ABSTRACT
Background
Myelodysplastic syndrome (MDS) is a haematopoietic malignancy that is characterized by a heterogeneous clinical course and dysplastic maturation of blood lineages. Immune dysregulation has gained attention as one of the fundamental mechanisms responsible for the development of MDS. This study aimed to screen immune-related biomarkers and pathways in MDS.
Methods
Differentially expressed mRNAs (DE-mRNAs) and differentially expressed microRNAs (DE-miRNAs) in different subtypes of MDS were sourced from the Gene Expression Omnibus (GEO) database. DE-mRNAs were intersected with immune-related gene sets to collect immune-related mRNAs, which were put into the Search Tool for the Retrieval of Interacting Genes (STRING) to construct protein–protein interaction (PPI) networks. Target mRNAs of DE-miRNAs were predicted using the miRDB database and intersected with screened immune-related mRNAs to construct miRNA-mRNA interaction networks. Topological analysis of constructed networks was applied to screen key molecules, which were assessed in independent datasets and previous literature. Enrichment analysis was applied to screen dysregulated pathways in MDS.
Results
Screened key mRNAs were mainly from the Toll-like receptor (TLR) family, including TLR2, TLR4, TLR7, and from the chemokine family, including C-X-C motif chemokine ligand 10 (CXCL10) and CC chemokine ligand 4 (CCL4). Cytokine-cytokine receptor interactions were among the major pathways in the enrichment analysis results. Hsa-miR-30b, hsa-miR-30e and hsa-miR-221 were validated as key miRNAs and modulate cytokine-cytokine receptor interactions by targeting immune-related mRNAs.
Conclusion
Dysregulated cytokines reflect the immunization status in MDS. Immune-related miRNA-mRNA interactions not only provide a perspective to our understanding of immunologic derangement in the pathogenesis of MDS but also provide new therapeutic opportunities.
Introduction
Myelodysplastic syndrome (MDS) is defined as a heterogeneous group of clonal bone marrow disorders that comprises the characteristics of bone marrow dysplasia with abnormal cell morphology, cytopenia and a tendency to transform to acute myeloid leukaemia [Citation1]. The French-American-British (FAB) Cooperative Study Group and World Health Organisation (WHO) defined and then refined the classification and subdivided MDS into different subtypes based on the percentage of immature blasts in the bone marrow, the presence of ringed sideroblasts and the degree of monocytosis, which are now regarded as the standard in diagnosing MDS and are widely used in clinical practice. Although classification based on morphology has made diagnosis and treatment more structural, the pathogenesis of MDS is complex and has not been fully elucidated. In addition to involving haematopoietic stem cells, the bone marrow microenvironment and the interaction between these two factors, emerging clinical and laboratory studies have underlined the role of immunity in pathological processes and clinical scenarios of MDS patients [Citation2–4]. A series of autoimmune manifestations have been reported in a large fraction of MDS patients, and patients suffering from autoimmune diseases are at a higher risk of developing MDS [Citation5–10]. Genetic studies of bone marrow cells also reveal that immune-related genes such as toll-interleukin 1 receptor domain containing adaptor protein (TIRAP), toll like receptor 4 (TLR4) and tumour necrosis factor receptor associated factor 6 (TRAF6) are overexpressed in MDS, activate the nuclear transcription factor-kappa B (NF-κB) signalling pathway and are implicated in numerous aspects of pathogenesis [Citation11].
Pathologies in MDS are inseparable from RNA interactions [Citation12]. An integrated analysis of protein-encoding RNAs and noncoding RNAs is favourable to provide a different perspective to understand the possible underlying mechanism. MicroRNAs (miRNAs) are a class of short noncoding RNA molecules that have been reported to contribute essential logic elements to immune responses [Citation13]. In the complex landscape of RNA, miRNA does not function alone but negatively regulates the expression of target genes by binding to complementary sites in the 3′ UTR of target messenger RNA (mRNA) at the posttranscriptional level. The dysregulated expression of miRNAs can lead to aberrant immune function and plays an indispensable role in the onset of MDS [Citation14–16]. For example, TRAF6 and TIRAP, genes that are in the MyD88-dependent pathway of innate immune signalling, are regulated by hsa-miR-146a and hsa-miR-145, respectively. These two miRNAs lose functionality in the deletion of chromosome 5q (del(5q)) subtype, which results in overaction of target genes and innate immune signalling, leading to a propensity to develop marrow failure or leukaemia [Citation14].
Bioinformatic methods are powerful tools to understand the pathological mechanisms of disease with the development of RNA sequencing techniques and computer biotechnology. Microarray technology has been widely used to compare genes that are differentially expressed in patients with multiple diseases, and it can provide updated knowledge about the pathogenesis of disease and alter both the research and clinical approach. In this study, microarrays of miRNAs and mRNAs based on a sample of CD34+ bone marrow were derived from the Gene Expression Omnibus (GEO) database. GEO2R analysis of the microarray was employed to obtain the differentially expressed mRNAs (DE-mRNAs) and differentially expressed miRNAs (DE-miRNAs). DE-mRNAs were intersected with gene sets obtained from the ImmPort immunity database to obtain immune-related mRNAs in different subtypes of MDS. Protein–protein interaction (PPI) networks of immune-related mRNAs were constructed to elucidate the interaction of screened molecules. Target immune-related mRNAs of DE-miRNAs were screened, and miRNA-mRNA interaction networks were constructed. Functional enrichment analysis of immune-related DE-mRNAs and DE-miRNAs was applied to explore significantly enriched pathways and biological processes. Topological analysis of constructed networks was applied to screen critical molecules and determine their values as hub biomarkers. The expression value of hub biomarkers was assessed in validation data sets and previous literature. We hope to offer a complementary perspective on understanding the underlying pathogenic mechanisms in MDS and provide innovative ideas for the management of MDS.
Methods and materials
Source of microarray data
The GEO database was screened to collect miRNA and mRNA expression profiles of MDS. The inclusion and exclusion criteria were as follows: (1) Patients with MDS complicated with other diseases were excluded; and (2) Samples were sourced from CD34+ bone marrow cells. Data analysed in this study were obtained from an open database, and ethics and patient consent were not required.
Data processing
DE-miRNAs and DE-mRNAs in different subtypes of MDS were analysed by comparing CD34+ bone marrow cells of MDS patients and normal control samples using the GEO2R tool. |Log2fold change (FC)| ≥ 1.5 and P value < 0.05 were set as the cut-off criteria.
Screen of immune-related mRNAs
ImmPort (https://www.immport.org/home) is an open repository of pooled immunology data from mechanistic studies on human subjects, immunology studies on model organisms and clinical settings [Citation17]. In this study, immune-related gene sets were collected from ImmPort and then intersected with DE-mRNAs for further analysis. The overlapping part between immune-related gene sets and DE-mRNAs was defined as immune-related mRNAs.
Construction of PPI networks and miRNA-mRNA interaction networks
Immune-related mRNAs in different subtypes of MDS were uploaded to the Search Tool for the Retrieval of Interacting Genes (STRING, https://string-db.org/) [Citation18] to construct PPI networks with confidence 0.4.
The miRDB database (http://mirdb.org/) [Citation19], an online database for miRNA target prediction and functional annotations, was screened to predict target mRNAs of DE-miRNAs. Immune-related mRNAs at the intersection of target mRNAs forecasted from miRDB were identified as the target immune-related mRNAs of DE-miRNAs, and immune-related miRNA-mRNA pairs were visualized in the form of miRNA-mRNA interaction networks using Cytoscape (version 3.6.1) [Citation20].
Hubs are crucial components for efficient communication in a network, and a high degree centrality (DC) is at the centre of the network and is regarded as an important research focus. CytoNCA [Citation21], a Cytoscape plugin for topological network analysis, was utilized to identify hub nodes in constructed networks based on DC. Molecules at DC ≥ median DC were defined as hub mRNAs and miRNAs in networks.
Functional enrichment analysis
To further comprehend the function of screened mRNAs and miRNAs, immune-related mRNAs and target immune-related mRNAs of screened DE-miRNAs were imported to KOBAS [Citation22] (http://kobas.cbi.pku.edu.cn/), a database to identify statistically significantly enriched pathways using a hypergeometric test, to perform Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis.
Hub molecules validation
Validation cohorts were screened from the GEO database to preliminarily validate screened hub mRNAs and miRNAs obtained from topological analysis of PPI networks and miRNA-mRNA interaction networks. Box plot analysis was used to depict the expression levels of screened molecules in MDS patients and healthy individuals, which are illustrated by the median and interquartile range. Additionally, hub mRNAs were evaluated by searching the existing literature to further assess alterations in expression.
Result
Screening of DE-mRNAs and DE-miRNAs in different subtypes of MDS
The miRNA expression profiles were obtained from GSE81372, which contains CD34+ bone marrow cells from 12 MDS patients and 6 healthy donors. The mRNA expression profiles were available from GSE18366, GSE4619, GSE19429 and GSE81173. Details of the screened datasets are summarized in . After data processing, 2374 DE-mRNAs and 145 DE-miRNAs in humans were screened to be differentially expressed between CD34+ bone marrow cells from MDS patients and normal controls. Among these, a total of 1579 upregulated and 795 downregulated DE-mRNAs were screened in patients with MDS subtypes refractory anaemia with excess blasts-1 (RAEB1), refractory anaemia with excess blasts-2 (RAEB2), refractory anaemia with ring sideroblasts (RARS), refractory anaemia (RA), refractory cytopenia with multilineage dysplasia (RCMD) and del(5q). These subtypes were compared with healthy control subjects, and the expression of 91 and 54 DE-miRNAs was upregulated and downregulated, respectively, in RAEB1, RAEB2, RCMD and del(5q) subtypes. The details of DE-mRNA and DE-miRNA in different MDS subtypes are shown in .
Figure 1. Details of DE-mRNAs and DE-miRNAs in different subtypes of MDS. DE-mRNAs and DE-miRNAs were screened with |log2fold change| ≥ 1.5 and P value < 0.05.
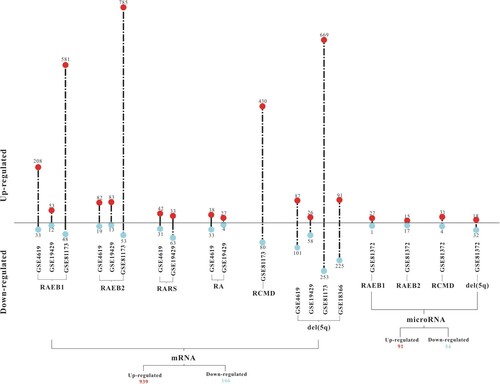
Table 1. Basic information of screened microarray data profiles.
Screening of immune-related mRNAs and construction of PPI networks and miRNA-mRNA interaction networks
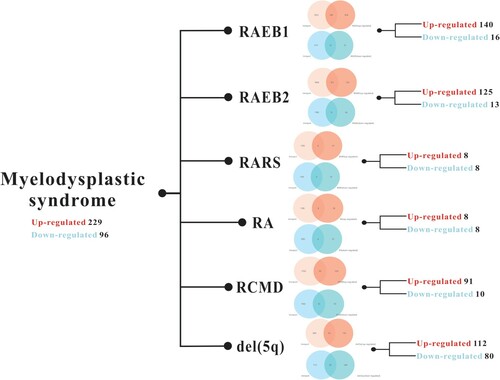
A total of 1793 genes were available from ImmPort and intersected with DE-mRNAs. Finally, 325 immune-related mRNAs were screened. Details of immune-related mRNAs in different subtypes of MDS are shown in .
Figure 2. Details of immune-related mRNAs in different subtypes of MDS are represented by a Venn diagram of mRNA intersections between DE-mRNAs and immune-related gene sets obtained from ImmPort.
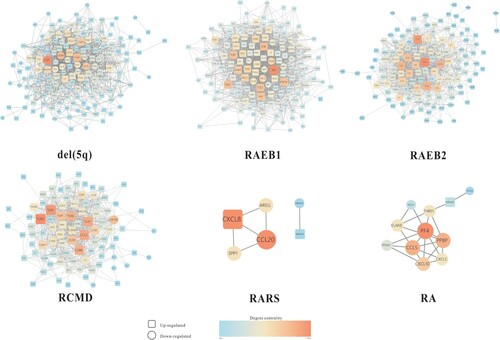
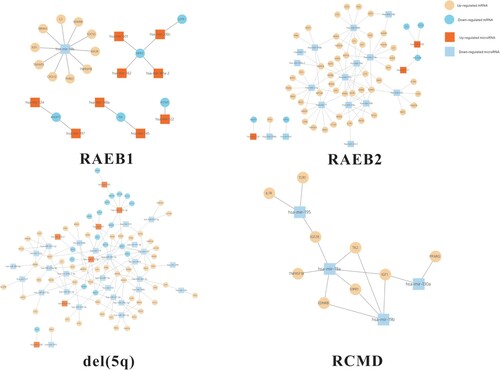
Constructed PPI networks of immune-related mRNAs in different subtypes of MDS are shown in . Nodes represent immune-related mRNAs, and edges represent the relationship. A total of 109 immune-related mRNAs were identified as the target of DE-miRNAs by screening miRDB. MiRNA-mRNA interaction networks in different subtypes of MDS were constructed and are shown in .
Functional analysis of immune-related mRNAs and miRNAs
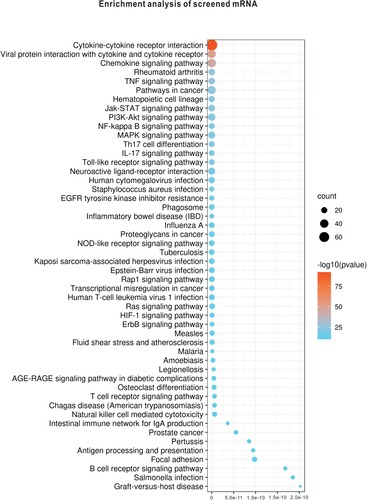
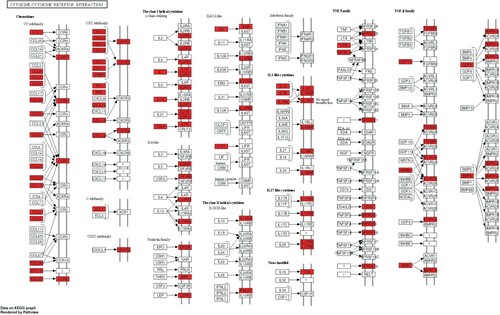
KEGG pathway enrichment analysis of screened immune-related mRNAs and DE-miRNAs was performed, and the top 50 significantly enriched pathways are shown in (A) and (B). Among these pathways, cytokine-cytokine receptor interactions were the most significantly enriched pathways of the immune-related mRNAs and represented the function of most of the DE-miRNAs, which are shown in a KEGG map (). Other significantly enriched pathways, such as the chemokine signalling pathway, NF-kB signalling pathway and Toll-like receptor (TLR) signalling pathway, were essential pathways that control cell growth, migration, proliferation, and metabolism and have been proven to be directly or indirectly involved in the initiation and progression of immune-related and tumour-related diseases.
Figure 5. KEGG pathway enrichment analysis was performed. (A) Results of KEGG pathway enrichment analysis of immune-related mRNAs are shown. (B) Results of KEGG pathway enrichment analysis of immune-related miRNAs are shown.
Topological analysis of constructed networks
Topological analysis of PPI networks and miRNA-mRNA interaction networks was performed to characterize topological attributes to screen hub nodes. According to the DC values, hub molecules mainly belong to the Toll-like receptor (TLR) family, chemokine family and interleukin family, most of which were upregulated DE-mRNAs. In miRNA-mRNA interaction networks, hub miRNAs regulated most of the immune-related mRNAs and mainly belonged to downregulated DE-miRNAs. Details of hub molecules and their DC values in different subtypes of MDS are shown in (A) and (B).
Figure 7. Screen of hub molecules in different networks was performed. (A) Hub mRNAs and the corresponding DC values were determined in PPI networks with DC ≥ median DC. (B) Hub miRNAs and the corresponding DC values were determined in miRNA-mRNA interaction networks with DC ≥ median DC.
Validation of the expression level of hub mRNAs in GSE58831 and the literature
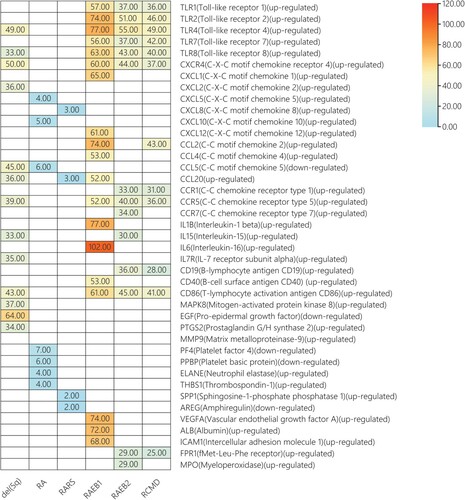
The expression level of hub mRNAs was preliminarily validated in cohort GSE58831. Box plots are shown in . Then, validated mRNAs were further validated by a literature search and are shown in . The results showed that TLR4 and TLR2 were significantly upregulated in RCMD and RAEB2, respectively. Additionally, upregulated mRNAs, including TLR2, TLR4, TLR7, CXCL10 and CCL4, were consistent with previous literature.
Figure 8. The expression value of screened hub mRNAs was determined in the GSE58831 validation dataset, *p<0.05, **p<0.01.
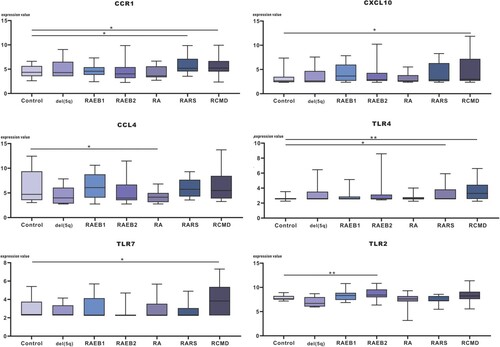
Table 2. The expression of validated mRNAs was described in previous literature.
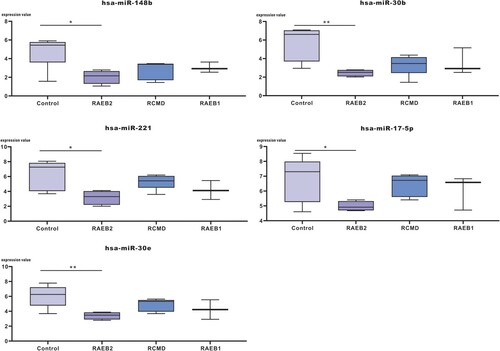
Validation of the expression level of DE-miRNAs in GSE76775 and functional analysis of validated miRNAs
The expression level of hub miRNAs was validated in GSE76775 and is shown in (A). The results showed that hub miRNAs (hsa-miR-148b, hsa-miR-30b, hsa-miR-30e, hsa-miR-221 and hsa-miR-17-5p) were downregulated in GSE76775, especially in RAEB2. Then, enrichment analysis was performed for validated miRNA immune-related mRNA targets to annotate the biological functions of miRNAs. Immune-related target mRNAs of validated miRNAs are shown in (B). The results of KEGG pathway enrichment analysis are shown in (C), and cytokine-cytokine receptor interactions were the significantly related pathways of hub miRNAs, including hsa-miR-30b, hsa-miR-30e and hsa-miR-221. Additionally, hsa-miR-30b and hsa-miR-30e were closely related to Th17 cell differentiation and the Jak-STAT signalling pathway.
Figure 9. Enrichment analysis of hub miRNAs validated in GSE76775 was performed. (A) The expression value of hub miRNAs was determined in the GSE76775 validation dataset, *p<0.05, **p<0.01. (B) The immune-related target mRNAs of the validated miRNAs are indicated. (C) KEGG pathway enrichment analysis of validated miRNAs was performed based on the theory that the function of miRNAs is manifested through the function of target mRNAs.
Discussion
Disorders of the immune process substantially impact the pathophysiology and progression of MDS [Citation29]. A series of innate and adaptive immunity-related signalling pathways are aberrantly activated, which results in clonal expansion of malignant cells and ineffective haematopoiesis. Genetic abnormalities, including gene mutations and gene expression abnormalities, are involved in the pathogenesis of multiple diseases. Studies at the genetic level are an entry point to usher in a paradigm shift in the recognition of this disease. For example, loss-of-function mutations in ten-eleven translocation 2 (TET2) were reported to foster unrestrained inflammation and inflammasome priming in response to TLR4 activation and were shown to increase interleukin 1 beta (IL-1β) elaboration, thereby augmenting the proinflammatory bone marrow niche [Citation30]. An available study that depicts the genomic landscape of MDS/CMML associated with systemic inflammatory and autoimmune diseases based on next-generation sequencing technology also revealed that common genetic susceptibilities with TET2/IDH mutations may arise in the stem cell compartment with myeloid and lymphoid progeny and participate in the onset and progression of both systemic inflammation and haematological malignancy [Citation31]. In addition to gene mutations, gene expression abnormalities also affect the functions of the corresponding encoded proteins and downstream signalling pathways, and it is already known that miRNA molecules regulate gene expression at the mRNA level. Our study investigated MDS pathogenesis at the transcriptional/posttranscriptional level by screening aberrantly expressed mRNAs and miRNAs and elucidated their potential roles in the dysregulation of immunity in MDS.
Considering the heterogeneous nature of MDS, DE-mRNAs and DE-miRNAs were analysed based on morphological subtypes of MDS. A total of 2374 DE-mRNAs intersected with immune-related gene sets obtained from ImmPort, and 325 immune-related mRNAs were obtained. Genes rarely function in an independent manner but fulfil their biological function by interacting with other molecules, and molecular interaction networks form the basis for understanding the mechanism of cell function and life processes. Based on this theory, immune-related mRNAs were uploaded to STRING, and PPI networks were constructed to elucidate the regulatory relationships of the screened molecules. MiRNA-mRNA interaction networks were constructed based on the principle that miRNAs bind to the 3′ untranslated region of mRNAs to silence genes by inhibiting translation and initiating degradation of target mRNAs. To further elucidate the function of the screened molecules, pathway enrichment analysis of immune-related mRNAs and DE-miRNAs was performed, and the results showed that cytokine-cytokine receptor interactions and chemokine signalling pathway were significantly enriched pathways. Topological analysis of PPI networks and miRNA-mRNA interaction networks was performed, and the results showed that most hub mRNAs belong to the TLR family, chemokine family and interleukin family, which were then assessed in GSE58831 and the literature. Hub mRNAs, including CCL4, CXCL10, TLR2, TLR4, and TLR7, were validated to be upregulated in the independent dataset GSE58831 and previous literature. The results of hub miRNAs validation showed that hsa-miR-148b, hsa-miR-30b, hsa-miR-30e, hsa-miR-221 and hsa-miR-17-5p were downregulated in GSE76775, which was in accordance with our screening results. Functional analysis revealed that cytokine-cytokine receptor interactions is a significant hub miRNA-regulated pathway, which was also consistent with the results of topological analysis and enrichment analysis in previous steps, confirming that dysregulated cytokines is a crucial theme in MDS pathogenesis.
Cytokines are hormone-like proteins and peptides whose principal function as secreted or released signalling molecules is to play a modulatory role in immunoinflammatory and haematopoietic homeostasis. Cytokines are important mediators of the immune system, and imbalance of the cytokine network may lead to autoimmune disease susceptibility [Citation32]. Within the haematopoietic system, cytokines stimulate the survival and proliferation of haematopoietic stem and progenitor cells and strongly influence their lineage production [Citation33]. Aberrant cytokines are a prominent feature of leukaemias and contribute to proliferation, survival, self-renewal and resistance to chemotherapy [Citation34]. Chemokines recruit and activate leucocytes and other cells at sites of inflammation, and aberrant release of chemokines can be observed in malignancies and autoimmune diseases. In MDS, ab
normal cytokine levels have been documented [Citation2,Citation26]. Some of the screened hub molecules, such as interleukin-15 (IL15), vascular endothelial growth factor A (VEGF), tumour necrosis factor α (TNF-α), interleukin-15 (IL15), pro-epidermal growth factor (EGF), CCL4 and CXCL10, have been found to be altered and reflect the inflammatory state of MDS to some extent [Citation27,Citation28]. The clinical significance of other members of the chemokine family in our screen is worth further investigation.
Other enriched pathways, such as the TLR signalling pathway and NF-κB signalling pathway, have been explored in MDS onset in previous research [Citation2,Citation23–25,Citation35]. There is crosstalk among these pathways, and they are also influenced by abnormal cytokine expression. TLRs are a family of pattern recognition receptors and play a central role in the innate immune response and pathogenesis of autoimmune diseases [Citation36]. Hub mRNAs, including TLR2, TLR4 and TLR7, were reported to be elevated in MDS patients, and enhanced TLR signalling is a common feature of MDS, which participates in increased death of bone marrow CD34+ cells in MDS [Citation37]. TLR-dependent signalling occurs mainly through the NF-κB signalling pathway [Citation38]. The NF-κB transcription factor is a critical regulator of immunity, stress responses, apoptosis and differentiation; it mediates the synthesis and release of a variety of cytokines, such as TNF-α, IL-1β, IL-6 and IL-8, and can be influenced by cytokines in turn [Citation38]. One example is that elevated levels of IL-8 and NF-κB have been reported to have a positive correlation in MDS patients and interact in a complex network of immune and inflammatory factors involved in pathogenesis [Citation39].
Cytokines perform pleiotropic functions through multiple overlapping signalling cascades in different cell types, constituting a key component of the immune system. The key role of deregulated cytokine production in autoimmunity underlies the rationale for therapeutic cytokine targeting with biologicals, an approach that has led to successes in the treatment of autoimmune diseases such as rheumatoid arthritis and psoriasis [Citation32]. Elevated expression of cytokines is a hallmark in the pathophysiology of MDS [Citation3], and regulation of cytokine signalling is considered a potential treatment strategy. For example, IL8 and C-X-C chemokine receptor type 2 (CXCR2) are overexpressed in stem cells from MDS, and CXCR2 expression is associated with worse prognosis. The IL8-CXCR2 pathway has been selected as a novel therapeutic target in MDS [Citation40]. Another example is luspatercept, a transforming growth factor beta (TGFβ) superfamily ligand trap, which can improve erythropoiesis in preclinical and early phase studies in MDS patients [Citation41,Citation42]. Additionally, a single miRNA can regulate different targets and potentially influence various biological and pathological processes. Altered miRNA levels play a relevant pathogenic role in autoimmunity by regulating the expression of cytokines [Citation43], which have the potential to be used as therapeutics to up- or downregulate protein cascade expression related to pathogenesis. In this study, hsa-miR-30b, hsa-miR-30e and hsa-miR-221 were screened and validated as hub miRNAs in MDS. They were all involved in cytokine-cytokine receptor interactions, and hsa-miR-30b and hsa-miR-30e may also play a potential role in Th17 cell differentiation and the Jak-STAT signalling pathway by targeting immune-related mRNAs. miRNA-mRNA interactions have been implicated in many fundamental biological processes, and the elucidation of miRNA-mRNA interactions not only reveals a possible pathogenesis mechanism but also provides a potential therapeutic strategy for controlling the production of pathogenic cytokines in autoimmune conditions of MDS.
These results indicate that immune-related mRNAs and miRNAs have the potential to be involved in the pathogenesis of MDS through a series of key signalling pathways. There are some limitations in this study. MDS is a highly heterogeneous disease with multiple subtypes and variable clinical processes. We performed analysis of different subtypes, including REAB1, RAEB2, RARS, RCMD, RA and del(5q), based on information on the clinical characteristics of patients obtained from the GEO database. However, raw datasets in the GEO database were not comprehensive enough, and we could not subgroup patients with therapeutic information, risk stratification or other clinical characteristics for a more nuanced analysis. Additionally, some of the screened mRNAs and miRNAs had been preliminarily validated to be aberrantly expressed in independent datasets or previous studies. Concentrated on collecting clinical samples and studying detailed mechanisms through in vitro and in vivo experiments, which are our important research directions in the future.
Conclusion
Dysregulated cytokine-cytokine receptor interactions are a crucial pathological condition in MDS. The re-establishment of cytokine homeostasis may benefit patients. Understanding the potential role of miRNAs in the regulation of cytokine signalling pathways is indispensable in the elucidation of immune dysregulation in MDS. Further, miRNA–mRNA interactions may offer a complementary theoretical perspective for MDS research.
Acknowledgements
The authors would like to acknowledge the principal investigators who made their data publicly accessible for research.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Ogawa S. Genetics of MDS. Blood. 2019;133:1049–1059.
- Ganan-Gomez I, Wei Y, Starczynowski DT, et al. Deregulation of innate immune and inflammatory signaling in myelodysplastic syndromes. Leukemia. 2015;29(7):1458–1469.
- Barreyro L, Chlon TM, Starczynowski DT. Chronic immune response dysregulation in MDS pathogenesis. Blood. 2018;132(15):1553–1560.
- Wang C, Yang Y, Gao S, et al. Immune dysregulation in myelodysplastic syndrome: clinical features, pathogenesis and therapeutic strategies. Crit Rev Oncol Hematol. 2018;122:123–132.
- Giannouli S, Voulgarelis M, Zintzaras E, et al. Autoimmune phenomena in myelodysplastic syndromes: a 4-yr prospective study. Rheumatology (Oxford). 2004;43(5):626–632.
- Taki M, Fukutake K, Matsushita T, et al. Correction to: inhibitor development, safety, and efficacy of Advate(R) in previously untreated patients with hemophilia A in a postmarketing surveillance in Japan. Int J Hematol. 2019;109(2):241.
- Ramadan SM, Fouad TM, Summa V, et al. Acute myeloid leukemia developing in patients with autoimmune diseases. Haematologica. 2012;97(6):805–817.
- Kristinsson S Y, Bjorkholm M, Hultcrantz M, et al. Chronic immune stimulation might act as a trigger for the development of acute myeloid leukemia or myelodysplastic syndromes. J Clin Oncol. 2011;29(21):2897–2903.
- Al Ustwani O, Ford LA, Sait SJ, et al. Myelodysplastic syndromes and autoimmune diseases–case series and review of literature. Leuk Res. 2013;37(8):894–899.
- Anderson LA, Pfeiffer RM, Landgren O, et al. Risks of myeloid malignancies in patients with autoimmune conditions. Br J Cancer. 2009;100(5):822–828.
- Datar GK, Goodel MA. Where there's smoke, there's fire: inflammation drives MDS. Trends Immunol. 2020;41(7):558–560.
- Kuang X, Chi J, Wang L. Deregulated microRNA expression and its pathogenetic implications for myelodysplastic syndromes. Hematology. 2016;21(10):593–602.
- Contreras J, Rao DS. MicroRNAs in inflammation and immune responses. Leukemia. 2012;26(3):404–413.
- Starczynowski DT, Kuchenbauer F, Argiropoulos B, et al. Identification of miR-145 and miR-146a as mediators of the 5q- syndrome phenotype. Nat Med. 2010;16(1):49–58.
- Lee JH, List A, Sallman DA. Molecular pathogenesis of myelodysplastic syndromes with deletion 5q. Eur J Haematol. 2019;102(3):203–209.
- Ganan-Gomez I, Wei Y, Yang H, et al. Overexpression of miR-125a in myelodysplastic syndrome CD34+ cells modulates NF-kappaB activation and enhances erythroid differentiation arrest. PLoS One. 2014;9(4):e93404.
- Bhattacharya S, Dunn P, Thomas CG, et al. Immport, toward repurposing of open access immunological assay data for translational and clinical research. Sci Data. 2018;5:180015.
- Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–D613.
- Wong N, Wang X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2015;43(Database issue):D146–D152.
- Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504.
- Tang Y, Li M, Wang J, et al. CytoNCA: a cytoscape plugin for centrality analysis and evaluation of protein interaction networks. Biosystems. 2015;127:67–72.
- Xie C, Mao X, Huang J, et al. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011;39(Web Server issue):W316–W322.
- Kuninaka N, Kurata M, Yamamoto K, et al. Expression of Toll-like receptor 9 in bone marrow cells of myelodysplastic syndromes is down-regulated during transformation to overt leukemia. Exp Mol Pathol. 2010;88(2):293–298.
- Maratheftis CI, Andreakos E, Moutsopoulos HM, et al. Toll-like receptor-4 is up-regulated in hematopoietic progenitor cells and contributes to increased apoptosis in myelodysplastic syndromes. Clin Cancer Res. 2007;13(4):1154–1160.
- Wei Y, Dimicoli S, Bueso-Ramos C, et al. Toll-like receptor alterations in myelodysplastic syndrome. Leukemia. 2013;27(9):1832–1840.
- Kornblau SM, McCue D, Singh N, et al. Recurrent expression signatures of cytokines and chemokines are present and are independently prognostic in acute myelogenous leukemia and myelodysplasia. Blood. 2010;116(20):4251–4261.
- Pardanani A, Finke C, Lasho TL, et al. IPSS-independent prognostic value of plasma CXCL10, IL-7 and IL-6 levels in myelodysplastic syndromes. Leukemia. 2012;26(4):693–699.
- Feng X, Scheinberg P, Wu CO, et al. Cytokine signature profiles in acquired aplastic anemia and myelodysplastic syndromes. Haematologica. 2011;96(4):602–606.
- Glenthoj A, Orskov AD, Hansen JW, et al. Immune mechanisms in myelodysplastic syndrome. Int J Mol Sci. 2016;17(6):944.
- Sallman DA, List A. The central role of inflammatory signaling in the pathogenesis of myelodysplastic syndromes. Blood. 2019;133(10):1039–1048.
- Zhao LP, Boy M, Azoulay C, et al. Genomic landscape of MDS/CMML associated with systemic inflammatory and autoimmune disease. Leukemia. 2021;Advance online publication.
- Nézondet A-LC, Poubelle EP, et al. The evaluation of cytokines to help establish diagnosis and guide treatment of autoinflammatory and autoimmune diseases. J Leukoc Biol. 2020;108(2):647–657.
- Endele M, Etzrodt M, Schroeder T. Instruction of hematopoietic lineage choice by cytokine signaling. Exp Cell Res. 2014;329(2):207–213.
- Van Etten RA. Aberrant cytokine signaling in leukemia. Oncogene. 2007;26(47):6738–6749.
- Pyatt DW, Stillman WS, Yang Y, et al. An essential role for NF-κB in human CD34+ bone marrow cell survival. Blood. 1999;93(10):3302–3308.
- Ji-Qing C, Szodoray P, Zeher M. Toll-like receptor pathways in autoimmune diseases. Clin Rev Allergy Immunol. 2016;50:1–17.
- Paracatu LC, Schuettpelz LG. Contribution of aberrant Toll like receptor signaling to the pathogenesis of myelodysplastic syndromes. Front Immunol. 2020;11:1236.
- Kulms D, Schwarz T. NF-κb and cytokines. Interleukins. 2006;74:283–300.
- de Matos AG, Ribeiro Junior HL, de Paula Borges D, et al. Interleukin-8 and nuclear factor kappa B are increased and positively correlated in myelodysplastic syndrome. Med Oncol. 2017;34(10):168.
- Schinke C, Giricz O, Li W, et al. IL8-CXCR2 pathway inhibition as a therapeutic strategy against MDS and AML stem cells. Blood. 2015;125(20):3144–3152.
- Fenaux P, Kiladjian JJ, Platzbecker U. Luspatercept for the treatment of anemia in myelodysplastic syndromes and primary myelofibrosis. Blood. 2019;133(8):790–794.
- Markham A. Luspatercept: first approval. Drugs. 2020;80(1):85–90.
- Salvi V, Gianello V, Tiberio L, et al. Cytokine targeting by miRNAs in autoimmune diseases. Front Immunol. 2019;10:15.