ABSTRACT
Objectives
Previous study (Br. J. Haematol. 2017; 176:498) reported that CD56 positive is associated with poor prognosis of patients with intermediate-risk acute myeloid leukemia (IR-AML). However, our data were inconsistent with the finding. Thus, in this study, we provided the different results to discuss.
Methods
A total of 262 bone marrow transcriptomic data of IR-AML in the GSE12417-GPL96 and GSE71014-GPL-10558 from the Gene Expression Omnibus database (GEO) database, and 92 IR-AML patients from the cancer genome atlas (TCGA) database were obtained for prognostic analysis and validation.
Results
Compared with low CD56 expression, IR-AML patients with high CD56 expression had a longer overall survival (OS) time and restricted mean survival time (RMST) and favorable OS rate in the GSE12417-GPL96 dataset. These results were confirmed in both GSE71014-GPL-10558 and TCGA datasets. Importantly, the inconsistency between our findings and the previous finding may be due to the following reasons: different detection methods, age stratification, countries, treatment options etc.
Conclusions
The prognostic value of CD56 expression in IR-AML may need to be comprehensively evaluated based on different detection methods, age stratification, countries, treatment options, and other factors. If confirmed, CD56 may be a biomarker for further risk stratification for IR-AML patients.
Introduction
Acute myeloid leukemia (AML) is a genetically heterogeneous malignancy. Cytogenetics and gene mutations provide the most important prognostic information for risk stratification at the time of diagnosis [Citation1–4], among them, 50% to 65% of patients are classified as intermediate-risk AML (IR-AML) [Citation5,Citation6]. Currently, cytogenetically normal chromosomes, NPM1 and CEBPA mutations, and FLT3-ITD are biomarkers for IR-AML [Citation7–9]. However, IR-AML is quite heterogeneous, and the 4-year overall survival (OS) rate is no greater than 45% [Citation10,Citation11]. Therefore, it is worth exploring novel biomarkers to further refine risk stratification for IR-AML. CD56, also known as neural cell adhesion molecule 1 (NCAM1), is mainly expressed in natural killer (NK) cells, but it is also expressed in T and B cells. As a co-activator, CD56 can stimulate NK cells to secrete a large number of cytokines and chemokines to kill target or tumor cells [Citation12]. Nevertheless, high expression of CD56 antigen is often associated with poor prognosis for AML patients, particularly those with t(8; 21) or t(15; 17) [Citation13,Citation14] alterations. Furthermore, Coelho-Silva et al. [Citation15] examined 64 bone marrow (BM) samples from IR-AML patients in a single clinical center and found that patients with high expression of CD56 antigen had poor OS and disease-free survival (DFS). This finding suggests that CD56 detection may be a cheap and effective alternative for the management of IR-AML patients. Interesting, in this study, when we used 357 BM samples from patients with IR-AML in three different datasets from the Gene Expression Omnibus database (GEO) and the Cancer Genome Atlas (TCGA) databases for survival analysis and validation, we had the inconsistent results [Citation15].
Materials and methods
Patient information
Microarray data including 161 bone marrow samples in the GSE12417-GPL96 [Citation16] dataset and 104 bone marrow samples in the GSE71014-GPL-10558 [Citation17] were downloaded from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/). Patients with cytogenetically normal acute myeloid leukemia (CN-AML) in these two datasets were classified as an intermediate-risk cohort. After excluding patients with incomplete prognostic information and an overall survival time of 0, the bone marrow transcriptomic data of 92 intermediate-risk AML patients in the cancer genome atlas (TCGA) database (https://cancergenome.nih.gov/) [Citation18,Citation19] were obtained using the UCSC-XENA platform (https://xenabrowser.net/datapages/) [Citation6,Citation20]. The GEO and TCGA databases are publicly available; thus, approval from the local ethics committee was not required.
Statistical analysis
All statistical analysis was conducted by R (version 4.0.2, https://www.r-project.org/). The optimal cut-off was obtained by the ‘maxstat’ package [Citation21,Citation22]. Differences in Kaplan-Meier curves were compared by the log-rank test. The ‘survRM2’ package was used to confirm the restricted mean survival time (RMST). A two-tailed P-value < 0.05 and a P-value < 0.1 were considered statistically significant and a clear trend, respectively.
Results and discussion
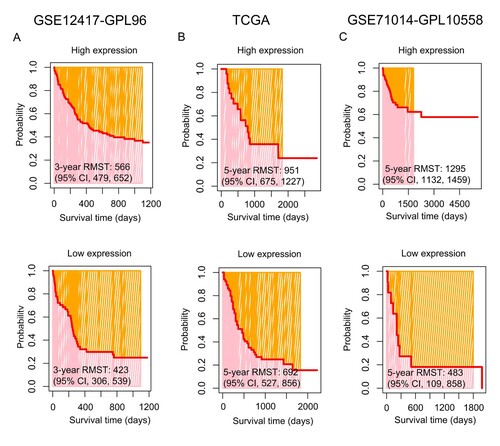
According to an optimal cut-off value, IR-AML patients were divided into two groups of CD56 expression: high and low (, left panel). In comparison with low CD56 expression, IR-AML patients with high CD56 expression had a longer OS time in the GSE12417-GPL96 dataset (high vs. low expression, median time: 480 vs. 210 days) ((A), middle panel), similar result could be found in the TCGA dataset (high vs. low expression, median time: 579 vs. 335 days) ((B), middle panel). Moreover, this result was again confirmed in the GSE71014-GPL-10558 dataset (high vs. low expression, median time: 321 vs. 239 days) ((C), middle panel). Notably, there was a clear trend where IR-AML patients with high but not low CD56 expression were associated with favorable OS in the GSE12417-GPL96 dataset (3-year OS rate, 37% vs. 25%, P = 0.066) ((A), right panel). This finding was confirmed with the TCGA dataset (5-year OS rate: 24% vs. 15%, P = 0.107) ((B), right panel). Interestingly, with an extension of the follow-up time i.e. up to 5556 days, IR-AML patients with high CD56 expression had more significantly favorable OS compared to those with low CD56 expression in the GSE71014-GPL-10558 dataset (5-year OS rate: 62% vs. 18%, P < 0.001) ((C), right panel). Furthermore, the RMST was used to evaluate the performance of the Kaplan-Meier curves. As shown in (A), the 3-year restricted mean survival time (RMST) for high and low CD56 expression was 566 [95% confidence interval (CI), 479, 652] and 423 (95% CI, 306, 539) days, respectively, in the GSE12417-GPL96 dataset. This result was also confirmed by the TCGA dataset [5-year RMST, high vs. low, 951 (95% CI: 675, 1,227) vs. 692 (95% CI: 527, 856) days] ((B)). Importantly, the findings above were again observed using the GSE71014-GPL-10558 dataset [5-year RMST, high vs. low, 1,295 (95% CI: 1132, 1459) vs. 483 (95% CI: 109, 858) days] ((C)). Overall, the results from all three datasets suggested that the Kaplan-Meier curves drawn according to the expression CD56 level had good distinction i.e. IR-AML patients with higher CD56 expression were associated with favorable OS.
Figure 1. Overall survival (OS) analysis of CD56 in intermediate-risk acute myeloid leukemia (IR-AML) in the GSE12417-GPL96 (A), the Cancer Genome Atlas (TCGA) (B) and GSE71014-GPL-10558 (C) datasets. The optimal cut-off values for CD56, and survival time distribution (middle panel) and Kaplan-Meier curves (right panel) based on the expression levels of CD56 are shown. CI, confidence interval.
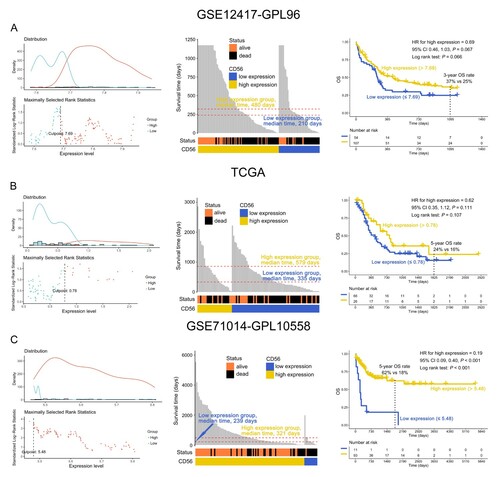
Figure 2. The restricted mean survival time (RMST) of IR-AML patients according to the expression levels of CD56 in the GSE12417-GPL96 (A), TCGA (B), and GSE71014-GPL-10558 (C) datasets.
The inconsistency between our findings and the results of Coelho-Silva et al. may be due to the following reasons (). First, the Coelho-Silva's study was based on only a single clinical center with 64 IR-AML patient samples to explore the prognostic importance of CD56; thus, the results may be limited to local findings, while the present study included a total of 357 IR-AML patients in three different datasets for OS analysis and validation, which may provide a more objectively result. Second, in the Coelho-Silva's study, the follow-up for IR-AML was short with a median follow-up of 208 days, and the longest follow-up was less than 2000 days. However, in this study, the follow-up for the three different datasets are 280, 395, and 440 days with the longest follow-up being 5556 days, suggesting that long-term follow-up may be better to confirm the biomarker value in AML. Third, in the Coelho-Silva's study, the ages of the IR-AML patients were young with a median age of 49 years, while in the present study, the age of the IR-AML patients is relatively older with median ages of 58 or 57 years in the GSE12417-GPL96 and TCGA datasets, respectively. It is thought that age may have influenced the OS results, which was based on the CD56 expression level. Fourth, the two studies have different detection methods for CD56 expression. Among these methods, Coelho-Silva et al. used flow cytometry to detect the expression of CD56 antigen, while the analysis of the current study was based on the data from microarray, which detected the global CD56 gene expression level in AML samples rather than the CD56 protein. Fifth, the treatments used for the IR-AML patients in the two studies may be different [Citation5,Citation16,Citation17,Citation23]. Finally, the IR-AML patients in the two studies were from different countries, which may be a reason why the results are inconsistent. Therefore, combining microarray, quantitative real-time polymerase chain reaction (qRT-PCR), and flow cytometry analysis of CD56 expression in BM samples from multiple clinical centers may be better for further confirming the role of CD56 expression in AML.
Table 1. Clinical information of patients with intermediate-risk acute myeloid leukemia.
Conclusions
The prognostic importance of CD56 expression in IR-AML may need to be comprehensively evaluated according to different detection methods, age stratification, countries, treatment options, and other factors. If confirmed, the expression of CD56 may be a biomarker for further risk stratification for IR-AML patients.
Authorship contributions
CTC performed the statistical analyses, interpreted the data, and drafted the manuscript. CLC helped to analyze the data. HZ reviewed and edited the manuscript. YQL contributed to the concept development and study design and reviewed the manuscript. All authors read and approved the final manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The datasets used and analyzed during this study are available from UCSC XENA platform (https://xenabrowser.net/datapages/) and GEO database (https://www.ncbi.nlm.nih.gov/geo/). The data that support the findings of this study are available from the corresponding author upon reasonable request.
Additional information
Funding
References
- Lin Y, Wang Y, Zheng Y, et al. Clinical characteristics and prognostic study of adult acute myeloid leukemia patients with ASXL1 mutations. Hematology. 2020;25(1):446–456.
- Wang K, Zhou F, Cai X, et al. Mutational landscape of patients with acute myeloid leukemia or myelodysplastic syndromes in the context of RUNX1 mutation. Hematology. 2020;25(1):211–218.
- Yu J, Li Y, Zhang D, et al. Clinical implications of recurrent gene mutations in acute myeloid leukemia. Exp Hematol Oncol. 2020;9:4.
- Bera R, Chiu M, Huang Y, et al. RUNX1 mutations promote leukemogenesis of myeloid malignancies in ASXL1-mutated leukemia. J Hematol Oncol. 2019;12(1):104.
- Lima A, de Mello M, Fernandes E, et al. Clinical outcomes of patients with acute myeloid leukemia: evaluation of genetic and molecular findings in a real-life setting. Blood. 2015;126(15):1863–1865.
- Chen C, Liang C, Wang S, et al. Expression patterns of immune checkpoints in acute myeloid leukemia. J Hematol Oncol. 2020;13(1):28.
- Becker H, Marcucci G, Maharry K, et al. Favorable prognostic impact of NPM1 mutations in older patients with cytogenetically normal de novo acute myeloid leukemia and associated gene- and microRNA-expression signatures: a cancer and leukemia Group B study. J Clin Oncol. 2010;28(4):596–604.
- Pastore F, Kling D, Hoster E, et al. Long-term follow-up of cytogenetically normal CEBPA-mutated AML. J Hematol Oncol. 2014;7:55.
- Solovey M, Wang Y, Michel C, et al. Nuclear factor of activated T-cells, NFATC1, governs FLT3(ITD)-driven hematopoietic stem cell transformation and a poor prognosis in AML. J Hematol Oncol. 2019;12(1):72.
- Schlenk R, Döhner K, Krauter J, et al. Mutations and treatment outcome in cytogenetically normal acute myeloid leukemia. N Engl J Med. 2008;358(18):1909–1918.
- Therneau T. (2020). A Package for Survival Analysis in R. R package version 32-7, https://CRANR-projectorg/package = survival.
- Kim N, Lee H, Lee H, et al. Natural killer cells as a promising therapeutic target for cancer immunotherapy. Arch Pharm Res. 2019;42(7):591–606.
- Iriyama N, Hatta Y, Takeuchi J, et al. CD56 expression is an independent prognostic factor for relapse in acute myeloid leukemia with t(8;21). Leuk Res. 2013;37(9):1021–1026.
- Montesinos P, Rayón C, Vellenga E, et al. Clinical significance of CD56 expression in patients with acute promyelocytic leukemia treated with all-trans retinoic acid and anthracycline-based regimens. Blood. 2011;117(6):1799–1805.
- Coelho-Silva J, Carvalho L, Oliveira M, et al. Prognostic importance of CD56 expression in intermediate risk acute myeloid leukaemia. Br J Haematol. 2017;176(3):498–501.
- Metzeler K, Hummel M, Bloomfield C, et al. An 86-probe-set gene-expression signature predicts survival in cytogenetically normal acute myeloid leukemia. Blood. 2008;112(10):4193–4201.
- Chuang M, Chiu Y, Chou W, et al. An mRNA expression signature for prognostication in de novo acute myeloid leukemia patients with normal karyotype. Oncotarget. 2015;6(36):39098–39110.
- Qin T, Wu S, Zhao H, et al. Molecular predictors of post-transplant survival in acute myeloid leukemia. Blood Cancer J. 2017;7(12):641.
- Chen C, Wang P, Mo W, et al. Expression profile analysis of prognostic long non-coding RNA in adult acute myeloid leukemia by weighted gene co-expression network analysis (WGCNA). J Cancer. 2019;10(19):4707–4718.
- Wang P, Fu Y, Chen Y, et al. Nomogram Personalizes and Visualizes the overall survival of patients with Triple-Negative Breast cancer based on the Immune genome. Biomed Res Int. 2020;2020:1–16.
- Seckinger A, Meissner T, Moreaux J, et al. Clinical and prognostic role of annexin A2 in multiple myeloma. Blood. 2012;120(5):1087–1094.
- Delgado J, Pereira A, Villamor N, et al. Survival analysis in hematologic malignancies: recommendations for clinicians. Haematologica. 2014;99(9):1410–1420.
- Raspadori D, Damiani D, Lenoci M, et al. CD56 antigenic expression in acute myeloid leukemia identifies patients with poor clinical prognosis. Leukemia. 2001;15(8):1161–1164.
