miR-146a and MALAT1 expression by qRT-PCR assay. Then we used luciferase reporter assay to further support the direct binding of miR-146a to MALAT1. Our results were consistent with previous reporters. They showed that MALAT1 contributed to the suppression of proliferation by upregulating miR-146a in LPS-induced inflammatory [Citation21,Citation22,Citation36]. Here we first verified that MALAT1 acted as ceRNA for miR-146a in AML.
What is the molecular mechanism by which MALAT1 regulates AML cell migration, proliferation and apoptosis? We previously reported that CXCR4 promoted AML cell migration, adhesion and drug resistance, which might be regulated by miR-146a. We supposed that MALAT1/miR-146a/CXCR4 axis might regulate migration, proliferation and apoptosis in AML cells. In luciferase reporter assay, we affirmed that CXCR4 was a target gene of miR-146a. In functional assays, we further supported that miR-146a regulated AML cell migration, proliferation and apoptosis by directly targeting CXCR4 expression. Then we used si-MALAT1 to knockdown MALAT1 expression. Finally, we observed a significant upregulation of miR-146a, an obvious downregulation of CXCR4 mRNA and protein. Interestingly, when using anti-miR-146a to block miR-146a expression induced by MALAT1 knockdown, the inhibitory effect of MALAT1 knockdown on CXCR4 expression could be partially restored. These data indicated that MALAT1 could regulate AML cell migration, proliferation and apoptosis by targeting CXCR4 expression through miR-146a; that is to say, there is a MALAT1/miR-146a/CXCR4 axis in AML.
In conclusion, the present study provides evidence that lncRNA MALAT1 modulates cell migration, proliferation and apoptosis by sponging miR-146a to regulate CXCR4 expression in AML, providing novel insights into a critical role of lncRNA and lncRNA-targeted diagnostics and therapeutics in AML.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Bi L, Sun L, Jin Z, et al. MicroRNA-10a/b are regulators of myeloid differentiation and acute myeloid leukemia. Oncol Lett. 2018;15(4):5611–5619.
- Tawfik B, Pardee TS, Isom S, et al. Comorbidity, age, and mortality among adults treated intensively for acute myeloid leukemia (AML). J Geriatr Oncol. 2016;7(1):24–31.
- Almeida AM, Ramos F. Acute myeloid leukemia in the older adults. Leuk Res Rep. 2016;6:1–7.
- Mambet C, Chivu-Economescu M, Matei L, et al. Murine models based on acute myeloid leukemia-initiating stem cells xenografting. World J Stem Cells. 2018;10(6):57–65.
- Nobili L, Lionetti M, Neri A. Long non-coding RNAs in normal and malignant hematopoiesis. Oncotarget. 2016;7(31):50666–50681.
- Wang L, Zhang H, Lei D. microRNA-146a Promotes Growth of acute leukemia cells by Downregulating Ciliary Neurotrophic Factor Receptor and Activating JAK2/STAT3 Signaling. Yonsei Med J. 2019;60(10):924–934.
- Garzon R, Volinia S, Liu CG, et al. MicroRNA signatures associated with cytogenetics and prognosis in acute myeloid leukemia. Blood. 2008;111(6):3183–3189.
- Starczynowski DT, Morin R, McPherson A, et al. Genome-wide identification of human microRNAs located in leukemia-associated genomic alterations. Blood. 2011;117(2):595–607.
- Hasani SS, Hashemi M, Eskandari-Nasab E, et al. A functional polymorphism in the miR-146a gene is associated with the risk of childhood acute lymphoblastic leukemia: a preliminary report. Tumour Biol. 2014;35(1):219–225.
- Moran VA, Perera RJ, Khalil AM. Emerging functional and mechanistic paradigms of mammalian long non-coding RNAs. Nucleic Acids Res. 2012;40(14):6391–6400.
- Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38.
- Barangi S, Hayes AW, Reiter R, et al. The therapeutic role of long non-coding RNAs in human diseases: A focus on the recent insights into autophagy. Pharmacol Res. 2019;142:22–29.
- Zhao C, Wang S, Zhao Y, et al. Long noncoding RNA NEAT1 modulates cell proliferation and apoptosis by regulating miR-23a-3p/SMC1A in acute myeloid leukemia. J Cell Physiol. 2019;234(5):6161–6172.
- Cheng P, Lu P, Guan J, et al. LncRNA KCNQ1OT1 controls cell proliferation, differentiation and apoptosis by sponging miR-326 to regulate c-Myc expression in acute myeloid leukemia. Neoplasma. 2020;67(2):238–248.
- Wang SL, Huang Y, Su R, et al. Silencing long non-coding RNA HOTAIR exerts anti-oncogenic effect on human acute myeloid leukemia via demethylation of HOXA5 by inhibiting Dnmt3b. Cancer Cell Int. 2019;19:114.
- Tripathi V, Ellis JD, Shen Z, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39(6):925–938.
- Ronchetti D, Agnelli L, Taiana E, et al. Distinct lncRNA transcriptional fingerprints characterize progressive stages of multiple myeloma. Oncotarget. 2016;7(12):14814–14830.
- Ahmadi A, Kaviani S, Yaghmaie M, et al. Altered expression of MALAT1 lncRNA in chronic lymphocytic leukemia patients, correlation with cytogenetic findings. Blood Res. 2018;53(4):320–324.
- Wen F, Cao YX, Luo ZY, et al. LncRNA MALAT1 promotes cell proliferation and imatinib resistance by sponging miR-328 in chronic myelogenous leukemia. Biochem Biophys Res Commun. 2018;507(1–4):1–8.
- Pouyanrad S, Rahgozar S, Ghodousi ES. Dysregulation of miR-335-3p, targeted by NEAT1 and MALAT1 long non-coding RNAs, is associated with poor prognosis in childhood acute lymphoblastic leukemia. Gene. 2019;692:35–43.
- Feng LL, Xin WN, Tian XL. MALAT1 modulates miR-146′s protection of microvascular endothelial cells against LPS-induced NF-kappaB activation and inflammatory injury. Innate Immun. 2019;25(7):433–443.
- Li H, Xie S, Li H, Zhang R, Zhang H. LncRNA MALAT1 mediates proliferation of LPS treated-articular chondrocytes by targeting the miR-146a-PI3K/Akt/mTOR axis. Life Sci. 2020;1(254):116801.
- Tan X, Huang Z, Li X. Long Non-coding RNA MALAT1 Interacts With miR-204 to modulate human Hilar Cholangiocarcinoma proliferation, migration, and invasion by targeting CXCR4. J Cell Biochem. 2017;118(11):3643–3653.
- Labbaye C, Spinello I, Quaranta MT, et al. A three-step pathway comprising PLZF/miR-146a/CXCR4 controls megakaryopoiesis. Nat Cell Biol. 2008;10(7):788–801.
- Braess J. [Acute myeloid leukemia]. Dtsch Med Wochenschr. 2016;141(24):1748–1751.
- Schwarzer A, Emmrich S, Schmidt F, et al. The non-coding RNA landscape of human hematopoiesis and leukemia. Nat Commun. 2017;8(1):218.
- <Prediction of competing endogenous RNA coexpression network as prognostic markers in AML.pdf>.
- Garzon R, Volinia S, Papaioannou D, et al. Expression and prognostic impact of lncRNAs in acute myeloid leukemia. Proc Natl Acad Sci U S A. 2014;111(52):18679–18684.
- Yoshimoto R, Mayeda A, Yoshida M, et al. MALAT1 long non-coding RNA in cancer. Biochim Biophys Acta. 2016;1859(1):192–199.
- Guo F, Guo L, Li Y, et al. MALAT1 is an oncogenic long non-coding RNA associated with tumor invasion in non-small cell lung cancer regulated by DNA methylation. Int J Clin Exp Pathol. 2015;8(12):15903–15910.
- Arun G, Spector DL. MALAT1 long non-coding RNA and breast cancer. RNA Biol. 2019;16(6):860–863.
- Si Y, Yang Z, Ge Q, et al. Long non-coding RNA Malat1 activated autophagy, hence promoting cell proliferation and inhibiting apoptosis by sponging miR-101 in colorectal cancer. Cell Mol Biol Lett. 2019;24:50.
- Sun Y, Ma L. New insights into long Non-coding RNA MALAT1 in cancer and metastasis. Cancers (Basel). 2019;11(2):216.
- Lu H, He Y, Lin L, et al. Long non-coding RNA MALAT1 modulates radiosensitivity of HR-HPV+ cervical cancer via sponging miR-145. Tumour Biol. 2016;37(2):1683–1691.
- Zhang Y, Chen Z, Li MJ, et al. Long non-coding RNA metastasis-associated lung adenocarcinoma transcript 1 regulates the expression of Gli2 by miR-202 to strengthen gastric cancer progression. Biomed Pharmacother. 2017;85:264–271.
- Dai L, Zhang G, Cheng Z, et al. Knockdown of LncRNA MALAT1 contributes to the suppression of inflammatory responses by up-regulating miR-146a in LPS-induced acute lung injury. Connect Tissue Res. 2018;59(6):581–592.
