ABSTRACT
Objective:
This study was to comprehensively clarify the associations between single nucleotide polymorphisms (SNPs) in inflammatory genes and the susceptibility to childhood leukemia.
Methods:
Eligible articles were collected from the databases of PubMed, EMBASE, Cochrane Library, CNKI and Wan Fang. The pooled odds ratios (ORs) and 95% confidence intervals (95% CIs) were calculated to estimate the association strength by using the STATA 15.0 software.
Results:
Sixteen studies were enrolled. These studies mainly evaluated SNPs in 13 genes, including C-X-C motif chemokine ligand 12 (CXCL12), toll-like receptor (TLR)−4, TLR6, TLR9, CD14, interleukin (IL)−1β, NLR family pyrin domain containing 3, IL-4, interleukin 4 receptor, IL-10, IL-13, macrophage migration inhibitory factor (MIF) and tumor necrosis factor-α. The meta-analysis indicated that CXCL12 rs1801157 (AG vs GG: OR = 1.99; 95%CI = 1.20–3.30; p = 0.008; AA + AG vs GG: OR = 1.92; 95%CI = 1.18–3.12; p = 0.009), TLR6 rs5743810 (TC vs TT: OR = 0.58; 95%CI = 0.39–0.85; p = 0.005), IL-10 rs1800871 (TC vs CC: OR = 1.19; 95%CI = 1.01–1.41; p = 0.044), rs1800872 (AC vs AA: OR = 1.53; 95%CI = 1.22–1.92; p < 0.001) and MIF rs755622 (CG versus GG: OR = 1.33; 95%CI = 1.07–1.67; p = 0.012) polymorphisms were associated with the risk of childhood leukemia. No significant correlations were found between SNPs in other genes and the childhood leukemia risk. Subgroup analyses of rs1800871 and rs1800872 confirmed the conclusions obtained in their overall meta-analytical processes.
Conclusion:
CXCL12 rs1801157, TLR6 rs5743810, IL-10 rs1800871, rs1800872 and MIF rs755622 polymorphisms may represent candidate biomarkers for the risk prediction of childhood leukemia.
Introduction
Leukemia, characterized by abnormal hematopoietic function and clonal proliferation of immature lymphocytes (ALL) or myeloid cells (AML) in the bone marrow and peripheral blood, is the leading malignancy in children, with an estimated 65,111 cases diagnosed in 2018 worldwide [Citation1]. Although advances in treatment options (e.g. chemoradiotherapy, immunotherapy and hematopoietic stem cell transplantation) have led to an improved survival rate in children, leukemia remains an incurable disease where a substantial proportion of cases succumb to subsequent relapses and eventual deaths [Citation1–4]. Therefore, it is necessary to identify biomarkers that early predict the risk for developing childhood leukemia and disease recurrence, and then guide individualized prevention strategies and treatments to reduce possible burdens on individuals, families and health care systems.
Although the etiology of childhood leukemia is complex, genetic susceptibility has been considered as an important contributor [Citation5]. Single nucleotide polymorphism (SNP) that involves a single base-pair substitution to influence the expression or function of genes is the most common type for genetic susceptibility [Citation6]. There is mounting evidence to support the important role that inflammation plays in the development and progression of leukemia [Citation7]. The levels of several cytokines, including tumor necrosis factor α (TNF-α), interleukin (IL)-1, IL-6, IL-8, interferon-γ, IL-12 and IL-10 were found to be significantly higher in children with ALL compared with the controls [Citation8,Citation9]. Elevated IL-4, IL-10, IL-2, TNF-α and macrophage migration inhibitory factor (MIF) were shown to be significantly associated with higher relapse rates or/and lower survival rates in pediatric leukemia patients [Citation10–12]. IL-7 was demonstrated to mediate the viability, growth and proliferation of T-ALL cells via activation of the STAT5 pathway [Citation13]. The highly expressed IL-10 may help AML cells to evade immune surveillance by driving their stemness via activation of the PI3 K/AKT signal pathway [Citation14]. Hence, functional SNPs in inflammatory genes may represent candidate biomarkers for the risk prediction of childhood leukemia.
Currently, there had been several studies which reported the associations of SNPs in inflammatory genes with the risk of childhood leukemia. For example, Alves et al. identified that the IL-1β CC genotype of rs19644 was associated with a significantly increased risk for pediatric ALL [CC vs. CT + TT: odds ratio (OR), 2.48; 95% confidence interval (CI), 1.26–4.88, p = 0.006; CC vs. CT: OR, 2.74; 95% CI: 1.37–5.51, p = 0.003] [Citation15]. Lo et al. found that compared with those harboring the TT genotype of polymorphic loci rs1800871 in IL-10, populations carrying CC (OR, 0.33; 95% CI: 0.16–0.68, p = 0.0034) genotype had decreased risks for the development of childhood ALL [Citation16]. Heterozygosity (AC: OR, 1.73; 95% CI: 1.19–2.51, p = 0.0053) and homozygosity (CC: OR, 6.34; 95% CI: 3.16–12.72, p = 0.0001) of rs1800872 for IL-10 were also reported to confer increased risks of childhood ALL [Citation16]. However, controversial results were revealed in some studies: no significant associations existed between the risk of childhood leukemia and rs19644, rs1800871 and rs1800872 polymorphisms regardless of the genetic models analyzed (p > 0.05) [Citation17–19]. These differences may result from the small sample size and population heterogeneity in individual studies.
In the present study, we aimed to conduct a comprehensive meta-analysis that integrated data from all published studies to confirm the associations between SNPs in inflammation-related genes and the susceptibility to childhood leukemia.
Materials and methods
Search strategy
This meta-analysis was conducted in accordance with the preferred reporting items for systematic reviews and meta-analysis (PRISMA) 2020 checklist. Electronic databases of PubMed, EMBASE, Cochrane Library, China National Knowledge Internet (CNKI) and Wan Fang were searched to screen relevant studies published up to November 2022. The combinations of the search keywords included (‘inflammation’ OR ‘inflammasome’ OR ‘immune’ OR ‘cytokine’ OR ‘chemokine’ OR ‘interleukin’ OR ‘tumor necrosis factor-α’ OR ‘interferon’ OR ‘transforming growth factor-beta’ OR ‘toll-like receptor’) AND (‘polymorphism’ OR ‘variant’ OR ‘variation’ OR ‘mutation’ OR ‘genetic association’) AND (‘leukemia’ OR ‘hematologic malignancy’) AND (‘pediatric’ OR ‘childhood’ OR ‘children’). The reference lists of all relevant studies and previous reviews were also checked to identify potentially eligible articles.
Inclusion and exclusion criteria
The studies that met the following inclusion criteria were included: (1) evaluated the associations of SNPs in inflammation-related genes and the susceptibility to childhood leukemia and clinical outcomes (such as relapse or survival); (2) in a case–control or cohort design; (3) provided the genotype frequencies of cases and controls for estimation of ORs and 95%CIs; and (4) the full-texts published in English or Chinese were available. The exclusion criteria were: (1) repeated publications; (2) case reports, reviews, conference abstracts, animal or cell studies; (3) adult leukemia samples were mixed; (4) polymorphisms were examined in less than two articles; (5) data could not be retrievable; and (6) studies did not focus on the related topic.
Data extraction and quality assessment
Data were extracted from the included studies by two authors independently and any disagreements were resolved through discussion with the third author. The following information were retrieved from the included studies: first author, year of publication, country of origin, study design, subtype of acute leukemia, genotyping method, sample size, matched condition in age and frequencies of genotypes and alleles in case and control groups. The Newcastle-Ottawa scale (NOS) [Citation20] was used to evaluate the quality of included studies. NOS scores ranged from zero to nine and studies with NOS scores ≥ 6 were considered to be of high quality.
Statistical analysis
All the statistical analyses were performed using STATA version 15.0 (Stata Corp., College Station, TX, USA). The pooled ORs and 95%CIs were calculated to estimate the relationships between genetic polymorphisms in inflammation-related genes and the risk of childhood leukemia or clinical outcomes under five genetic models, including allelic (B vs A), homozygous (BB vs AA), heterozygous (BA vs AA; BA vs. BB), dominant (BB + BA vs AA) and recessive (BB vs BA + AA). The major (A) and minor (B) alleles were defined according to the SNP database (http://www.ncbi.nlm.nih.gov/snp). The Z-test was conducted to determine the significance of the accumulated ORs, with a p-value < 0.05 set as the statistical threshold. The heterogeneity among studies was measured by Cochrane’s Q-square test and I2 statistics. A random-effect model (DerSimonian-Laird method) was used to combine the results of eligible studies if there was statistical heterogeneity (p < 0.1 and I2 > 50%); otherwise, a fixed-effect model (Mantel-Haenszel method) was utilized. Subgroup analyses stratified by ethnicity (Asian and non-Asian), sample size (< 200 and < 200), genotyping method (PCR and high-throughput) or study design (single center and multiple-center) were conducted for SNPs with the number of included studies larger than 5 to investigate their influences on the results. Publication bias was assessed by Egger’s linear regression test. The trim-and-fill method was applied to adjust the pooled effects when there was evidence of publication bias (p < 0.05). A sensitivity analysis was performed to confirm the stability of the results by omitting one study at a time and pooling the data of other studies.
Results
Literature search
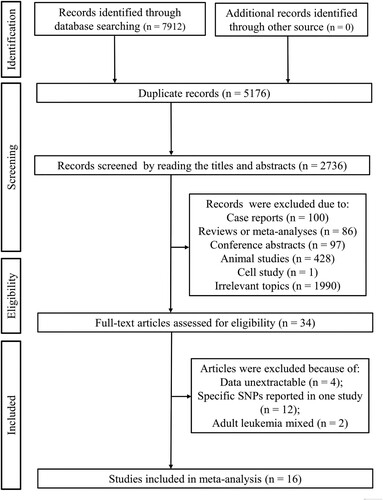
The flow chart of the literature search is shown in . A total of 7912 records were retrieved from the initial search of the electronic databases. Of them, 5176 articles were duplicates and excluded. Next, the title and abstract were read, followed by which 2702 were removed because they were case reports (n = 100), reviews (n = 86), conference abstracts (n = 97), animal studies (n = 428), cell study (n = 1) and irrelevant topics (n = 1990). Subsequently, the full-texts of the left 34 studies were downloaded and entered into the final review to assess their eligibility. As a result, 18 studies were further excluded due to the following reasons: data unextractable (n = 4), specific SNPs reported only in one study (n = 12) and adult leukemia samples mixed (n = 2). Finally, 16 studies were included in this meta-analysis [Citation15–19, Citation21–31].
Study characteristics
The main characteristics of included studies are summarized in . These 16 studies were published from 2001 to 2022. Three studies recruited childhood leukemia patients from Asian countries (mainland of China, n = 2; Taiwan of China, n = 1) and the remaining 13 studies analyzed the non-Asian populations (Germany, n = 3; Brazil, n = 4; Netherlands, n = 1; Greece, n = 1; France, n = 2; Egypt, n = 1; Italy, n = 1). Fourteen studies investigated the differences in the genotype and allele distributions of SNPs between childhood leukemia cases (n = 2771) and healthy controls (n = 3753); two studies compared the SNPs between childhood leukemia patients with (n = 104) and without relapses (n = 156). The controls were healthy children in nine studies (that is, age-matched), while in other studies, the control samples were collected from blood donors, including children and adults. Genetic polymorphisms in 13 inflammation-related genes, including C-X-C motif chemokine ligand 12 (CXCL12), toll-like receptor (TLR)−4, TLR6, TLR9, CD14, IL-1β, NLR family pyrin domain containing 3 (NLRP3), IL-4, interleukin 4 receptor (IL-4R), IL-10, IL-13, MIF and tumor necrosis factor (TNF)-α, were analyzed in childhood ALL patients. The main genotyping method was PCR (based on sequence-specific primers, restriction fragment length polymorphism or competitive allele-specific); also, high-throughput techniques (including an Illumina 370k Quad platform, Mass Array, and a custom platform of 96 SNPs) were used in three studies. The NOS score of all studies was higher than 6, indicating they were of high quality ().
Table 1. Characteristics of the included studies.
Meta-analysis of CXCL12 polymorphisms and the risk of childhood leukemia
Two studies [Citation22, Citation25] evaluated the relationship between CXCL12 rs1801157 polymorphism and childhood leukemia. Using a fixed-effects model (I2 = 0, p > 0.05), the pooled results showed that CXCL12 rs1801157 polymorphism was significantly associated with the risk of childhood ALL under heterozygous (AG vs GG: OR = 1.99; 95%CI = 1.20–3.30; p = 0.008) and dominant (AA + AG vs GG: OR = 1.92; 95%CI = 1.18–3.12; p = 0.009) models. No significant associations were observed in other comparisons (p > 0.05; ).
Table 2. Meta-analysis results to estimate the association of polymorphisms in inflammation-related genes with the susceptibility to childhood leukemia.
Meta-analysis of TLR6 polymorphisms and the risk of childhood leukemia
Two studies [Citation23, Citation24] investigated the association between TLR6 rs5743810 polymorphism and childhood leukemia. The pooled results showed that relative to the carriers with the TT genotype, populations with the TC genotype had a decreased risk to develop childhood ALL (OR = 0.58; 95%CI = 0.39–0.85; p = 0.005), which was estimated using a fixed-effects model (I2 = 0, p = 0.976). No significant association was found in the analysis of other genetic models (p > 0.05; ).
Meta-analysis of IL-10 polymorphisms and the risk of childhood leukemia
Six studies [Citation16–19, Citation21, Citation26] were included for the meta-analysis of rs1800896 polymorphism and five studies [Citation16–19, Citation21] were eligible for the meta-analysis of rs1800871 and rs1800872 polymorphisms in IL-10. The overall results indicated that there were no significant associations between IL-10 rs1800896 polymorphism and the susceptibility to childhood leukemia under any genetic models, while rs1800871 (TC vs CC: OR = 1.19; 95%CI = 1.01–1.41; p = 0.044) () and rs1800872 (AC vs AA: OR = 1.53; 95%CI = 1.22–1.92; p < 0.001) () polymorphisms had significant relationships with the risk of childhood ALL under a heterozygous model ().
Figure 2. Forest plot of the association between IL-10 rs1800871 polymorphism and childhood ALL patients under the heterozygous (TC vs CC) model. OR, odds ratio; CI, confidence interval.
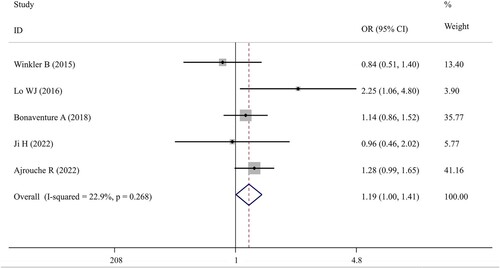
Figure 3. Forest plot of the association between IL-10 rs1800872 polymorphism and childhood ALL patients under the heterozygous (AC vs AA) model. OR, odds ratio; CI, confidence interval.
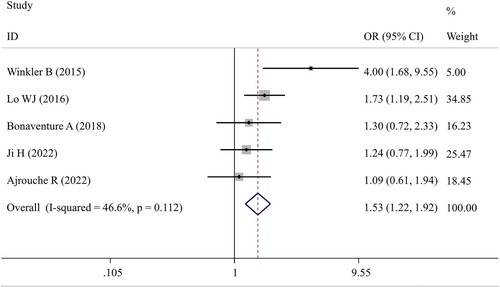
The negative associations between rs1800896 polymorphism and childhood leukemia were confirmed in all subgroups stratified by ethnicity, sample size, genotyping method or study design. In line with the overall analysis, positive results were detected for the rs1800871 polymorphism under the heterozygous model in the subgroup with the samples collected from multiple centers (TC vs CC: OR = 1.26; 95%CI = 1.05–1.52; p = 0.013) (). Additionally, the recessive model (TT vs TC + CC: OR = 1.44; 95%CI = 1.05–1.98; p = 0.025) () was revealed to be significantly associated with an increased risk of childhood ALL in the subgroup analysis restricted to studies with PCR as the genotyping method. These two results indicated that the populations with the TT genotype may have the highest risk to develop childhood ALL, followed by TC and then CC. For the rs1800872 polymorphism, subgroup analysis results demonstrated that the heterozygous genotype AC may confer an increased risk of developing childhood ALL compared to AA genotype, especially for Asian populations (OR = 1.51; 95%CI = 1.10–2.08; p = 0.01), PCR-based genotyping groups (OR = 2.39; 95%CI = 1.07–5.34; p = 0.034) or multiple-center trials (OR = 1.46; 95%CI = 1.11–1.92; p = 0.007) (). Moreover, subgroup analysis of studies with genotyping performed by high-throughput platforms found that the childhood ALL risk of genotype CC was significantly lower than that of genotype AC (OR = 1.24; 95%CI = 1.03–1.49; p = 0.025) (). More interestingly, AA genotype seemed to exert a protective effect against childhood ALL (vs CC determined via PCR-based genotyping methods: OR = 0.18; 95%CI = 0.11–0.31; p < 0.001), which was also indicated according to the reduced significance in the dominant model analysis (AA + AC vs CC: p = 0.042 following high-throughput genotyping) ().
Table 3. Subgroup analysis for the association of IL-10 polymorphisms and childhood leukemia.
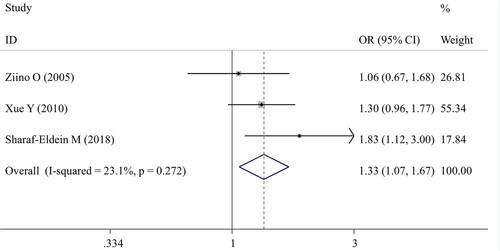
Meta-analysis of MIF polymorphisms and the risk of childhood leukemia
Three studies [Citation27, Citation28, Citation31] examined the effects of MIF rs755622 polymorphism on the development of childhood leukemia. The pooled results showed a significant association in the CG versus GG genotype (OR = 1.33; 95%CI = 1.07–1.67; p = 0.012) (; ). The comparisons of other models did not detect any statistical associations.
Meta-analysis of other polymorphisms and the risk of childhood leukemia or outcomes
There were two studies to explore the associations between TLR4 rs4986790 [Citation23, Citation24], TLR9 rs187084 [Citation23,Citation24], CD14 rs187084 [Citation23,Citation24], IL-1β rs16944 [Citation15,Citation18], NLRP3 rs10754558 [Citation15,Citation18], IL-4 rs2070874 [Citation17, Citation32], rs2243250[Citation17, Citation32], IL-13 rs20541 [Citation17, Citation32], IL-4R rs1801275 [Citation17, Citation32] polymorphisms and the risk of childhood leukemia, respectively. The pooled meta-analyses failed to find any significant associations between all these polymorphisms and the risk of childhood leukemia under various genetic models (p > 0.05; ).
Two studies reported the association of TNF-α rs1800629 polymorphisms with the risk of relapses in childhood leukemia cases [Citation29, Citation30]. The integrated analysis indicated no evidence of correlations between TNF-α rs1800629 and the relapses of childhood leukemia patients under all genetic models (p > 0.05; ).
Publication bias and sensitivity analysis
Egger’s test was used to examine the publication bias for the IL-10 and MIF polymorphisms because they were analyzed with the number of studies ≥ 3. The results implied that there was no significant publication bias for all three polymorphic loci in IL-10 (rs1800896, rs1800871 and rs1800872) and rs755622 polymorphism in MIF under all genetic models (p > 0.05). The sensitivity analysis also suggested that the removal of any single study did not influence the pooled ORs materially ().
Discussion
The present meta-analysis explored the associations of polymorphisms in inflammation-related genes with the risk of childhood leukemia and clinical outcomes by pooling the data from 16 studies. The integrated analysis results showed that CXCL12 rs1801157, TLR6 rs5743810, IL-10 rs1800871, rs1800872 and MIF rs755622 polymorphisms were significantly associated with the risk of childhood ALL. The trends of their associations remained unchanged after the removal of any one study in sensitivity analyses, suggesting our meta-analysis results were robust.
Among significant polymorphisms, the associations of IL-10 rs1800871 and rs1800872 with leukemia were previously investigated by meta-analyses [Citation33–35]. However, these published meta-analyses mainly focused on adult leukemia [Citation33–35] and no subgroup analysis was performed to distinguish the association difference between adult and childhood leukemia if both of them were included [Citation34, Citation35]. In the present study, we, for the first time, specifically analyzed the relationships of IL-10 rs1800871 and rs1800872 with the susceptibility to childhood ALL. Also, compared with the studies of Gao et al. [Citation35] and Hong et al. [Citation34], we further included two novel studies published in 2022 and a large sample size was involved in these two studies (n = 1115 in the study of Ajrouche et al. [Citation32]; n = 340 in the study of Ji et al. [Citation18]). Thus, our study may be innovative and the obtained conclusions may be more credible. The novelty of our study was also reflected by the different results between our study and published meta-analyses: that is, our overall meta-analysis showed that IL-10 rs1800871 and rs1800872 were significantly associated with childhood ALL under TC vs CC (OR = 1.19; p = 0.044) and AC vs AA (OR = 1.53; p < 0.001) heterozygous models, but the published meta-analyses did not detect any significant associations under various models [Citation33–35]. Our subgroup analysis further confirmed the overall analysis conclusions of rs1800871 and added a new result (that is, also significant under TT vs TC + CC model: OR = 1.44; p = 0.025); however, negative conclusions were still achieved in the subgroup analyses of published meta-analyses [Citation34, Citation35]. The heterozygous (AC vs AA) model in IL-10 rs1800872 polymorphism was indicated to mainly influence the susceptibility to childhood leukemia in Asians, which was not reported in published meta-analyses [Citation34,Citation35]. Similar to the results of Gao et al. [Citation35], heterozygous (AC vs CC) and dominant (AA + AC vs CC) models were shown to confer increased susceptibility to childhood ALL; nevertheless, our results were not obtained in subgroups stratified by race like Gao et al. [Citation35] (mainly significant for non-Chinese populations), although the significance was seen to be relatively obvious in non-Asians (p < 0.1) than Asians (p > 0.1) in our study.
Both rs1800871 and rs1800872 polymorphisms belonged to single nucleotide substitutions in the upstream of the transcriptional start site in the promoter region; thus, they were believed to alter the transcriptional activity of IL-10. This hypothesis had been demonstrated by previous studies: Kumari et al. found that serum IL-10 level was significantly higher for the variant genotypes TC (6.01 ± 3.20) and TT (6.53 ± 2.74) at rs1800871 relative to genotype CC (4.83 ± 2.72) [Citation36]. Xia et al. also detected a higher protein level of IL-10 in the TT genotype compared to the CC + CT genotype (10.82 ± 7.33 vs 7.62 ± 6.96 pg/ml) at rs1800871 [Citation37]. IL-10 rs1800872 (−592 C/A) polymorphism analysis revealed that the IL-10 level was higher in cervical cancer (22.26 ± 11.19 vs 20.67 ± 10.67) [Citation38] and pituitary adenoma (0.392 ± 1.665 vs 0.171 ± 0) [Citation39] patients carrying the AC genotype compared with AA genotype carriers. Moreover, the IL-10 level of genotype CC was found to be significantly lower than that of the other genotype (AA + AC) in the HPV-infected cervical cancer group [Citation40]. From these findings, we speculated that the increased risk to develop childhood leukemia in IL-10 rs1800871 TC and TT genotypes or rs1800872 AC genotype carriers may be attributed to the high expression of IL-10. This speculation was indirectly confirmed according to the results obtained from previous studies which indicated the level of IL-10 was significantly increased in pediatric leukemia patients compared with the healthy controls and elevated IL-10 predicted poor response to therapy, relapses and deaths from leukemia [Citation8, Citation12, Citation41]. Also, in vitro studies showed that exposure to IL-10 stimulated the clonogenic growth of leukemia cells [Citation42]. Inhibition of IL-10 production drove innate immunity-mediated clearance of leukemic cells and lengthened the survival of leukemic model mice [Citation43].
Since rare studies reported the associations of CXCL12 rs1801157, TLR6 rs5743810 and MIF rs755622 polymorphisms with childhood leukemia, no published meta-analysis was found until now. Although two or three studies were included in our study, the sample size of them was considerable, especially MIF (case, n = 677; control, n = 1021), indicating our conclusions may be believable. In line with the analysis of chronic adult myeloid leukemia [Citation44] and other cancers [Citation45], there was a significant association between the CXCL12 (also known as SDF1) rs1801157 polymorphism and the risk of childhood leukemia under the heterozygous (AG vs GG: OR = 1.99; p = 0.008) and dominant (AA + AG vs GG: OR = 1.92; p = 0.009) models. Compared to G, the A mutation in the 3’-untranslated region of CXCL12 (rs1801157) led to an upregulated expression level of CXCL12 [Citation46], while the high concentrations of CXCL12 could promote the proliferation [Citation47] and mediate trans-endothelial migration of leukemia cells [Citation48] via binding with its receptor CXCR4. The SNP rs5743810 (745 C > T) is located in the exonic region of chromosome 4 and the C-to-T mutation resulted in a change from serine to proline at the position 249 in the extracellular domain of the protein. This non-synonymous mutation (allele 745 T) was reported to significantly reduce the NF-κB signaling activity in comparison to allele 745C and cause the low production of downstream IL-6 [Citation49]. The expression of IL-6 was detected to be upregulated in childhood ALL patients [Citation8,Citation9]. Thus, theoretically, populations with TC or TT genotype may have a decreased risk for developing childhood ALL (especially TC), which was confirmed in the study of Xabregas et al. [Citation23]. The MIF rs755622 polymorphism is located in the binding site of the response element of transcription factor activator protein 4 (AP4). The MIF - 173C mutational allele was reported to create a putative binding site for AP4 and then enhance the transcription of the MIF gene [Citation50]. Multivariate cox-regression analysis identified high MIF expression as the only independent predictor for poor prognosis of childhood ALL patients [Citation11]. The use of MIF inhibitor inhibited bone marrow mesenchymal stromal cell-induced leukemia survival [Citation51]. Thus, subjects with CG genotype had a higher risk to develop childhood leukemia than those with GG genotype, which was demonstrated in our study.
Some limitations should be acknowledged to explain the results. First, the number of included studies for most of the polymorphic loci (except of IL-10) was small, which may lead to an underestimation or overestimation of their associations with childhood leukemia. Second, only ALL cases were investigated in our included studies. Whether there were associations of CXCL12 rs1801157, TLR6 rs5743810, IL-10 rs1800871, rs1800872 and MIF rs75562 with childhood AML patients, was unclear. Third, the associations of these SNPs with patients’ prognoses were also unknown. Fourth, it is generally believed that childhood leukemia is a disease resulting from complex interactions between genetic and environmental factors. However, our pooled results were calculated only based on the primary data which were not adjusted by environmental factors because of lacking relevant data. Therefore, more well-designed studies (including sex – and age-matched controls, different ethnic backgrounds or environmental exposures) with larger sample sizes are still needed to further validate the associations of SNPs in various inflammation-related genes and the vulnerability to childhood leukemia.
Conclusions
This meta-analysis provides evidence that the heterozygous genotypes of CXCL12 rs1801157 (AG), TLR6 rs5743810 (TC), IL-10 rs1800871 (TC), rs1800872 (AC) and MIF rs755622 (CG) polymorphisms may be particularly associated with the risk of childhood ALL. Populations with these genotypes should be periodically monitored and be given some anti-inflammatory diets (or drugs as a combined therapy) to prevent the development of childhood leukemia and poor prognosis. However, future analysis with more included studies is necessary to confirm our conclusions.
Availability of data, code and other materials
All data can be obtained in published articles.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Sharma R. A systematic examination of burden of childhood cancers in 183 countries: estimates from GLOBOCAN 2018. Eur J Cancer Care (Engl). 2021;30(5):e13438.
- Dufort YAG, Castiglioni M, Pagés C, et al. Treatment of childhood acute myeloid leukemia in Uruguay: results of 2 consecutive protocols over 20 years. J Pediatr Hematol. 2020;42(5):359–366.
- Gottschalk Højfeldt S, Grell K, Abrahamsson J, et al. Relapse risk following truncation of pegylated asparaginase in childhood acute lymphoblastic leukemia. Blood. 2021;137(17):2373–2382.
- Eckert C, Parker C, Moorman AV, et al. Risk factors and outcomes in children with high-risk B-cell precursor and T-cell relapsed acute lymphoblastic leukaemia: combined analysis of ALLR3 and ALL-REZ BFM 2002 clinical trials. Eur J Cancer. 2021;151:175–189.
- Bloom M, Maciaszek JL, Clark ME, et al. Recent advances in genetic predisposition to pediatric acute lymphoblastic leukemia. Expert Rev Hematol. 2020;13(1):55–70.
- Gutierrez-Camino A, Martin-Guerrero I, García-Orad A. Genetic susceptibility in childhood acute lymphoblastic leukemia. Med Oncol. 2017;34(10):179.
- Urwanisch L, Luciano M, Horejs-Hoeck J. The NLRP3 inflammasome and Its role in the pathogenicity of leukemia. Int J Mol Sci. 2021;22(3):1271.
- Pérez-Figueroa E, Sánchez-Cuaxospa M, Martínez-Soto KA, et al. Strong inflammatory response and Th1-polarization profile in children with acute lymphoblastic leukemia without apparent infection. Oncol Rep. 2016;35(5):2699–2706.
- Latoch E, Konończuk K, Konstantynowicz-Nowicka K, et al. Asymptomatic survivors of childhood acute lymphoblastic leukemia demonstrate a biological profile of inflamm-aging early in life. Cancers (Basel). 2022;14(10):2522.
- Kaya Z, Yüce D, Kirkiz S, et al. Prognostic role of serum cytokines and soluble HLA-G levels in children with leukemia who undergo allogeneic stem cell transplantation. Cytokine. 2022;153:155869.
- Sharaf-Eldein M, Elghannam D, Elderiny W, et al. Prognostic implication of MIF gene expression in childhood acute lymphoblastic leukemia. Clin Lab. 2018;64(9):1429–1437.
- Stevens AM, Horton TM, Glasser CL, et al. IL-10 and TNFα are associated with decreased survival in low-risk pediatric acute myeloid leukemia; a children's oncology group report. Pediatr Hematol Oncol. 2023;40(2):147–158.
- Ribeiro D, Melão A, van Boxtel R, et al. STAT5 is essential for IL-7-mediated viability, growth, and proliferation of T-cell acute lymphoblastic leukemia cells. Blood Adv. 2018;2(17):2199–2213.
- Xu Y, Mou J, Wang Y, et al. Regulatory T cells promote the stemness of leukemia stem cells through IL10 cytokine-related signaling pathway. Leukemia. 2022;36(2):403–415.
- Alves FS, Xabregas LA, Kerr MWA, et al. Genetic polymorphisms of inflammasome genes associated with pediatric acute lymphoblastic leukemia and clinical prognosis in the Brazilian Amazon. Sci Rep. 2021;11(1):9869.
- Lo WJ, Chang WS, Hsu HF, et al. Significant Association of Interleukin-10 Polymorphisms with Childhood Leukemia Susceptibility in Taiwan. In vivo. 2016;30(3):265–269.
- Bonaventure A, Orsi L, Rudant J, et al. Genetic polymorphisms of Th2 interleukins, history of asthma or eczema and childhood acute lymphoid leukaemia: Findings from the ESCALE study (SFCE). Cancer Epidemiol. 2018;55:96–103.
- Ji H, Li F, Song L, et al. Inflammation-related gene polymorphisms associated With childhood acute lymphoblastic leukemia. J Pediatr Hematol Oncol. 2023;45(1):e9–e13.
- Ajrouche R, Chandab G, Petit A, et al. Allergies, genetic polymorphisms of Th2 interleukins, and childhood acute lymphoblastic leukemia: The ESTELLE study. Pediatr Blood Cancer. 2022;69(3):e29402.
- Lo CK, Mertz D, Loeb M. Newcastle-Ottawa Scale: comparing reviewers’ to authors’ assessments. BMC Med Res Methodol. 2014;14:45.
- Winkler B, Taschik J, Haubitz I, et al. TGFβ and IL10 have an impact on risk group and prognosis in childhood ALL. Pediatr Blood Cancer. 2015;62(1):72–79.
- de Lourdes Perim A, Guembarovski RL, Oda JM, et al. CXCL12 and TP53 genetic polymorphisms as markers of susceptibility in a Brazilian children population with acute lymphoblastic leukemia (ALL). Mol Biol Rep. 2013;40(7):4591–4596.
- Xabregas LA, Hanna FSA, Magalhães-Gama F, et al. Association of Toll-like receptors polymorphisms with the risk of acute lymphoblastic leukemia in the Brazilian Amazon. Sci Rep. 2022;12(1):15159.
- Miedema KG, Tissing WJ, Poele T, et al. Polymorphisms in the TLR6 gene associated with the inverse association between childhood acute lymphoblastic leukemia and atopic disease. Leukemia. 2012;26(6):1203–1210.
- Kampouraki E, Lourou M, Zervou MI, et al. Role of CXCL12, TP53 and CYP1A1 gene polymorphisms in susceptibility to pediatric acute lymphoblastic leukemia. Oncol Lett. 2021;22(3):659.
- Hiroki CH, Amarante MK, Petenuci DL, et al. IL-10 gene polymorphism and influence of chemotherapy on cytokine plasma levels in childhood acute lymphoblastic leukemia patients. Blood Cells Mol Dis. 2015;55(2):168–172.
- Xue Y, Xu H, Rong L, et al. The MIF −173G/C polymorphism and risk of childhood acute lymphoblastic leukemia in a Chinese population. Leuk Res. 2010;34(10):1282–1286.
- Sharaf-Eldein M, Elghannam D, Abdel-Malak C. MIF-173G/C (rs755622) polymorphism as a risk factor for acute lymphoblastic leukemia development in children. J Gene Med. 2018;20(9):e3044.
- Stanulla M, Schrauder A, Welte K, et al. Tumor necrosis factor and lymphotoxin-alpha genetic polymorphisms and risk of relapse in childhood B-cell precursor acute lymphoblastic leukemia: a case-control study of patients treated with BFM therapy. BMC Blood Disord. 2001;1(1):2.
- Lauten M, Matthias T, Stanulla M, et al. Association of initial response to prednisone treatment in childhood acute lymphoblastic leukaemia and polymorphisms within the tumour necrosis factor and the interleukin-10 genes. Leukemia. 2002;16(8):1437–1442.
- Ziino O, D'Urbano LE, De Benedetti F, et al. The MIF-173G/C polymorphism does not contribute to prednisone poor response in vivo in childhood acute lymphoblastic leukemia. Leukemia. 2005;19(12):2346–2347.
- Ajrouche R, Chandab G, Petit A, et al. Allergies, genetic polymorphisms of Th2 interleukins, and childhood acute lymphoblastic leukemia: The ESTELLE study. Pediatr Blood Cancer. 2022;69(3):e29402.
- Ahadi HR, Sadrabadi AE, Jalili A, et al. Association of Interleukin 10 (IL-10) Gene Polymorphism (819 T > C) with Susceptibility to Acute Myeloid Leukemia: A Meta-Analysis. Iran J Public Health. 2022;51(1):19–26.
- Hong P, Feng WY, Fu LH, et al. Associations between genetic polymorphisms in interleukin-10 and hematological oncology: evidence from a meta-analysis. Cancer Biol Ther. 2020;21(4):372–378.
- Gao S, Tang K, Chen J, et al. The single nucleotide polymorphisms of interleukin-10 are associated with the risk of leukaemia: Evidence from 18 case-control studies. Medicine (Baltimore). 2020;99(44):e22929.
- Kumari R, Kumar S, Ahmad MK, et al. Promoter variants of TNF-α rs1800629 and IL-10 rs1800871 are independently associated with the susceptibility of coronary artery disease in north Indian. Cytokine. 2018;110:131–136.
- Xia L, Chen M, Bi D, et al. Combined analysis of interleukin-10 gene polymorphisms and protein expression in children With cerebral palsy. Front Neurol. 2018;9:182.
- Zidi S, Gazouani E, Stayoussef M, et al. IL-10 gene promoter and intron polymorphisms as genetic biomarkers of cervical cancer susceptibility among Tunisians. Cytokine. 2015;76(2):343–347.
- Palivonaite M, Gedvilaite G, Glebauskiene B, et al. IL-10 gene Rs1800871, Rs1800872, and Rs1800896 polymorphisms and IL-10 serum levels association with pituitary adenoma. Biomedicines. 2022;10(8):1921.
- Berti FCB, Pereira APL, Trugilo KP, et al. IL-10 gene polymorphism c.−592C > A increases HPV infection susceptibility and influences IL-10 levels in HPV infected women. Infect Genet Evol. 2017;53:128–134.
- Bien E, Balcerska A, Adamkiewicz-Drozynska E, et al. Pre-treatment serum levels of interleukin-10, interleukin-12 and their ratio predict response to therapy and probability of event-free and overall survival in childhood soft tissue sarcomas, Hodgkin's lymphomas and acute lymphoblastic leukemias. Clin Biochem. 2009;42(10-11):1144–1157.
- Nishioka C, Ikezoe T, Yang J, et al. CD82 regulates STAT5/IL-10 and supports survival of acute myelogenous leukemia cells. Int J Cancer. 2014;134(1):55–64.
- El Hajj H, Hleihel R, El Sabban M, et al. Loss of interleukin-10 activates innate immunity to eradicate adult T-cell leukemia-initiating cells. Haematologica. 2021;106(5):1443–1456.
- Zhang X, Fan Y, Li Z. SDF1-3'A polymorphism is associated with increased risk of hematological malignancy: a meta-analysis. Onco Targets Ther. 2017;10:1575–1583.
- Tong X, Ma Y, Deng H, et al. The SDF-1 rs1801157 Polymorphism is Associated with Cancer Risk: An Update Pooled Analysis and FPRP Test of 17,876 Participants. Sci Rep. 2016;6:27466.
- Garcia-Moruja C, Rueda P, Torres C, et al. Molecular phenotype of CXCL12beta 3'UTR G801A polymorphism (rs1801157) associated to HIV-1 disease progression. Curr HIV Res. 2009;7(4):384–389.
- Mowafi F, Cagigi A, Matskova L, et al. Chemokine CXCL12 enhances proliferation in pre-B-ALL via STAT5 activation. Pediatr Blood Cancer. 2008;50(4):812–817.
- Cui XY, Tjønnfjord GE, Kanse SM, et al. Tissue factor pathway inhibitor upregulates CXCR7 expression and enhances CXCL12-mediated migration in chronic lymphocytic leukemia. Sci Rep. 2021;11(1):5127.
- Shey MS, Randhawa AK, Bowmaker M, et al. Single nucleotide polymorphisms in toll-like receptor 6 are associated with altered lipopeptide- and mycobacteria-induced interleukin-6 secretion. Genes Immun. 2010;11(7):561–572.
- Donn R, Alourfi Z, Benedetti FD, et al. Mutation screening of the macrophage migration inhibitory factor gene: Positive association of a functional polymorphism of macrophage migration inhibitory factor with juvenile idiopathic arthritis. Arthritis Rheum. 2002;46(9):2402–2409.
- Abdul-Aziz AM, Shafat MS, Mehta TK, et al. MIF-Induced stromal PKCβ/IL8 Is essential in human acute myeloid leukemia. Cancer Res. 2017;77(2):303–311.