ABSTRACT
Background
T-cell acute lymphoblastic leukemia (T-ALL) is a highly proliferative hematologic malignancy. Circular RNA hsa_circ_0000745 (circ_0000745) has been reported as an oncogene in acute lymphoblastic leukemia (ALL). However, whether circ_0000745 can drive T-ALL progression by controlling notch receptor 1 (NOTCH1) expression is unclear.
Methods
Relative expression of circ_0000745 and NOTCH1 in bone marrow (BM) samples and T-ALL cells was detected by real-time quantitative polymerase chain reaction (RT-qPCR). Loss- and gain-of-function experiments were executed to evaluate the effects of circ_0000745 and NOTCH1 on T-ALL cell proliferation and apoptosis. The microRNAs (miRs) that might jointly interact with circ_0000745 and NOTCH1 were predicted by bioinformatics analysis and verified by dual-luciferase reporter and RNA immunoprecipitation (RIP) assays.
Results
Circ_0000745 and NOTCH1 were overexpressed in T-ALL BM and T-ALL cells. Functionally, both circ_0000745 and NOTCH1 overexpression promoted T-ALL cell proliferation and curbed T-ALL cell apoptosis. In contrast, both circ_0000745 and NOTCH1 silencing restrained T-ALL cell proliferation and induced T-ALL cell apoptosis. Furthermore, circ_0000745 could control T-ALL cell proliferation and apoptosis through regulating NOTCH1 expression. Importantly, circ_0000745 regulated NOTCH1 expression by sponging miR-193b-3p.
Conclusion
Our findings proposed a novel model in which circ_0000745 promoted cell proliferation and curbed cell apoptosis via upregulating NOTCH1 through serving as a miR-193b-3p sponge in T-ALL.
Introduction
Acute lymphoblastic leukemia (ALL), a malignant disease of the hematopoietic system that originates from the precursors of the lymphatic lineage, is more likely to occur in children [Citation1]. T-cell acute lymphoblastic leukemia (T-ALL), which belongs to ALL, results from the malignant transformation of T cell precursors, and it accounts for approximately 15% of pediatric cases [Citation2]. Most T-ALL patients respond to conventional chemotherapy, but about 15% to 20% of children with T-ALL will relapse and have a low cure rate [Citation3,Citation4]. Accordingly, exploring the progression mechanism of pediatric T-ALL is crucial to discovering new treatment methods.
The NOTCH transmembrane receptor family transduces extracellular signals [Citation5]. Moreover, NOTCH signals control a variety of cell fate decisions, such as immune regulation and proliferation [Citation6]. Recent studies have shown the involvement of notch receptor 1 (NOTCH1) in many malignancies [Citation7–9]. Notably, NOTCH1 signaling is necessary for pluripotent progenitor cells to commit to T cell fate and normal T cell development [Citation10]. Also, NOTCH1 has become the main regulator of T-ALL cell metabolism and growth [Citation11]. However, the network regulation mechanism related to NOTCH1 dysregulation in T-ALL is still unclear.
Based on the competing endogenous RNA (ceRNA) hypothesis, circular RNAs (circRNAs) distributed in the cytoplasm can regulate gene expression through functioning as microRNA (miR) sponges [Citation12,Citation13]. CircRNAs are produced by a splicing event that covalently connects the downstream splice donor site with the upstream splice acceptor site [Citation14,Citation15]. They are emerging as potential diagnostic biomarkers and therapeutic targets, including blood malignancies [Citation16]. A series of studies conducted in leukemia reported that circRNAs are implicated in ALL progression [Citation17]. As an example, circRNA-PVT1 played a promoting influence on ALL cell growth [Citation18]. Another example was that circRNA-0000097 repressed T-ALL cell growth arrest via adsorbing miR-223-3 p and increasing FBW7 expression [Citation19]. CircRNA hsa_circ_0000745 (circ_0000745), derives from the SPECC1 gene, is identified as a possible marker for gastric cancer diagnosis [Citation20]. Furthermore, circ_0000745 has been uncovered to exert a promoting influence on ALL cell proliferation [Citation21]. However, the relationship between circ_0000745 and NOTCH1 in T-ALL progression is unclear.
Accordingly, the purport of the research was to investigate the involvement of circ_0000745 and NOTCH1 in T-ALL.
Materials and methods
Human bone marrow (BM) samples
BM specimens were collected after informed consent, with approval of the Ethics Committee of Xi’an Gaoxi Hospital. 42 patients were diagnosed with T-ALL at Xi’an Gaoxi Hospital, and 19 healthy volunteers were recruited as controls. Details of the clinic-pathological parameters were shown in .
Table 1. The correlation between circ_0000745 expression and pathological parameters of T-ALL clinical samples.
Cell culture
Human primary peripheral blood mononuclear cells (PBMC) (#CP-H158, Procell, Wuhan, China) and 2 T-ALL cell lines CCRF-CEM (#CL-0058) and Jurkat (#CL-0129) (Procell) were cultivated in Roswell Park Memorial Institute-1640 Medium (Procell) supplemented with1% penicillin/streptomycin (Procell) and 10% fetal bovine serum (Procell). Another T-ALL cell line TALL-104 (#CRL-11386™) (ATCC, Manassas, VA, USA) was cultivated in Iscove’s Modified Dulbecco’s Medium (ATCC) with 100 IU/mL interleukin-2 (Procell), 20% fetal bovine serum (Procell), 0.5 µg/mL D-mannitol (Sigma, St. Louis, MO, USA), and 2.5 µg/mL human albumin (Sigma). All cells were cultivated at 37˚C with 5% carbon dioxide.
Plasmid construction and RNA oligonucleotides
Full-length sequences of circ_0000745 and NOTCH1 were cloned into the empty pCD5-ciR vector (Vector) (Geneseed, Guangzhou, China) at EcoRI and BamHI sites and pcDNA vector (Thermo Fisher, Waltham, MA, USA) at HindIII and EcoRV sites, respectively, to construct pCD5-ciR-circ_0000745 (circ_0000745) and pcDNA-NOTCH1 (NOTCH1) plasmids. Small interfering (si) RNAs targeting NOTCH1 (si-NOTCH1#1 and si-NOTCH1#2) and circ_0000745 (si-circ_0000745#1 and si-circ_0000745#2) were used to silence circ_0000745 and NOTCH1, and si-NC and si-con were utilized as controls for siRNA targeting NOTCH1 and circ_0000745. The above oligonucleotides and miR-193b-3p inhibitor (in-miR-193b-3p), inhibitor negative control (in-miR-NC), miR-193b-3p mimic (miR-193b-3p), as well as mimic negative control (miR-NC) were synthesized by Sangon Biotech Co., Ltd. (Shanghai, China). Transfection of T-ALL cells (CCRF-CEM and Jurkat) with plasmids and/or oligonucleotides was performed using Lipofectamine 3000 (Thermo Fisher).
RNA isolation, complementary DNA synthesis, and real-time quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated from BM specimens and cultured cells using TRIzol™ Reagent (Thermo Fisher), followed by purification using the Purelink™ RNA Mini Kit (Thermo Fisher). Isolation of cytoplasmic and nuclear RNA from CCRF-CEM and Jurkat cells was performed as directed by the protocols of the PARIS kit (Thermo Fisher). To verify the circular form of circ_0000745, total RNA derived from CCRF-CEM and Jurkat cells were digested with 3 U/μg RNase R (Thermo Fisher) or DEPC-treated water (Thermo Fisher). Complementary DNA was synthesized from purified RNA using the HiScript III 1st Strand cDNA Synthesis Kit (Vazyme, Nanjing, China) or miRNA 1st Strand cDNA Synthesis Kit (Vazyme). For quantitative analysis of RNA expression, complementary DNA was mixed with AceQ SYBR qPCR Master Mix (Vazyme) and run on Roche Applied Science LightCyclerTM 480 (Roche, Basel, Switzerland). Data were evaluated using the 2-ΔΔCt method and normalized to the housekeeping gene U6 or GAPDH. Primer sequences were listed in .
Table 2. Primers for RT-qPCR used in this study
Cell counting kit-8 (CCK-8) assay
About 2×104 transfected T-ALL cells were allowed to grow in 96-well plates. After incubating for a period of time, 10 μL of CCK-8 reagent (Sigma) was added to each well. 4 h later, the absorbance was measured at 450 nm using an enzyme immunoassay analyzer (Bio-Tek, Winooski, VT, USA).
5-ethynyl-2’- deoxyuridine (EdU) assay
The proliferative activity of CCRF-CEM and Jurkat cells was analyzed using an EdU detection kit (Ribobio, Guangzhou, China). In brief, about 5×104 cells were allowed to grow for 48 h and then incubated with 20 nM EdU for 4 h, followed by treating with 0.5% Triton X-100 (Sigma), 4% paraformaldehyde (Sigma), and anti-EdU working solution. Staining of the nucleus was performed with the 4’,6-diamidino-2-phenylindole (DAPI) solution, and the proportion of EdU-positive cells was observed using fluorescence microscopy (Olympus, Tokyo, Japan).
Flow cytometry for cell cycle analysis
The cells were collected after transfection for 48 h, followed by fixed in pre-cold 70% ethanol. Subsequently, the cells were incubated with 10 mg/mL RNase, stained with 1 mg/mL propidium iodide (PI) and then subjected to a flow cytometer (BD Biosciences, San Jose, CA, USA).
Flow cytometry for cell apoptosis analysis
Cell apoptosis was analyzed using the Annexin V-FITC Cell Apoptosis Detection Kit (Beyotime, Shanghai, China) on a flow cytometer (BD Biosciences) based on the manufacturer’s descriptions. The apoptotic rate was the sum of the early and late apoptotic rates.
Western blotting
BM specimens and cultured cells were lysed in RIPA buffer. Following centrifugation (10000 g, 10 min), the supernatants were collected and whole extracts were separated by 10% SDS-PAGE. Proteins were transferred onto nitrocellulose membranes (Thermo Fisher) and then blocked in 5% milk in TBST. Incubation with antibodies recognizing NOTCH1 (ab52627, 1:1000, Abcam, Cambridge, MA, USA), Bcl-2 (ab32124, 1:1000, Abcam), Bax (ab182733, 1:2000, Abcam), and GAPDH (ab128915, 1:10000, Abcam) was carried for 16 h at 4˚C. Membranes were then incubated with a secondary antibody (ab97051, 1:5000, Abcam). Immunoreactive bands were detected using a chemiluminescence system (Thermo Fisher), followed by imaging using the Gel Doc system (Bio-Rad, Hercules, CA, USA).
Dual-luciferase reporter assay
To generate luciferase reporter for circ_0000745 and NOTCH1, fragments of wild type circ_0000745, mutant circ_0000745, wild type 3’UTR of NOTCH1 mRNA, mutant 3’UTR of NOTCH1 mRNA were cloned into the empty psiCHECK-2 vector (Promega, Madison, WI, USA). CCRF-CEM and Jurkat cells were co-transfected with a recombinant psiCHECK-2 vector and miR-NC or miR-193b-3p mimic. 2 days later, luciferase activities were measured with a dual-luciferase assay system (Promega).
RNA immunoprecipitation (RIP)
An RNA-Binding Protein Immunoprecipitation Kit (Millipore, Billerica, MA, USA) was utilized for RIP analysis. The cell lysates were incubated with RIP buffer containing magnetic beads conjugated with beads coated with Anti-Ago2 (Millipore) or IgG antibody (Abcam). After extraction and purification, the immunoprecipitated RNAs were subjected to RT-qPCR.
Statistical analysis
Each experiment was performed with 3 biological replicates and data were represented as mean ± standard deviation. All statistical tests were conducted using GraphPad Prism 8 (GraphPad Software, San Diego, CA, USA). Unpaired Student’s t-tests were used to assess significance when comparing data between 2 variances. Analysis of variance (ANOVA) was used to determine significance when comparing data with 3 or more variances. Differences were considered statistically significant at a value of P < 0.05 (*P < 0.05, **P < 0.01, and ***P < 0.001).
Results
Circ_0000745 expression had a positive correlation with NOTCH1 mRNA
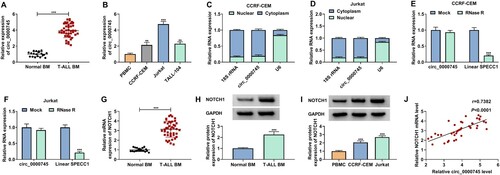
To investigate the involvement between circ_0000745 and NOTCH1 in T-ALL progression, we detected the changes of circ_0000745 and NOTCH1 mRNA in T-ALL BM and T-ALL cell lines. RT-qPCR presented higher expression levels of circ_0000745 in T-ALL BM and T-ALL cell lines compared to their respective control (A and B). There was a significant association between circ_0000745 expression and pathological data, including LDH level (P = 0.0205) and risk stratification (P = 0.0063) (). To determine the intracellular distribution of circ_0000745, we performed the nuclear-cytoplasmic fractionation and observed that approximately 80.67% of circ_0000745 was distributed in the cytoplasm (C and D). RT-qPCR exhibited that circ_0000745 had a stronger resistibility to RNase R, bur linear SPECC1 was digested sharply (E and F). We then verified that the mRNA and protein levels of NOTCH1 in T-ALL BM. As expected, the mRNA and protein levels of NOTCH1 were highly expressed in T-ALL BM (G and H). Similar findings were observed with NOTCH1 protein in T-ALL cells (I). Moreover, circ_0000745 expression in T-ALL BM had a positive correlation with NOTCH1 mRNA (J). Collectively, these findings suggested that there was a positive correlation between circ_0000745 and NOTCH1 mRNA in T-ALL.
Figure 1. Circ_0000745 expression had a positive correlation with NOTCH1 mRNA in T-ALL BM. (A) RT-qPCR analysis of circ_0000745 expression in T-ALL BM (n = 42) and normal BM (n = 19). (B) RT-qPCR analysis of circ_0000745 expression in T-ALL cell lines, and the PBMC cell line was used a negative control. (C and D) Subcellular localization of circ_0000745 in T-ALL cells was analyzed. 18S rRNA acted as a positive control for cytoplasmic RNA, and U6 served as a positive control for nuclear RNA. (E and F) RNase R digestion was performed to verify the circular form of circ_0000745. (G) RT-qPCR analysis of the expression of NOTCH1 mRNA in T-ALL BM (n = 42) and normal BM (n = 19). (H) Western blotting analysis of the protein level of NOTCH1 in T-ALL BM (n = 6) and normal BM (n = 6). (I) Western blotting analysis of the protein level of NOTCH1 in T-ALL cell lines. (J) The correlation between circ_0000745 and NOTCH1 mRNA in T-ALL BM was assessed by Pearson’s correlation analysis. **P < 0.01 and ***P < 0.001.
Circ_0000745 facilitated T-ALL cell proliferation and repressed T-ALL cell apoptosis
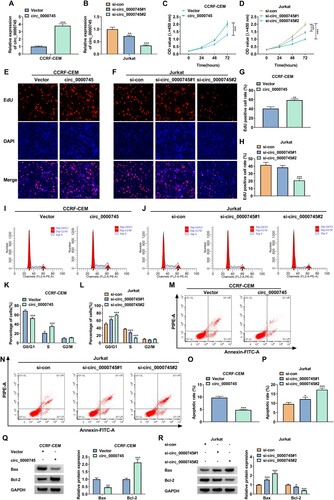
To illustrate the effects of circ_0000745 overexpression and silencing on T-ALL cell proliferation and apoptosis, we selected to overexpress circ_0000745 in CCRF-CEM cells and silence circ_0000745 in Jurkat cells. To change the expression of circ_0000745, we designed circ_0000745 overexpression plasmids and two siRNAs spanning the circ_0000745 junction. After transfection, circ_0000745 expression was markedly increased in CCRF-CEM cells (A) and decreased in Jurkat cells (B). Upregulation of circ_0000745 overtly promoted CCRF-CEM cell proliferation and the silencing of circ_0000745 repressed Jurkat cell proliferation, as confirmed in the CCK-8 assay (C and D). The results of EdU assay presented that circ_0000745 overexpression resulted in an increase in the number of proliferating CCRF-CEM cells, and circ_0000745 inhibition caused a decrease in the number of proliferating Jurkat cells, especially in the si-circ_0000745#2 group (E-H). We observed a substantial promotion of cell cycle under ectopic circ_0000745 expression in CCRF-CEM cells. On the contrary, there was a substantial repression of cell cycle in circ_0000745-inhibited Jurkat cells (I-L). Also, circ_0000745 overexpression repressed CCRF-CEM cell apoptosis, but circ_0000745 silencing facilitated Jurkat cell apoptosis (M-P). In addition, ectopic circ_0000745 expression decreased the protein level of Bax and elevated the protein level of Bcl-2 in CCRF-CEM cells (Q). Conversely, circ_0000745 knockdown increased the protein level of Bax and decreased the protein level of Bcl-2 in Jurkat cells (R). In sum, these finding revealed that circ_0000745 promoted cell proliferation and repressed cell apoptosis in T-ALL cells.
Figure 2. Circ_0000745 constrained cell proliferation and suppressed cell apoptosis in T-ALL cells. (A-R) CCRF-CEM cells were transfected with Vector or circ_0000745 and Jurkat cells were transfected with si-con, si-circ_0000745#1, or si-circ_0000745#2. (A and B) Relative expression of circ_0000745 in CCRF-CEM cells (A) and Jurkat cells (B) was analyzed by RT-qPCR. (C-H) Analysis of the proliferation of CCRF-CEM and Jurkat cells by CCK-8 and EdU assays. (I-P) Flow cytometry assays were executed to analyze cell cycle and apoptosis in CCRF-CEM and Jurkat cells. (Q and R) Relative protein levels of Bax and Bcl-2 in CCRF-CEM and Jurkat cells were evaluated using western blotting. *P < 0.05, **P < 0.01, and ***P < 0.001.
NOTCH1 promoted T-ALL cell proliferation and inhibited T-ALL cell apoptosis
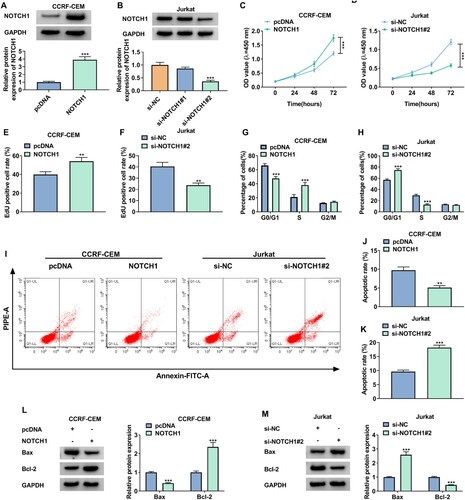
To explore the function of NOTCH1, the NOTCH1 overexpression plasmid was transfected into CCRF-CEM cells and two siRNAs targeting NOTCH1 were transfected into Jurkat cells. Also, the protein level of NOTCH1 was markedly elevated in CCRF-CEM cells after transfection with NOTCH1 overexpression plasmids and reduced in Jurkat cells after transfection with si-NOTCH1#2 (A and B). Moreover, NOTCH1 upregulation promoted the proliferation of CCRF-CEM cells, but NOTCH1 silencing curbed the proliferation of Jurkat cells (C-F). As expected, ectopic NOTCH1 expression induced cell cycle progression and constrained cell apoptosis in CCRF-CEM cells. On the contrary, NOTCH1 downregulation inhibited cell cycle progression and induced cell apoptosis in Jurkat cells (G-K). Additionally, the protein level of Bax was decreased in NOTCH1-overexpressed CCRF-CEM cells and increased in NOTCH1-inhibiting Jurkat cells. However, the protein level of Bcl-2 was elevated in NOTCH1-overexpressed CCRF-CEM cells and reduced in NOTCH1-inhibiting Jurkat cells (3L and M). Together, these results suggested that NOTCH1 promoted cell proliferation and suppressed cell apoptosis in T-ALL cells.
Figure 3. NOTCH1 facilitated cell proliferation and inhibited cell apoptosis in T-ALL cells. (A-M) CCRF-CEM cells were transfected with pcDNA or NOTCH1 and Jurkat cells were transfected with si-NC, si-NOTCH1#1, or si-NOTCH1#2. (A and B) Relative protein level of NOTCH1 in CCRF-CEM cells (A) and Jurkat cells (B) was analyzed by western blotting. (C-F) The proliferation of CCRF-CEM and Jurkat cells was determined by CCK-8 and EdU assays. (G-K) Flow cytometry assays were carried out to assess cell cycle and apoptosis in CCRF-CEM and Jurkat cells. (L and M) Relative protein levels of Bax and Bcl-2 in CCRF-CEM and Jurkat cells were detected using western blotting. **P < 0.01 and ***P < 0.001.
Circ_0000745 acted as a miR-193b-3p sponge
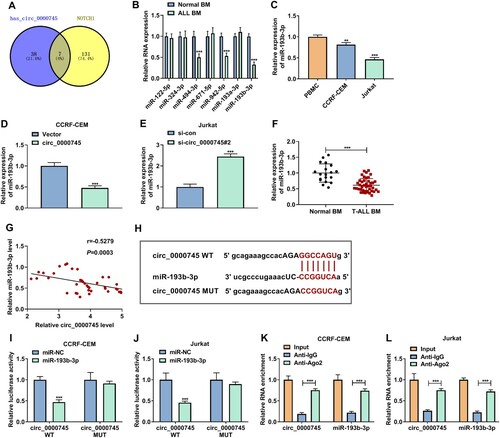
Given circ_0000745 mainly distributed in the cytoplasm, we further analyzed whether circ_0000745 functioned as a miR sponge and regulated NOTCH1 expression by binging to miR. Through starBase prediction, we discovered that there were 38 miRs that might interact with circ_0000745 and 131 miRs that might target NOTCH1. Cross-crossing analysis showed that circ_0000745 and NOTCH1 might share the miR response elements of 7 miRs (miR-122-5p, miR-324-3p, miR-494-3p, miR-671-5p, miR-942-5p, miR-193a-3p, and miR-193b-3p) among these miRs (A). Also, 3 miRs (miR-494-3p, miR-942-5p, and miR-193b-3p) were overtly downregulated in 6 randomly T-ALL BM, especially the expression of miR-193b-3p (B). Similar findings were observed with miR-193b-3p in T-ALL cells (C). As expected, circ_0000745 upregulation decreased miR-193b-3p expression in CCRF-CEM cells (D). However, circ_0000745 silencing increased miR-193b-3p expression in Jurkat cells (E). In addition, miR-193b-3p expression was lower in 42 T-ALL BM than that in 19 normal BM (F). Furthermore, the expression of miR-193b-3p and circ_0000745 had a negative correlation in T-ALL BM (G). To verify the hypothesis that circ_0000745 was a miR-193b-3p sponge, we constructed a luciferase plasmid containing the circ_0000745 WT sequence or its mutant sequence circ_0000745 MUT (H). Dual-luciferase reporter assay showed that miR-193b-3p mimic decreased the luciferase activity of the luciferase plasmid containing the circ_0000745 WT sequence, but the luciferase activity of the luciferase plasmid containing the circ_0000745 MUT sequence did not change (I and J). RIP assay displayed that the abundance of circ_0000745 and miR-193b-3p was higher in the Ago2-precipitates (K and L). Collectively, these findings suggested that circ_0000745 acted as a miR-193b-3p sponge.
Figure 4. Circ_0000745 was identified as a miR-193b-3p sponge. (A) Venn diagram showed overlapping miRs that might jointly interact with circ_0000745 and NOTCH1. (B) RT-qPCR analysis of 7 miRs in 6 randomly T-ALL BM and normal BM. (C) RT-qPCR analysis of miR-193b-3p in PBMC and T-ALL cells lines. (D) Impact of circ_0000745 overexpression on miR-193b-3p expression in CCRF-CEM cells was evaluated by RT-qPCR. (E) Impact of circ_0000745 downregulation on miR-193b-3p expression in Jurkat cells was evaluated by RT-qPCR. (F) RT-qPCR analysis of miR-193b-3p in T-ALL BM (n = 42) and normal BM (n = 19). (G) The correlation between miR-193b-3p and circ_0000745 expression in T-ALL BM was determined by Pearson’s correlation analysis. (H) The alignment of circ_0000745 with miR-193b-3p. (I and J) Dual-luciferase reporter assay was executed to verify the relationship between circ_0000745 and miR-193b-3p. (K and L) RIP assay was performed to verify the interaction between circ_0000745 and miR-193b-3p. **P < 0.01 and ***P < 0.001.
NOTCH1 acted as a miR-193b-3p target
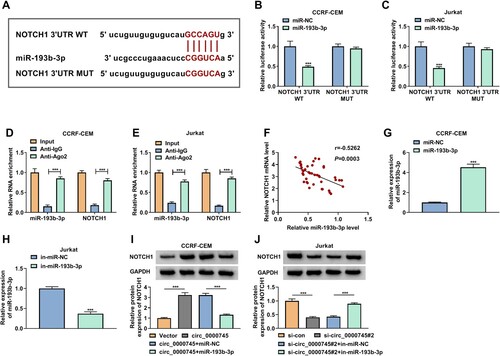
We then sought to identify the relationship between NOTCH1 and miR-193b-3p. The conserved binding sites of miR-193b-3p in the 3’UTR of NOTCH1 mRNA were exhibited in A. Also, miR-193b-3p could suppress NOTCH1 3’UTR luciferase activity, whereas the mutation of the binding-site canceled the inhibiting impact of miR-193b-3p on NOTCH1 3’UTR luciferase activity (B and C). Moreover, a conspicuous enrichment with NOTCH1 mRNA and miR-193b-3p was observed in the Ago2-precipitates (D and E). Furthermore, there was a negative correlation between the expression of miR-193b-3p and NOTCH1 mRNA in T-ALL BM (F). In addition, the overexpression efficiency of miR-193b-3p mimic in CCRF-CEM cells and the interference efficiency of in-miR-193b-3p in Jurkat cells were verified and showed in G and H. As expected, exogenetic overexpression of miR-193b-3p reversed the promoting influence of circ_0000745 upregulation on the protein level of NOTCH1 in CCRF-CEM cells (I). And miR-193b-3p inhibitor counteracted the inhibiting effect of circ_0000745 downregulation on the protein level of NOTCH1 in Jurkat cells (J). In sum, miR-193b-3p targeted NOTCH1, and circ_0000745 regulated NOTCH1 expression by serving as a miR-193b-3p decoy.
Figure 5. Circ_0000745 regulated NOTCH1 expression by adsorbing miR-193b-3p. (A) The conserved binding sites of miR-193b-3p in the 3’UTR of NOTCH1 mRNA. (B and C) Dual-luciferase reporter assay was performed to analyze the relationship between miR-193b-3p and NOTCH1. (D and E) The enrichment of miR-193b-3p and NOTCH1 mRNA in Ago2-precipitates and IgG-precipitates was analyzed RT-qPCR. (F) The correlation between miR-193b-3p and NOTCH1 mRNA in T-ALL BM. (G and H) RT-qPCR analysis of the overexpression efficiency of miR-193b-3p mimic in CCRF-CEM cells and the interference efficiency of in-miR-193b-3p in Jurkat cells. (I and J) Western blotting analysis of the protein level of NOTCH1 in CCRF-CEM cells transfected with Vector, circ_0000745, circ_0000745+miR-NC, or circ_0000745+miR-193b-3p as well as Jurkat cells transfected with si-con, si-circ_0000745#2, si-circ_0000745#2+in-miR-NC, or si-circ_0000745#2+in-miR-193b-3p. ***P < 0.001.
Circ_0000745 regulated T-ALL cell proliferation and apoptosis via regulating NOTCH1 expression
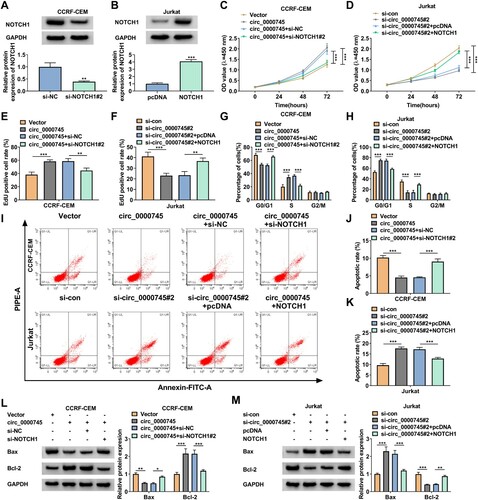
Considering the above findings, we further validated whether circ_0000745 controlled T-ALL cell proliferation and apoptosis via regulating NOTCH1 expression. To silence NOTCH1 in CCRF-CEM cells and overexpress NOTCH1 in Jurkat cells, si-NOTCH1#2 and NOTCH1 overexpression plasmids were transfected into CCRF-CEM and Jurkat cells, respectively. The results exhibited that the protein level of NOTCH1 was prominently decreased in si-NOTCH1#2-transfected CCRF-CEM cells and elevated in NOTCH1-transfected Jurkat cells (A and B). Moreover, NOTCH1 silencing offset the promoting effect of circ_0000745 overexpression on CCRF-CEM cell proliferation, but NOTCH1 upregulation counteracted the inhibitory effect of circ_0000745 knockdown on Jurkat cell proliferation (C-F). Also, knockdown of NOTCH1 reversed the suppressive effect of circ_0000745 upregulation on cell cycle arrest and apoptosis in CCRF-CEM cells, but overexpression of NOTCH1 overturned the promoting effect of circ_0000745 inhibition on cell cycle arrest and apoptosis in Jurkat cells (G-K). In addition, NOTCH1 downregulation overturned circ_0000745 overexpression-mediated effects on the protein levels of Bax and Bcl-2 in CCRF-CEM cells (L). And the inhibition of NOTCH1 reversed circ_0000745 silencing-mediated effects on the protein levels of Bax and Bcl-2 in Jurkat cells (M). Taken together, circ_0000745 regulated T-ALL cell proliferation and apoptosis via modulating NOTCH1 expression.
Figure 6. Circ_0000745 regulated cell proliferation and apoptosis through modulating NOTCH1 expression in T-ALL cells. (A and B) Western blotting analysis of the protein level of NOTCH1 in CCRF-CEM cells transfected with si-NC or si-NOTCH1#2 as well as Jurkat cells transfected with pcDNA or NOTCH1. (C-M) CCRF-CEM cells were transfected with Vector, circ_0000745, circ_0000745+si-NC, or circ_0000745+si-NOTCH1#2 and Jurkat cells were transfected with si-con, si-circ_0000745#2, si-circ_0000745#2+pcDNA, or si-circ_0000745#2+NOTCH1. (C-F) The proliferation of CCRF-CEM and Jurkat cells was analyzed using CCK-8 and EdU assays. (G-K) Cell cycle and apoptosis were analyzed using flow cytometry assays in CCRF-CEM and Jurkat cells. (L and M) Western blotting analysis of the protein levels of Bax and Bcl-2 in CCRF-CEM and Jurkat cells. *P < 0.05, **P < 0.01, and ***P < 0.001.
Discussion
Recent findings from scholars in different fields have shown that the deregulation of circRNA is associated with the occurrence of many diseases, including T-ALL. Buratin et al. reported that large-scale circRNAs in T-ALL were deregulated, and circRNA-ZNF609 silencing decreased T-ALL cell viability [Citation22]. Furthermore, the deficiency of circRNA-000094 played a promoting effect on T-ALL cell malignant phenotypes, which was achieved via adsorbing miR-223-3p and downregulating FBW7 [Citation19]. At present, the regulatory mechanisms governed by circRNAs are still largely unexplored in T-ALL.
Circ_0000745, located at chr17:20107645-20109225, is a circRNA from the SPECC1 gene. It was uncovered that the redundancy of circ_0000745 facilitated cervical cancer tumorigenesis via upregulation of miR-409-3p [Citation23]. Also, circ_0000745 silencing induced ALL cell apoptosis and constrained ALL cell proliferation via blocking the ERK pathway [Citation21]. Our results revealed that circ_0000745 was overexpressed in T-ALL BM. Circ_0000745 knockdown induced T-ALL apoptosis and repressed T-ALL cell proliferation, whereas circ_0000745 overexpression restrained T-ALL cell apoptosis and promoted T-ALL cell proliferation. These data manifested that circ_0000745 acted as an oncogene in T-ALL.
NOTCH1 is a crucial oncogenic driver in T-ALL [Citation24]. Also, the activation of NOTCH1 signaling can induce T-ALL in a mouse model [Citation25]. The degradation of NOTCH1 mediated by ARRB1 could inhibit T-ALL cell proliferation [Citation26]. Luo et al. reported that NOTCH1 had higher levels in T-ALL BM, and NEAT1 facilitated T-ALL cell proliferation by upregulating NOTCH1 [Citation27]. Our results also exhibited higher NOTCH1 levels in T-ALL BM than normal BM, and knockdown of NOTCH1 constrained T-ALL cell proliferation and induced T-ALL cell apoptosis. These data suggested that NOTCH1 served as an oncogene in T-ALL.
Based on the ceRNA hypothesis and circ_0000745 was mainly distributed in the cytoplasm, we suspected that circ_0000745 might regulate NOTCH1 expression by binging to miRs. After bioinformatics analysis, luciferase reporter and RIP assays validated that circ_0000745 served as a decoy of miR-193b-3p, which targeted NOTCH1. Previous findings revealed that miR-193b-3p had anti-tumor activity in multiple cancers [Citation28–30]. In leukemia, miR-193b-3p acted as a potential biomarker for patients with chronic myeloid leukemia to respond to imatinib treatment [Citation31]. Furthermore, miR-193b-3p exerted a tumor-inhibiting influence on T-ALL [Citation32]. Herein, miR-193b-3p had lower levels in T-ALL BM. Also, circ_0000745 regulated NOTCH1 expression via sponging miR-193b-3p. Moreover, circ_0000745 controlled T-ALL cell proliferation and apoptosis by modulating NOTCH1 expression. Thus, we concluded that circ_0000745 promoted T-ALL cell proliferation and repressed T-ALL cell apoptosis by elevating NOTCH1 expression via functioning as a miR-193b-3p sponge.
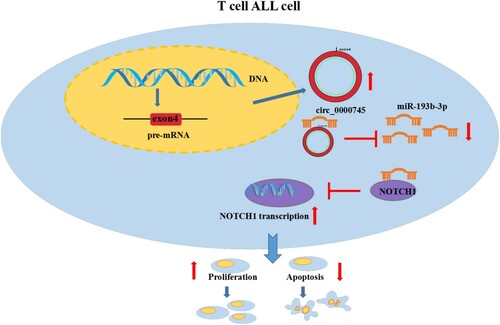
Our results led us to propose a model in which elevated circ_0000745 adsorbs miR-193b-3p and prevents miR-193b-3p from interacting with NOTCH1 mRNA, thereby increasing the level of NOTCH1, which in turn promotes T-ALL cell proliferation and inhibits T-ALL cell apoptosis (). The research offers a novel mechanism by which circ_0000745 controls T-ALL cell proliferation and apoptosis.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Malard F, Mohty M. Acute lymphoblastic leukaemia. Lancet. 2020;395:1146–1162.
- Karrman K, Johansson B. Pediatric T-cell acute lymphoblastic leukemia. Genes Chromosomes Cancer. 2017;56:89–116.
- Hunger SP, Mullighan CG. Acute lymphoblastic leukemia in children. N Engl J Med. 2015;373:1541–1552.
- McMahon CM, Luger SM. Relapsed T cell ALL: current approaches and New directions. Curr Hematol Malig Rep. 2019;14:83–93.
- Zanotti S, Canalis E. Notch signaling and the skeleton. Endocr Rev. 2016;37:223–253.
- Aster JC, Pear WS, Blacklow SC. The varied roles of notch in cancer. Annu Rev Pathol. 2017;12:245–275.
- Jackstadt R, van Hooff SR, Leach JD, et al. Epithelial NOTCH signaling rewires the tumor microenvironment of colorectal Cancer to drive poor-prognosis subtypes and metastasis. Cancer Cell. 2019;36:319–36.e7.
- Rice MA, Hsu EC. Loss of Notch1 activity inhibits Prostate Cancer growth and Metastasis and sensitizes Prostate Cancer cells to antiandrogen therapies. Mol Cancer Ther. 2019;18:1230–1242.
- Aslan M, Ghoochani A, Su A, et al. IMPAD1 functions as mitochondrial electron transport inhibitor that prevents ROS production and promotes lung cancer metastasis through the AMPK-Notch1-HEY1 pathway. Mol Cancer Ther. 2020;485:27–37.
- Deftos ML, Bevan MJ. Notch signaling in T cell development. Curr Opin Immunol. 2000;12:166–172.
- Sanchez-Martin M, Ferrando A. The NOTCH1-MYC highway toward T-cell acute lymphoblastic leukemia. Blood. 2017;129:1124–1133.
- Salmena L, Poliseno L, Tay Y, et al. A ceRNA hypothesis: the rosetta stone of a hidden RNA language? Cell. 2011;146:353–358.
- Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. 2016;17:272–283.
- Li X, Yang L, Chen LL. The biogenesis, functions, and challenges of Circular RNAs. Mol Cell. 2018;71:428–442.
- Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338.
- Perez de Acha O, Rossi M, Gorospe M. Circular RNAs in Blood malignancies. Front Mol Biosci. 2020;7:109.
- Jamal M, Song T, Chen B, et al. Recent progress on Circular RNA research in acute myeloid leukemia. Front Oncol. 2019;9:1108.
- Hu J, Han Q, Gu Y, et al. Circular RNA PVT1 expression and its roles in acute lymphoblastic leukemia. Epigenomics. 2018;10:723–732.
- Hou Y, Sun J, Huang J, et al. Circular RNA circRNA_0000094 sponges microRNA-223-3p and up-regulate F-box and WD repeat domain containing 7 to restrain T cell acute lymphoblastic leukemia progression. Hum Cell. 2021;34:977–989.
- Huang M, He YR, Liang LC, et al. Circular RNA hsa_circ_0000745 may serve as a diagnostic marker for gastric cancer. World J Gastroenterol. 2017;23:6330–6338.
- Liu X, Zhou C, Li Y, et al. Upregulation of circ-0000745 in acute lymphoblastic leukemia enhanced cell proliferation by activating ERK pathway. Gene. 2020;751:144726.
- Buratin A, Paganin M, Gaffo E, et al. Large-scale circular RNA deregulation in T-ALL: unlocking unique ectopic expression of molecular subtypes. Blood Adv. 2020;4:5902–5914.
- Cui X, Chen J, Zheng Y, et al. Circ_0000745 promotes the progression of cervical Cancer by regulating miR-409-3p/ATF1 axis. Cancer Biother Radiopharm. 2020. (Online ahead of print)
- Hales EC, Taub JW, Matherly LH. New insights into Notch1 regulation of the PI3K-AKT-mTOR1 signaling axis: targeted therapy of γ-secretase inhibitor resistant T-cell acute lymphoblastic leukemia. Cell Signal. 2014;26:149–161.
- Saccomani V, Grassi A. miR-22-3p negatively affects tumor progression in T-cell acute lymphoblastic leukemia. Cells. 2020;9:1726.
- Shu Y, Wang Y, Lv WQ, et al. ARRB1-Promoted NOTCH1 degradation Is suppressed by OncomiR miR-223 in T-cell acute lymphoblastic leukemia. Cancer Res. 2020;80:988–998.
- Luo YY, Wang ZH, Yu Q, et al. LncRNA-NEAT1 promotes proliferation of T-ALL cells via miR-146b-5p/NOTCH1 signaling pathway. Pathol Res Pract. 2020;216:153212.
- Zhang J, Qin J, Su Y. miR-193b-3p possesses anti-tumor activity in ovarian carcinoma cells by targeting p21-activated kinase 3. Biomed Pharmacother. 2017;96:1275–1282.
- Wang H, Chen W, Yang P, et al. Knockdown of linc00152 inhibits the progression of gastric cancer by regulating microRNA-193b-3p/ETS1 axis. Cancer Biol Ther. 2019;20:461–473.
- Yang Z, Zhuang Q, Hu G, et al. MORC4 is a novel breast cancer oncogene regulated by miR-193b-3p. J Cell Biochem. 2019;120:4634–4643.
- Ramachandran SS, Muiwo P, Ahmad HM, et al. miR-505-5p and miR-193b-3p: potential biomarkers of imatinib response in patients with chronic myeloid leukemia. Leuk Lymphoma. 2017;58:1981–1984.
- Mets E, Van der Meulen J, Van Peer G, et al. MicroRNA-193b-3p acts as a tumor suppressor by targeting the MYB oncogene in T-cell acute lymphoblastic leukemia. Leukemia. 2015;29:798–806.