ABSTRACT
Objectives
Hemophilia A (HA, OMIM: 306700) is an X-linked recessive bleeding disorder, caused by defects of the F8 gene which encodes the coagulation factor VIII (FVIII). F8 intron 22 and intron 1 inversion (Inv22 and Inv1) account for ∼45% and 1–5% of severe HA cases, respectively. We herein described an aberrant Inv1 with concomitant large duplication and deletion in a Chinese severe HA patient.
Methods
Long distance PCR and multiplex PCR were used to detect Inv22 and Inv1. Multiplex ligation-dependent probe amplification (MLPA) was applied to examine exonic duplication and deletion of the F8 gene. Coverage analysis of read depth data from whole-genome sequencing (WGS) was used to analyze the intronic duplication and deletion of the F8 gene.
Results
We have identified an aberrant F8 Inv1 in a 1-year-old Chinese severe HA patient showing inversed int1h-1 and normal int1h-2. Coverage analysis of WGS data further illustrated the aberrant Inv1 with concomitant a duplication of 117 kb and a deletion of 1.8 kb.
Conclusion
In conclusion, we reported an aberrant Inv1 with concomitant large duplication and deletion in a severe Chinese HA patient. Moreover, WGS provides rapid genetic diagnosis of hereditary disorders with point mutations, deletions, insertions and CNVs.
1. Introduction
Hemophilia A (HA, OMIM: 306700) is an X-linked recessive bleeding disorder with a prevalence of 1:5000 new-born male. HA is caused by quantitative or functional defects of the coagulation factor VIII (FVIII) encoded by the F8 gene [Citation1]. The severity of HA is classified into three categories according to the plasma FVIII activity (FVIII:C): severe (FVIII:C <1%), moderate (FVIII:C 1%–5%), and mild (FVIII:C 5-40%) [Citation2]. F8 intron 22 and intron 1 inversion (Inv22 and Inv1) account for ∼45% and 1–5% of severe HA cases, respectively. The frequency of Inv1 was 2.94% in a Chinese population with severe HA [Citation3].
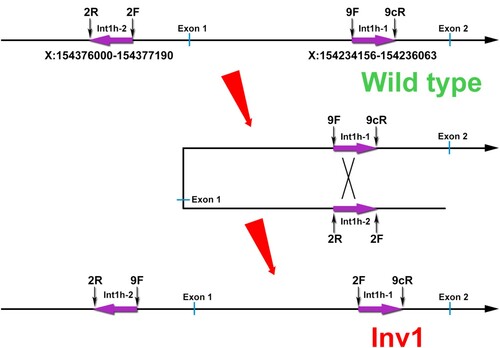
Inv1 is derived from an intra-chromosomal homologous recombination crossing over two almost identical 1040-bp sequences between int1h-1 within intron 1 and int1h-2 located in approximately 125-kb telomeric distal from the F8 gene, in opposite orientation to inth-1. Inv1 disrupts F8 gene coding sequence and results in severe HA [Citation4]. Inv1 could be detected using two polymerase chain reactions (PCRs) reactions capable of identifying both HA patients and carriers.
2. Methods
DNA sample was extracted from the peripheral blood mononuclear cells (PBMCs) and used for Inv22, Inv1, multiplex ligation-dependent probe amplification (MLPA), and WGS. The int1h-1 is amplified using primers specific for int1h-1 (9F, 9cR) and the primer 2F. Healthy controls show a 1.5-kb fragment and Inv1 patients harbor a 1.0-kb product, while HA carriers have both fragments. The int1h-2 is amplified using primers specific for int1h-2 (2F and 2R) and the primer 9F, yielding a 1.0-kb and a 1.5-kb products from healthy controls and Inv1 patients respectively (), while HA carriers have both fragments [Citation4]. Coverage analysis of read depth data from whole-genome sequencing (WGS) was used to analyze the intronic duplication and deletion of the F8 gene. This study was approved by the ethnic committee of Tongji Hospital, and informed consent was obtained from the parents.
3. Results and discussion
A 1-year-old boy with severe HA (FVIII:C <0.2%) was admitted, and his parents came for genetic test and counseling for further child. As Inv22 and Inv1 account for near 50% of all severe HA cases, Inv22 and Inv1 were performed for the boy and his mother. Inv22 showed same bands with healthy control (data not shown), however, aberrant patterns of Inv1 were found in this boy and his mother. The boy had a 1.0-kb band in int1h-1, while his mother had both 1.0-kb and 1.5-kb fragments. For int1h-2, both the boy and his mother carried a 1.0-kb product. They showed aberrant patterns of Inv1 compared with healthy control, Inv1 patient, or Inv1 carrier (A).
Figure 2. The aberrant patterns of Inv1 in a 1-year-old boy with severe HA. A, The int1h-1 is amplified using int1h-1 specific primers (9F, 9cR) and the int1h-2 specific primer 2F, while the int1h-2 is amplified using int1h-2 specific primers (2F and 2R) and the primer 9F. The boy showed similar int1h-1 band with Inv1 patient, while had similar int1h-2 band with healthy control. His mother had similar int1h-1 bands with Inv1 carrier, while had similar int1h-2 band with healthy control. B, The int1h-1 was amplified in three PCR tubes including different primers, 9F+9cR+2F, 9F+9cR (for wild type sequence), and 9cR+2F (for Inv1 sequence). The int1h-2 was amplified in three PCR tubes including different primers, 2F+2R+9F, 2F+2R (for wild type sequence), and 2R+9F (for Inv1 sequence). The boy showed same pattern of int1h-1 with Inv1 patient, but had same pattern of int1h-2 with healthy control. This result confirmed the presence of inversed int1h-1. C, MLPA was performed to detect duplication and deletion within the exonic regions of the F8 gene. The ratio between 0.75 and 1.25 was considered as normal.
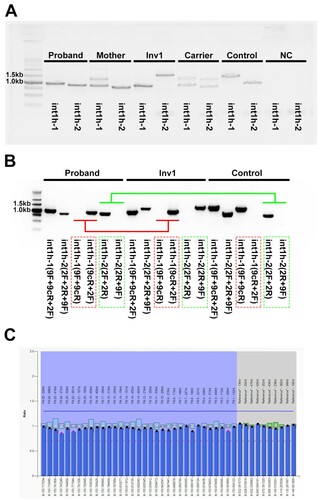
The int1h-1 is amplified using int1h-1 specific primers (9F, 9cR) and the int1h-2 specific primer 2F, while the int1h-2 is amplified using int1h-2 specific primers (2F and 2R) and the primer 9F. To clearly identify the product of int1h-1 and int1h-2, we amplified int1h-1 and int1h-2 using diverse primer pairs. The int1h-1 was amplified in three PCR tubes including different primers, 9F+9cR+2F (mixed), 9F+9cR (for wild type sequence), and 9cR+2F (for Inv1 sequence). The int1h-2 was amplified in three PCR tubes including different primers, 2F+2R+9F (mixed), 2F+2R (for wild type sequence), and 2R+9F (for Inv1 sequence). The boy showed same pattern of int1h-1 (amplified using 9cR+2F primer pair for Inv1 sequence) with Inv1 patient, but had same pattern of int1h-2 (amplified using 2F+2R primer pair for wild type sequence) with healthy control (B). The presence of inversed int1h-1 in the boy suggests the occurrence of Inv1. However, the wild-type in1h-2 deserved further investigation.
To date, aberrant patterns of Inv1 have been reported involving complex duplication/deletion rearrangements near the F8 gene, leading to the presence of zero or two copies of int1h-1/int1h-2 [Citation5,Citation6]. To detect the complex duplication/deletion rearrangements of the F8 gene, MLPA was applied, and no duplication/deletion was found in the exonic regions of the F8 gene in the boy (C). However, the intronic regions, 5'- untranslated region (5'-UTR) and 3'-UTR of the F8 gene, were not covered by MLPA due to the lack of probes for these regions.
Previously, we have applied coverage analysis of next-generation sequencing (NGS) data involving the F8 gene to detect large exon deletion in patients of HA, and the results were consistent with MLPA [Citation7]. Moreover, coverage analysis of read depth from whole-exome sequencing (WES) data has been used in the detection of copy-number variations (CNVs) [Citation8]. Compared to WES, whole-genome sequencing (WGS) could detect both exonic regions and intronic sequences. WGS has been also used for CNV analysis [Citation9,Citation10]. Therefore, WGS was performed for the boy on NovaSeq6000 platform by Aegicare Biotech (Shenzhen, China) with a mean coverage of 30X. A large deletion crossing X:154235303-154237171 (hg19) and a large duplication including X:154259274-154376426 (hg19) were identified by the coverage analysis of read depth (A and B). The genomic region of int1h-1 amplified using 9F+9cR was located in X:154234156-154236063 and int1h-2 amplified using 2F+2R was located in X:154376000-154377190 respectively (C).
Figure 3. Coverage analysis of the WGS data. A, A large deletion crossing X:154235303-154237171 (hg19) was found in the boy. B, A large duplication including X:154259274-154376426 (hg19) was identified by the coverage analysis. C, The genomic regions of int1h-1 and int1h-2 amplified using 9F+9cR and 2F+2R respectively. The red dotted line labeled int1h-1 or int1h-2, and the bottom region was the almost identical region.
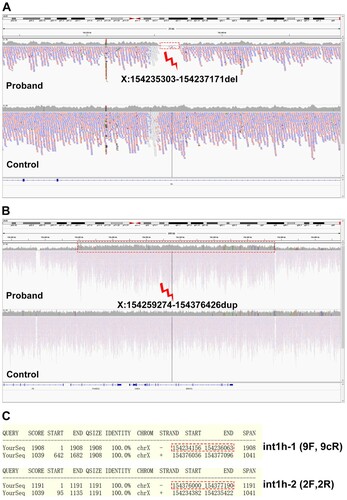
The presence of inversed int1h-1 (amplified using 9cR+2F primer pair) in the boy (B) confirmed the occurrence of Inv1. The large deletion region (X:154235303-154237171, hg19) included the homologous region of int1h-2 (X:154234382-154235422, hg19) within int1h-1, which might result in the absence of inversed int1h-2 (amplified using 2R+9F primer pair). Moreover, the large duplication region (X:154259274-154376426, hg19) crossed the int1h-2 (X:154376000-154377190, hg19) partially. This duplication might lead to two copies of int1h-2, and one copy was inversed with inth1-1, while the other copy could be still detected using 2F+2R primer pair showing similar bands with healthy control. Taken together, the genomic rearrangements were caused by an aberrant F8 Inv1 with concomitant a duplication of 117-kb and a deletion of 1.8-kb at Xq28 ().
Figure 4. Hypothetical arrangement of the X chromosome in the patient. The deletion occurred in X:154235303-154237171 involving int1h-1. The duplication occurred in X:154259274-154376426 involving int1h-2.

With the exception of classic F8 Inv1, aberrant Inv1 with concomitant duplication/deletion have been found in severe HA patients. Sanna et al. reported aberrant F8 Inv1 with concomitant a 19.32-kb duplication involving a part of the int1h-1 and extending through intron 6, as well as a 41.87-kb extragenic deletion within the telomeric position in a severe HA patient from Southern Italy [Citation5]. You et al. reported an aberrant pattern of Inv1 with a duplication of 227.3 kb and a deletion of 2.56 kb within the F8 gene in a Chinese pedigree with severe HA [Citation6].
In conclusion, we reported an aberrant Inv1 with concomitant large duplication and deletion in a severe Chinese HA patient. Moreover, WGS provides rapid genetic diagnosis of hereditary disorders with point mutations, deletions, insertions, and CNVs.
Acknowledgement
This work was partially supported by the Natural Science Foundation of Hubei Province (No. 2015CFB725 for Huijun Li), Shiyan Pilot Project (18Y77) and Hubei Provincial Youth Talent Project (Q20162113) for Hongmei Wang.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Correction Statement
This article has been republished with minor changes. These changes do not impact the academic content of the article.
Additional information
Funding
References
- Berntorp E, Shapiro AD. Modern haemophilia care. Lancet. 2012;379:1447–1456.
- White GC, Rosendaal F, Aledort LM, et al. Definitions in hemophilia. Recommendation of the scientific subcommittee on factor VIII and factor IX of the scientific and standardization committee of the International Society on Thrombosis and Haemostasis. Thromb Haemost. 2001;85:560.
- Lu YL, Wang XF, Ding QL, et al. Prevalence of the factor 8 gene intron 1 inversion in Chinese haemophiliacs and its application to carrier detection and prenatal diagnosis in haemophilia A families. Haemophilia Official J World Federation Hemophil. 2011;17:541–542.
- Bagnall RD, Waseem N, Green PM, et al. Recurrent inversion breaking intron 1 of the factor VIII gene is a frequent cause of severe hemophilia A. Blood. 2002;99:168–174.
- Sanna V, Ceglia C, Tarsitano M, et al. Aberrant F8 gene intron 1 inversion with concomitant duplication and deletion in a severe hemophilia A patient from Southern Italy. J Thromb Haemos. 2013;11:195–197.
- You G, Chi K, Lu Y, et al. Identification and characterisation of a novel aberrant pattern of intron 1 inversion with concomitant large insertion and deletion within the F8 gene. Thromb Haemost. 2014;112:264–270.
- He X, Xiong Z, Shen N, et al. Performance of next-generation sequencing in the detection of large exon deletion in patients of haemophilia A. Haemophilia Official J World Federation Hemophilia. 2018;24:e296–e300.
- Quenez O, Cassinari K, Coutant S, et al. Detection of copy-number variations from NGS data using read depth information: a diagnostic performance evaluation. Eur J Hum Genetics. 2020. DOI:https://doi.org/10.1038/s41431-020-0672-2.
- Xu J, Wu J, Teng X, et al. Large duplication in LMBR1 gene in a large Chinese pedigree with triphalangeal thumb polysyndactyly syndrome. Am J Med Genet A. 2020;182(9):2117–2123.
- Bobyn A, Zarrei M, Zhu Y, et al. Ancestry and frequency of genetic variants in the general population are confounders in the characterization of germline variants linked to cancer. BMC Med Genet. 2020;21:92.