ABSTRACT
Objective:
The lives and safety of humans are significantly threatened by acute myeloid leukemia (AML), which is proven to be the most prevalent acute leukemia. This work is therefore intended to investigate and analyze the expressions of miR-361-3p and Histone Lysine Methyltransferase 2A (KMT2A) in tissues and cell lines of AML and identify an advanced and novel target for the therapy of AML.
Methods:
The qRT-PCR and western blot assays were conducted to find expressions of miR-361-3p/KMT2A in AML PB and cell lines. After then, tests using CCK-8 and EdU were run to see how KMT2A affected the growth of AML cells. Transwell migration and invasion assay was conducted to evaluate KMT2A's contribution to the migration and invasion of AML cells. ENCORI and miRWalk predicted the association between KMT2A and miR-361-3p, and the dual-luciferase reporter experiment verified it. Furthermore, rescue studies were used to ascertain how KMT2A affected the miR-361-3p-regulated AML cells’ abilities to proliferate, migrate, and invade.
Results:
miR-361-3p was poorly expressed while KMT2A was abundantly expressed. Additionally, KMT2A downregulation prevented AML cells from proliferating. PCNA and Ki-67 protein levels fell when KMT2A was silent. Furthermore, AML cells’ motility, invasion, and metastasis were inhibited by low KMT2A expression. KMT2A was also identified as a direct target of miR-361-3p and negatively correlated with miR-361-3p. Finally, the over-expression of KMT2A partially reversed the inhibitory effects of up-regulation of miR-361-3p.
Conclusion:
A potential therapeutic candidate target for the treatment of AML may be miR-361-3p/KMT2A.
1. Introduction
The most prevalent form of leukemia in adults, acute myeloid leukemia (AML), has been proven to be a very diverse hematological cancer [Citation1]. The occurrence of AML is related to uncontrolled progressions of proliferating, apoptosis, and active invasion and migrating of leukemia cells [Citation2]. These days, chemotherapy and hematopoietic stem cell transplantation are the two major approaches for treating AML, but only a small number of patients complete hematopoietic stem cell transplantation, leaving the overall five-year survival rate at less than 20%, which is not the ideal outcome [Citation3,Citation4]. With advancing age, the incidence rate of AML is also increasing [Citation5]. For patients under 60, chemotherapy is currently advised. Patients over 60 rarely opt for chemotherapy due to its low rate of complete remission and harm to normal cells [Citation6]. Therefore, there was an urgent need for alternative therapy, and so far, much attention had been attached to targeted therapy.
According to studies and reports, the expression of microRNAs (miRNAs) is changed after post-transcriptional regulation in AML, and these miRNAs exert certain functional roles through cascade with downstream target genes, further leading to the occurrence of AML [Citation7]. For instance, the down-regulation of miR-1290 adversely regulates FOXG1 and SOCS3, which in turn suppresses proliferation and causes death in AML cells [Citation8]. Additionally, miR-34 has an inducing effect on apoptosis and prevents HL-60 and THP-1 cells from autophagy by targeting HMGB1, while miR-34a is less abundant in AML cells [Citation9]. Lung cancer, prostate cancer, pancreatic ductal adenocarcinoma, and other malignancies have all been linked to miR-361-3p, which is created from the miR-361 precursor [Citation10–12]. The expressions of miR-361-3p in AML and its potential function, however, are yet unclear. miRNAs have a strong correlation with genetic and chromosomal alterations as AML progresses [Citation13]. A comprehensive understanding of cancer-related miRNAs and their integrated networks with downstream target genes effectively guided the development of new targeted therapeutic strategies for AML.
Histone lysine methyltransferase 2A (KMT2A), as a member of the KMT family, is located on chromosome 11 and consists of 37 exons with a total length of about 89 Kb [Citation14]. Previous research has displayed that KMT2A has a close correlation with AML [Citation15]. For example, abnormal chromosome rearrangement of KMT2A produces MLL-AF9 fusion protein, causing AML in mice [Citation16]. Besides, Chen et al. confirm that the KMT2A fusion protein drives the expression of HOX, inducing leukemia transformation of hematopoietic progenitor cells [Citation17]. However, the precise roles and potential regulatory mechanisms of KMT2A in AML remain unknown.
Our research aimed to provide light on KMT2A's biological roles in AML cells as well as their related regulatory mechanisms. As anticipated, KMT2A was blatantly up-regulated while miR-361-3p was noticeably down-regulated in both AML PB and cell lines. Down-regulation of KMT2A significantly reduced AML cells’ abilities to proliferate, migrate and invade, whereas up-regulation of KMT2A had the opposite effect. Additionally, a dual-luciferase reporter experiment verified the targeted relationship between KMT2A and miR-361-3p which was anticipated by a bioinformatics investigation. Furthermore, as over-expression of KMT2A partially reversed the inhibitory effects of miR-361-3p mimic on proliferation, migration, and invasion of AML cells, miR-361-3p/KMT2A may offer a novel therapeutic potential target in the treatment of AML.
2. Materials and methods
2.1. Collection of AML samples
Peripheral blood (PB) samples were collected from 30 newly diagnosed KMT2A rearranged AML patients (14 females, 16 males) in Funing People's Hospital from June 3rd, 2020 to June 4th, 2021. And 30 healthy individuals (15 females, 15 males) were set up as controls. The samples collected were all kept at −80°C condition. All patients were approved by the medical ethics committee of Funing People's Hospital and signed the consent of patients before collection. According to French America British (FAB) and World Health Organization (WHO), AML was diagnosed. According to FAB classification, 30 AML patients include 6 M2 patients, 7 M3 patients, 9 M4 patients, and 8 M5 patients. The number of lymphocytes was 23–82 × 109/L. Leukocyte numbers of 18–45 × 109/L. They were all newly diagnosed patients with no history of other malignant tumors and did not receive any anti-tumor drug treatment.
2.2. Cell line and cell culture
The human AML cell lines (HL-60, KG-1A, KO52, and THP-1) were purchased from Beijing Zhongke Quality Inspection Biotechnology Co., Ltd., And bone marrow stromal cell line HS-5 was acquired from the American type culture collection (ATCC, Manassas, VA). In an incubator set at 37°C with 5% CO2 concentration, cells were grown in DMEM media containing 10% fetal bovine serum (FBS; Invitrogen; Grand Island, NY), 100 μU/mL penicillin; and 100 g/mL streptomycin.
2.3. Cell transfection
The Shanghai Institute of Nutrition and Health provided the miR-361-3p mimic and NC mimic, shRNA for KMT2A knockdown (sh-KMT2A) and negative control (sh-NC), and pcDNA3.1 for KMT2A overexpression (pcDNA-KMT2A) and negative control (pcDNA-NC) (Shanghai, China). The temporary transfection of AML cells was carried out using Lipofectamine 2000 (Invitrogen, USA).
2.4. CCK-8 assay
According to the procedure, the CCK-8 assay was conducted to detect cell proliferation. It was assessed using a CCK-8 kit (Beyotime Biotechnology, China) after transfected HL-60 and KO52 cells (1 × 104 cells per well) were grown in 96-well plates for 0, 24, 48, and 72 h, respectively. At 490 nm, the optical density was discovered.
2.5. Edu assay
After transfection, the EdU assay was performed using the EdU assay kit (RiboBio, China) to detect the proliferation of AML cells. HL-60 or KO52 cells (5 × 104/well) were inoculated into a 24-well plate and incubated with EdU (50 µM) for 2 h. Then, it was fixed with 4% neutral paraformaldehyde (Sigma) for 30 min at room temperature. Cells were permeabilized with 0.5% Triton X-100 (Sigma) for 20 min and then washed three times with PBS. Following the manufacturer's instructions, HL-60 and KO52 cells were also stained. The proportion of cells being positive for EdU was analyzed using the ImageJ software (NIH).
2.6. Transwell migration and invasion assays
For cell invasion, serum-free DMEM media was diluted with frozen BD Matrigel at a 1: 2 ratio before being deposited at 4°C. The transwell top chamber was then filled with the diluted Matrigel (100 μL per well), and it was incubated at 37°C for 4 h. HL-60 and KO52 cells were resuspended in serum-free DMEM media, adjusted to a concentration of 5 × 105/mL, and injected with 200 μL each well in the top chamber to study cell migration and invasion. There were 600 L of medium with 20% FBS in the bottom chambers. The non-migrated or invaded cells were removed after a 48-hour incubation period, and the migrated or invaded cells were counted under a Nikon microscope (Tokyo).
2.7. qRT-PCR analysis
AML Each group's cells were collected, given two rounds of PBS washing, and then total RNA was extracted with the assistance of the Trizol reagent. The PrimeScript reverse transcription reagent kit's instructions were followed while extracting 2 μL of total RNA for reverse transcription (Thermo Fisher Scientific, USA). Thermo Fisher Scientific and Applied Biosystems’ SYBR Green qPCR Master Mixes and Biosystems 7500 Sequence Detection System were used to carry out qRT-PCR. The normalization used U6 and -actin. In order to gauge the relative expression levels, the 2−ΔΔCT approach was employed. The reaction was conducted under the following conditions: prolonged at 72°C for 10 min after 30 cycles of 94°C for 3 min, 94°C for 45 s, 57°C for 45 s, and 72°C for 45 s. The primer sequences were KMT2A forward, 5’-CCGGGCA CTGTTAAACATTCCACTTCTCG-3’, and reverse, 5’-CTGTTAAACATTCCACTTCTCGAGAAGT G-3’; miR-361-3p forward, 5’-ACACTCCAGCTGGGTCCCCCAGGTGTGATTC-3’ and reverse, 5’-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGAAATCAGA-3’; β-actin forward, 5’-GTCACCTTCACCGTTCCAGTTTT-3’ and reverse, 5’-CTTAGTTGCGTTACACCCTTTCTT-3’; U6 forward, 5’-CTCGCTTCGGCAGCACA-3’, and reverse, 5’-AACGCTTCACGAATTTGCGT-3’.
2.8. Western blot analysis
AML cells in each group had their total protein removed using a RIPA kit. 10% SDS-PAGE was adopted to separate the protein, and next, it was transferred to a PVDF membrane. The first antibody was applied to the membrane at 4°C and incubated overnight. KMT2A (ab234435, 1: 1,000), PCNA (ab92552, 1: 1,000), Ki-67 (ab15580, 1: 1,000), MMP-2 (ab92536, 1: 1,000), MMP-9 (ab76003, 1: 1,000), and β-actin (ab8226, 1: 1,000) were among the primary antibodies purchased from Abcam. After that, PBST was applied after the membrane had been rinsed three times. For two hours, the membrane was treated. We utilized a room-temperature HRP-adjusted anti-rabbit secondary antibody.
2.9. Target prediction and dual-luciferase reporter assay
ENCORI, miRWalk, and TargetScan were adopted to predict the possible miRNAs targeting KAM2T, and miR-361-3p was chosen. The KAM2T 3'-UTR dual-luciferase reporter vectors for the wild-type and mutant KAM2T were created. Using the lipofectamine 2000, luciferase reporter vectors and miR-361-3p mimic were transfected into HL-60 and H562 cells (Invitrogen, CA, USA). Using a dual-luciferase reporter system (Berthold, Germany) and following the manufacturer's guidance and instructions, luciferase activity was assessed after 24 h.
2.10. Statistical analysis
Using Prism 6.0, statistical analysis was carried out (GraphPad, San Diego, CA, USA). The statistical outcomes were presented quantitatively as mean ± standard deviation (SD). Besides, student's t-test or one-way ANOVA was adopted to make a comparison between the results. P < 0.05 was considered statistically significant.
3. Results
3.1. KMT2A is up-regulated in AML PB and cell lines
Gathering peripheral blood samples from AML patients and using qRT-PCR to measure KMT2A mRNA expression were the first steps in figuring out how KMT2A influences the onset and progression of the disease. A shows that KMT2A was up-regulated in AML PB. Additionally, the expression of KMT2A in AML cells such as HL-60, KG-1a, KO52, and THP-1 was assessed using qRT-PCR and western blot tests. In instance, the HL-60 cells and KO52 cells depicted in B and C consistently demonstrated that KMT2A was highly expressed in AML cells. These results suggested that KMT2A could play an oncogenic role in the initiation and progression of AML.
Figure 1. KMT2A is up-regulated in AML PB as well as cell lines. (A) KMT2A expression in AML PB was discovered using a qRT-PCR assay. *P < 0.05 vs. control group. KMT2A mRNA, together with protein expressions, was found using (B) qRT-PCR as well as (C) western blot assays in AML cell lines. *P < 0.05, **P < 0.01 vs. HS-5 cells. All data were exhibited as mean ± SD. n = 3.
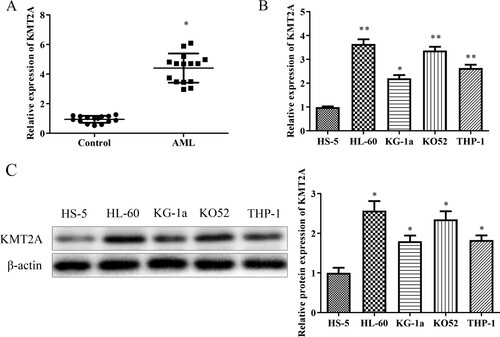
3.2. Down-regulation of KMT2A inhibits proliferation, migration and invasion of AML cells
To further understand the role of KMT2A in AML cells, HL-60 cells and KO52 cells were transfected with sh-KMT2A or sh-NC, and transfection effectiveness was evaluated using a qRT-PCR assay. A shows that sh-KMT2A transfected HL-60 cells and KO52 cells expressed KMT2A at considerably lower levels than the sh-NC group. The effects of KMT2A on the growth of HL-60 cells and KO52 cells were next examined using the CCK-8 and EdU assays. As expected, the CCK-8 results showed that downregulating KMT2A in a time-dependent manner prevented the development of HL-60 cells and KO52 cells (B). Additionally, the EdU data demonstrated that HL-60 cells and KO52 cells transfected with sh-KMT2A had a considerably smaller number of EdU-positive cells than the sh-NC group (C). A western blot test was utilized to evaluate the effect of KMT2A on the levels of expression of proliferation-related proteins such PCNA and Ki-67. KMT2A knockdown decreased PCNA and Ki-67 protein expression in HL-60 cells and KO52 cells, as shown by the results in D. The effects of KMT2A on HL-60 and KO52 cell migration and invasion were evaluated using transwell migration and invasion experiments. Findings from E show that compared to the sh-NC group, downregulating KMT2A dramatically decreased the migration and invasion of HL-60 cells and KO52 cells. Additionally, the effects of KMT2A on the expression levels of proteins related to metastasis, such as MMP-2 and MMP-9, were examined using a western blot assay. Data in F demonstrated that down-regulating KMT2A inhibited MMP-2 and MMP-9 protein expression in HL-60 cells and KO52 cells. According to these findings, KMT2A downregulation prevented AML cells’ abilities to proliferate, migrate and invade.
Figure 2. Down-regulation of KMT2A prevents AML cells from proliferating, migrating, and invading. Expression of KMT2A in HL-60 cells and KO52 cells which were transfected with sh-NC or sh-KMT2A was assessed using a qRT-PCR assay (A). (B) The CCK-8 assay was used to explore the vital capability of HL-60 cells and KO52 cells that had been transfected with sh-NC or sh-KMT2A at the specified times. (C) The EdU assay was used to detect the proliferating ability of HL-60 cells and KO52 cells that had been transfected with sh-NC or sh-KMT2A. (D) Western blot assay was used to assess PCNA as well as Ki-67 protein expressions in HL-60 cells and KO52 cells which were transfected with sh-NC or sh-KMT2A. (E) Transwell migration and invasion assays were adopted to evaluate the migration and invasion of HL-60 cells and KO52 cells which were transfected with sh-NC or sh-KMT2A. (F) Western blot assay was adopted to assess the protein expressions of MMP-2 as well as MMP-9 in HL-60 cells and KO52 cells which were transfected with sh-NC or sh-KMT2A. *P < 0.05, **P < 0.01 vs. sh-NC group. All data were exhibited as mean ± SD. n = 3.
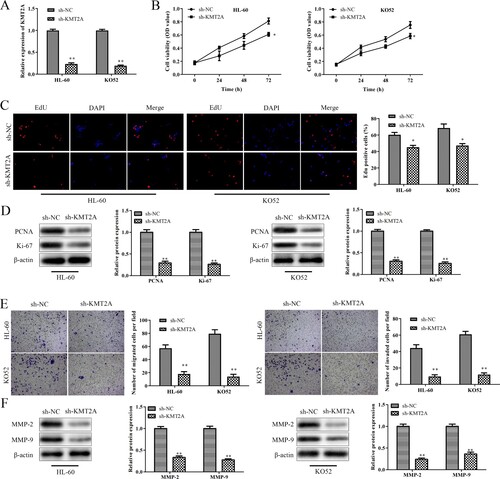
3.3. Up-regulation of kmt2a promotes proliferation, migration, and invasion of AML cells
To further highlight KMT2A's role in AML cells, HL-60 cells, and KO52 cells were transfected with pcDNA-KMT2A or pcDNA-NC, and the efficiency of the transfection was evaluated by qRT-PCR. A's results show that pcDNA-KMT2A transfected HL-60 cells and KO52 cells expressed KMT2A at levels that were significantly greater than those of the pcDNA-NC group. The CCK-8 data showed that up-regulation of KMT2A boosted the proliferation of HL-60 cells and KO52 cells in a time-dependent manner as compared to the pcDNA-NC group (B). Additionally, the EdU outcomes demonstrated that pcDNA-KMT2A-transfected HL-60 cells and KO52 cells had a significantly higher number of EdU-positive cells than the pcDNA-KMT2A group (C). Additionally, KMT2A over-expression significantly increased PCNA and Ki-67 protein expression in HL-60 cells and KO52 cells when compared to the pcDNA-NC group. Furthermore, data from transwell migration and invasion showed that up-regulation of KMT2A promoted migration and invasion of HL-60 cells and KO52 cells in comparison to the pcDNA-NC group. Additionally, over-expression of KMT2A dramatically raised the protein levels of MMP-2 and MMP-9 in HL-60 cells and KO52 cells as compared to the pcDNA-NC group. These results suggest that KMT2A overexpression promoted AML cell proliferation, invasion, and migration.
Figure 3. Up-regulation of KMT2A promotes AML cells’ abilities to proliferate, migrate and invade. (A) The expression of KMT2A in HL-60 cells and KO52 cells transfected with pcDNA-NC or pcDNA-KMT2A was assessed using a qRT-PCR experiment. (B) The CCK-8 test was used to determine the viability of HL-60 as well as KO52 cells that had been transfected with pcDNA-NC or pcDNA-KMT2A at the specified periods. (C) An EdU test was used to measure the proliferating ability of HL-60 cells and KO52 cells that had been transfected with either pcDNA-NC or pcDNA-KMT2A. (D) Western blot assay was used to evaluate PCNA and Ki-67 protein expressions in HL-60 cells and KO52 cells which were transfected with pcDNA-NC or pcDNA-KMT2A. (E) Transwell migration and invasion assays were used to determine the abilities to migrate and invade HL-60 cells and KO52 cells which were transfected with pcDNA-NC or pcDNA-KMT2A. (F) Western blot analysis was used to assess the protein expression of MMP-2 and MMP-9 in HL-60 cells and KO52 cells that were transfected with pcDNA-NC or pcDNA-KMT2A. *P < 0.05, **P < 0.01 vs. pcDNA-NC group. All data were exhibited as mean ± SD. n = 3.
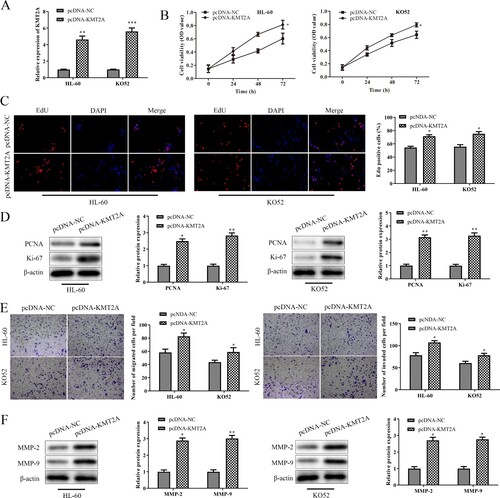
3.4. KMT2A is a target gene of miR-361-3p
The likely upstream miRNAs regulating KMT2A were examined using ENCORI and miRWalk, and the results pointed to miR-361-3p as a leading candidate. Using a dual-luciferase reporter experiment, the targeting relationship between miR-361-3p and KMT2A was verified. As expected, the inhibitory impact was abolished by a mutation in the predicted binding locations on the 3'-UTR of KMT2A. The 3'-UTR of KMT2A's luciferase activity was dramatically decreased by the exogenous synthesis of miR-361-3p. (B) Moreover, the expression of KMT2A in HL-60 cells and KO52 cells transfected with miR-361-3p mimic or NC mimic was evaluated using qRT-PCR and western blot assays. C and D show that miR-361-3p mimic-transfected HL-60 cells and KO52 cells expressed KMT2A at lower levels than the NC mimic. Finally, the qRT-PCR test was used to identify miR-361-3p expression in AML PB and cell lines. E and F show that MiR-361-3p was downregulated in AML PB and cell lines. These results demonstrated that KMT2A is a target gene of miR-361-3p in AML.
Figure 4. KMT2A is a target gene of miR-361-3p. (A) Using ENCORI and miRWalk, the potential upstream miRNAs of KMT2A were forecast. (B) The interactions between miR-361-3p and KMT2A were proved using a dual-luciferase reporter study. By using (C) qRT-PCR and (D) western blot tests, the mRNA and protein expression of KMT2A in HL-60 cells and KO52 cells that were transfected with NC mimic or miR-361-3p mimic was discovered. **P < 0.01 vs. NC mimic group. The qRT-PCR test was adopted to find miR-361-3p expression in (E) AML PB and (F) cell lines. *P < 0.05, **P < 0.01 vs. control group or HS-5 cells. All data were exhibited as mean ± SD. n = 3.
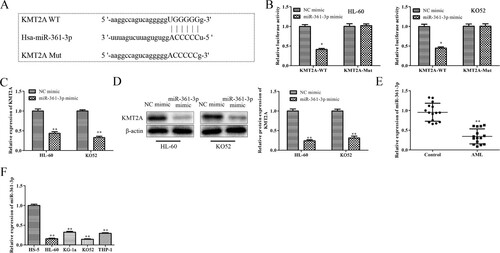
3.5. MiR-361-3p regulates proliferation, migration, and invasion of AML cells via targeting KMT2A
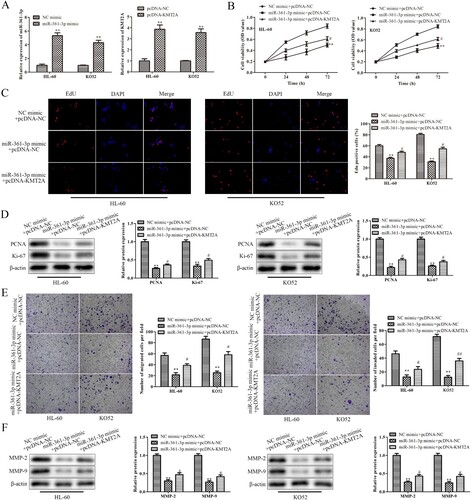
According to the results above, miR-361-3p specifically targeted KMT2A in AML cells. By using rescue studies, it was determined if miR-361-3p controlled AML cells’ abilities to proliferate, migrate and invade by targeting KMT2A. A qRT-PCR experiment was first used to evaluate the transfection effectiveness of HL-60 cells and KO52 cells transfected with miR-361-3p mimic or pcDNA-KMT2A. A's results show that pcDNA-KMT2A considerably increased KMT2A expression while miR-361-3p mimic dramatically increased miR-361-3p expression in HL-60 cells and KO52 cells. Data from CCK-8 and EdU subsequently showed that over-expression of KMT2A partially reversed the inhibitory effects of miR-361-3p mimic on the proliferation of HL-60 cells and KO52 cells, whereas miR-361-3p mimic decreased the proliferation of HL-60 cells and KO52 cells in a time-dependent manner (B and C). Similarly, in HL-60 cells and KO52 cells, miR-361-3p up-regulation significantly reduced PCNA and Ki-67 protein expressions, whereas miR-361-3p mimic co-transfected with pcDNA-KMT2A marginally increased PCNA and Ki-67 expressions (D). Additionally, miR-361-3p upregulation inhibited MMP-2 and MMP-9 protein expression in HL-60 cells and KO52 cells as well as migration and invasion. Co-transfection of the inhibitory effects with pcDNA-KMT2A and the miR-361-3p mimic, however, was only partially successful (E and 5F). According to these findings, miR-361-3p was a key mediator connecting KMT2A activity to AML cells’ abilities to proliferate, migrate and invade.
Figure 5. MiR-361-3p regulates the abilities to proliferate, migrate and invade of AML cells via targeting KMT2A. (A) The effectiveness of transfection in HL-60 cells and KO52 cells which were transfected with miR-361-3p mimic or pcDNA-KMT2A was assessed using a qRT-PCR test. **P < 0.01 vs. NC mimic group or pcDNA-NC group. (B) The CCK-8 test was used at the appropriate periods to determine the vitality of HL-60 cells and KO52 cells following transfection. (C) The EdU assay was used to gauge the proliferation of HL-60 cells and KO52 cells following transfection. (D) Western blot analysis was used to assess PCNA and Ki-67 protein expressions in HL-60 cells and KO52 cells after transfection. (E) Transwell migration and invasion assays were used to detect the migrating and invading abilities of HL-60 cells and KO52 cells following transfection. (F) Western blot analysis was used to assess the protein expression of MMP-2 and MMP-9 in HL-60 cells and KO52 cells after transfection. **P < 0.01 vs.NC mimic + pcDNA-NC group, #P < 0.05, ##P < 0.01 vs. miR-361-3p mimic + pcDNA-NC group. All data were presented as mean ± SD. n = 3.
4. Discussion
In addition to it being regarded as a gene with multiple conserved functional domains, KMT2A cooperates with a variety of complexes to promote accessibility and transcription of the genome [Citation18]. If a KMT2A mutation occurs, which causes dysfunction, further leading to abnormal cell development or loss of function [Citation19]. With the advancement of gene sequencing technologies, the KMT2A mutation has now been discovered in a variety of cancers. The SP1-KMT2A-ANO1 signaling pathway is abundantly expressed in gastric cancer tissues and cell lines, and overexpression of KMT2A greatly promotes gastric cancer cells’ abilities to migrate and invade [Citation20,Citation21]. In addition, Ballabio et al. find that KMT2A enhances the HGF-MET pathway through the ETS2-MLL complex, which directly activates MMP-1 and MMP-3 transcription to promote invasion and metastasis of hepatocellular carcinoma cells [Citation22]. Moreover, Svoboda et al. show that KMT2A is over-expressed in Ewing’s sarcoma, and functions as oncogenes [Citation23]. Furthermore, according to studies and reports, KMT2A has also been proven to be closely related to AML [Citation24]. The precise function of KMT2A in AML and any potential regulatory mechanisms are still unclear. The present study found that KMT2A's expression is extensively exhibited in AML PB and cell lines, as was predicted, suggesting that KMT2A may function as an oncogene in AML.
CCK-8 assay, EdU assay, transwell migration and invasion assay, and western blot assay for AML cells’ abilities to proliferate, migrate and invade were all used to investigate the impact of KMT2A on these processes. In line with expectations, down-regulation of KMT2A significantly reduced AML cells’ ability to proliferate, migrate, and invade, whereas up-regulation of KMT2A had the opposite results. There is some evidence that KMT2A has beneficial impacts on the development and metastasis of other cancers. For instance, Zhang et al. discover that since KMT2A triggers the hTERT signaling pathway and aids in the development of melanoma, it is conceivable to target KMT2A/hTERT for the treatment of melanoma [Citation25]. Besides, Sun et al. demonstrate that neuroblastoma overexpresses KMT2A and that KMT2A down-regulation inhibits neuroblastoma cell growth and metastasis by regulating the Myc signaling pathway [Citation26]. These results essentially corroborated those from this study, indicating that KMT2A's aberrant expression was crucial to the development of AML.
Based on these, it was difficult to identify the upstream miRNAs that control KMT2A. MiRNAs currently have a considerable impact on the onset and development of AML. For instance, MiR-342 expression is more common in AML PB than in PB from comparably healthy controls. Naa10p focuses on MiR-342 to significantly slow down AML cell proliferation and the G1/S transition in leukemia cells [Citation27]. Besides that, miR-183-5p is overexpressed in AML PB and cell lines, and miR-381-5p downregulation prevents AML cells from differentiating and inhibits proliferation by controlling Erbin [Citation28]. Furthermore, miR-345-5p is only weakly expressed in AML cells, and miR-345-5p inhibition promotes AML cell proliferation by targeting AKT1/2 [Citation29]. These findings demonstrated that miRNAs played a crucial role in RA development by regulating downstream target genes. By using a dual-luciferase reporter experiment and bioinformatic analysis, this miRNA (miR-361-3p) was discovered. In recent years, it has been discovered that miR-361-3p plays a variety of roles in different cancers. For instance, whereas miR-361-3p is expressed at low levels in colorectal cancer tissues, miR-361-3p amplification promotes cell death by targeting SH2B1 [Citation30]. Correspondingly, in tissues or cell lines derived from lung cancer, miR-361-3p is not strongly expressed. However, when it is overexpressed, it negatively modulates the expression of FRAT1, which prevents lung cancer cell motility and invasion [Citation31]. MiR-361-3p is also markedly overexpressed in tissues associated with breast cancer. Breast cancer cells that have miR-361-3p overexpression proliferate and are less likely to die by inhibiting the production of E2F1 [Citation32]. These discoveries revealed important cancer prognostic variables or therapeutic targets. Similarly, miR-361-3p was shown to be significantly down-regulated in AML PB and cell lines in our investigation. Additionally, miR-361-3p was dramatically downregulated, which prevented AML cells from proliferating, migrating, and invading. Through a rescue experiment, it was also determined if KMT2A had a part in the advancement of AML, which is controlled by miR-361-3p. As anticipated, miR-361-3p mimic's inhibitory effects on AML cells’ abilities to proliferate, migrate and invade were partially reversed by the over-expression of KMT2A.
In conclusion, miR-361-3p was little expressed in both AML PB and cell lines, but KMT2A was robustly expressed. AML cells’ ability to proliferate, migrate, and invade was inhibited when KMT2A was downregulated, whereas the opposite was true when KMT2A was upregulated. Furthermore, KMT2A was involved in how miR-361-3p affected AML cells’ abilities to proliferate, migrate and invade, clearly demonstrating how the miR-361-3p/KMT2A axis may help us better understand the oncogenic mechanisms and therapeutic approaches in AML.
Ethics approval and consent to participate
The work described has been carried out by the Declaration of Helsinki for experiments involving humans. The experimental protocol was approved by the Ethics Committee of Funing People's Hospital.
Consent for publication
All the participants approved the publication.
Authors’ contributions
CJ conceived and designed the study. DX and JJ conducted most of the experiments. GH analyzed the data. HZ performed the literature search and data extraction. DX drafted the manuscript. DX and CJ finalized the manuscript. All authors read and approved the final manuscript.
Availability of data and material
All data generated or analyzed during this study are included in this published article.
Acknowledgments
We deeply appreciate the support of all participants.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Kayser S, Levis MJ. Clinical implications of molecular markers in acute myeloid leukemia. Eur J Haematol. 2019;102:20–35.
- Yan R, et al. Ectopic expression of human airway trypsin-like protease 4 in acute myeloid leukemia promotes cancer cell invasion and tumor growth. Cancer Med. 2019;8:2348–2359.
- Takami A. Hematopoietic stem cell transplantation for acute myeloid leukemia. Int J Hematol. 2018;107:513–518.
- Langabeer SE. Screening for an underlying myeloproliferative neoplasm in patients with thrombocytosis post-induction chemotherapy for acute myeloid leukemia. Leuk Res Rep. 2020;14:100218. doi:10.1016/j.lrr.2020.100218
- Shallis RM, Wang R, Davidoff A, et al. Epidemiology of acute myeloid leukemia: recent progress and enduring challenges. Blood Rev. 2019;36:70–87.
- Chen KTJ, Gilabert-Oriol R, Bally MB, et al. Recent treatment advances and the role of nanotechnology, combination products, and immunotherapy in changing the therapeutic landscape of acute myeloid leukemia. Pharm Res. 2019;36:019–2654.
- Liao Q, Wang B, Li X, et al. miRNAs in acute myeloid leukemia. Oncotarget. 2017;8:3666–3682.
- Sun Y, et al. miR-1290 promotes proliferation and suppresses apoptosis in acute myeloid leukemia by targeting FOXG1/SOCS3. J Biol Regul Homeost Agents. 2019;33:1703–1713.
- Liu L, Ren W, Chen K. MiR-34a promotes apoptosis and inhibits autophagy by targeting HMGB1 in acute myeloid leukemia cells. Cell Physiol Biochem. 2017;41:1981–1992.
- Chen L, et al. Circular RNA 100146 functions as an oncogene through direct binding to miR-361-3p and miR-615-5p in non-small cell lung cancer. Mol Cancer. 2019 Jan 21;18(1):13. doi:10.1186/s12943-019-0943-0
- Fletcher CE, et al. Androgen receptor-modulatory microRNAs provide insight into therapy resistance and therapeutic targets in advanced prostate cancer. Oncogene. 2019;38:5700–5724.
- J. Hu, et al. MiR-361-3p regulates ERK1/2-induced EMT via DUSP2 mRNA degradation in pancreatic ductal adenocarcinoma. Cell Death Dis. 2018;9:018–0839.
- Wallace JA, O'Connell RM. MicroRNAs and acute myeloid leukemia: therapeutic implications and emerging concepts. Blood. 2017;130:1290–1301.
- Tkachuk DC, Kohler S, Cleary ML. Involvement of a homolog of Drosophila trithorax by 11q23 chromosomal translocations in acute leukemias. Cell. 1992;71:691–700.
- Mims AS, et al. A novel regimen for relapsed/refractory adult acute myeloid leukemia using a KMT2A partial tandem duplication targeted therapy: results of phase 1 study NCI 8485. Haematologica. 2018;103:982–987.
- Lee JW, et al. Regulation of HOXA9 activity by predominant expression of DACH1 against C/EBPα and GATA-1 in myeloid leukemia with MLL-AF9. Biochem Biophys Res Commun. 2012;426:299–305.
- Chen CW, Armstrong SA. Targeting DOT1L and HOX gene expression in MLL-rearranged leukemia and beyond. Exp Hematol. 2015;43:673–684.
- Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer. 2007;7:823–833.
- Ansari KI, Mandal SS. Mixed lineage leukemia: roles in gene expression, hormone signaling and mRNA processing. Febs J. 2010;277:1790–1804.
- Liu F, et al. TMEM16A overexpression contributes to tumor invasion and poor prognosis of human gastric cancer through TGF-β signaling. Oncotarget. 2015;6:11585–11599.
- Zeng X, et al. Transcriptional activation of ANO1 promotes gastric cancer progression. Biochem Biophys Res Commun. 2019;512:131–136.
- Ballabio E, Milne TA. Molecular and epigenetic mechanisms of MLL in human leukemogenesis. Cancers. 2012;4:904–944.
- Svoboda LK, et al. Tumorigenicity of Ewing sarcoma is critically dependent on the trithorax proteins MLL1 and menin. Oncotarget. 2017;8:458–471.
- Sakhdari A, et al. Homogeneously staining region (hsr) on chromosome 11 is highly specific for KMT2A amplification in acute myeloid leukemia (AML) and myelodysplastic syndrome (MDS). Cancer Genet. 2019;238:18–22.
- Zhang C, et al. KMT2A promotes melanoma cell growth by targeting hTERT signaling pathway. Cell Death Dis. 2017;8:285.
- Sun Y, et al. WDR5 supports an N-Myc transcriptional complex that drives a protumorigenic gene expression signature in neuroblastoma. Cancer Res. 2015;75:5143–5154.
- Wang H, He H, Yang C. miR-342 suppresses the proliferation and invasion of acute myeloid leukemia by targeting Naa10p. Artif Cells Nanomed Biotechnol. 2019;47:3671–3676.
- Zheng Z, et al. miR-183-5p inhibits occurrence and progression of acute myeloid leukemia via targeting erbin. Mol Ther. 2019;27:542–558.
- Ying X, Zhang W, Fang M, et al. miR-345-5p regulates proliferation, cell cycle, and apoptosis of acute myeloid leukemia cells by targeting AKT2. J Cell Biochem. 2018;2:27461.
- Liu J, Zhu J, Xiao Z, et al. BBOX1-AS1 contributes to colorectal cancer progression by sponging hsa-miR-361-3p and targeting SH2B1. FEBS Open Bio. 2020;27:2211–5463.
- Li X, Zheng H. LncRNA SNHG1 influences cell proliferation, migration, invasion, and apoptosis of non-small cell lung cancer cells via the miR-361-3p/FRAT1 axis. Thorac Cancer. 2020;11:295–304.
- Hua B, et al. MicroRNA-361-3p promotes human breast cancer cell viability by inhibiting the E2F1/P73 signalling pathway. Biomed Pharmacother. 2020;125(:25):109994. doi:10.1016/j.biopha.2020.109994
