ABSTRACT
Objective:
This study analyzed the relationship between bone marrow microvessel density (MVD) and the expression of four miRNAs with chronic myelogenous leukemia (CML) resistance after tyrosine kinase inhibitor (TKI) treatment.
Methods:
234 CML patients were divided into resistance and non-resistance groups in terms of the results of the 5-year follow-up. Patients were divided into the Optimum response group and the Warning/Failure group based on TKI response. MVD was determined by immunohistochemistry, and the expression levels of four miRNAs (miR-106a, miR-155, miR-146a, and miR-340) in bone marrow biopsy specimens were examined by qPCR. We evaluated the association of MVD with four miRNAs and them predictive value for CML resistance after TKI treatment.
Results:
The MVD and the levels of miR-106a, miR-155, and miR-146a were significantly higher while the miR-340 level was lower in the resistance group than the non-resistance group. Besides, MVD had a significant correlation with the levels of miR-340 and miR-155. According to the results of survival analysis, MVD as well as miR-340 and miR-155 levels were observably correlated with 5-year survival of patients without TKI resistance. The results of the ROC curve indicated that the MVD, miR-106a, miR-340, and miR-155 had good predictive accuracy for CML resistance after TKI treatment. As for the results of multivariate analysis, disease stage, risk level (high risk), high MVD, low miR-340 expression, and high miR-155 expression were all independent risk factors for CML resistance.
Conclusion:
MVD and the expression of miR-340 and miR-155 are closely associated with CML resistance after TKI treatment.
Introduction
Chronic myelogenous leukemia (CML), a malignancy caused by clonal proliferation of bone marrow hematopoietic stem cells, is characterized by leukocytosis and aggregation of granulocytes and their progenitors. CML affects 1–2 in every 100,000 adults and is becoming more common over time [Citation1]. It accounts for approximately 15% of newly diagnosed leukemia cases in adults [Citation2]. In 2022, approximately 1022 patients will die from CML in the United States [Citation3]. Imatinib, bosutinib, nilotinib, and dasatinib are the four tyrosine kinase inhibitors (TKIs) that are currently accepted as first-line treatments for CML both domestically and abroad [Citation4]. If a CML patient has developed resistance to TKIs and experienced failed treatment or the patient is in an advanced state, it is challenging to treat the disease. Therefore, the search for biomarkers of prognosis in CML patients is of great importance.
Angiogenesis is associated with tumor growth, invasion, and metastasis [Citation5]. Microvessel density (MVD) measurement is a quantitative method to assess angiogenesis [Citation6]. CML patients exhibit a significant increase in MVD compared to those with normal bone marrow [Citation7]. The best therapeutic response to TKI therapy in CML patients was investigated by Ćojbašić et al. [Citation8]. In their exploration, compared with controls, CML patients had significantly higher levels of angiogenesis parameters. Furthermore, low MVD, minor axis, area, circularity, and roundness and higher aspect ratio are positive prognostic factors for achievement of complete cytogenetic response (CCgR), while only low MVD for achievement of major molecular response (MMoR). However, there has not yet been any evidence of a connection between MVD and CML resistance.
MicroRNA (miRNA) is a small non-coding RNA that plays a role in the post-transcriptional regulation of gene expression [Citation9]. It can promote and regulate cell signaling, apoptosis, and cell development and proliferation [Citation10]. MiRNAs have great potential as therapeutic targets and prognostic markers in different types of cancer [Citation11]. Their roles in a variety of malignancies have also been widely documented [Citation12–14]. These RNAs are crucial in development and regulation of T cells as well as immune reconstitution after TKI treatment [Citation15]. Studies have shown that the development of TKIs greatly improves long-term survival in patients with CML, but amplification or mutation of the BCR-ABL1 oncogene and changes in the ATP-binding cassette (ABC) transporter can lead to drug intolerance or resistance in nearly a quarter of patients and the withdrawal of TKI therapy. Furthermore, miRNAs have also been shown to play a key role in different stages of CML [Citation9,Citation16]. To be more precise, miR-495-3p overexpression inhibits leukemic cell growth and TKI resistance in Imatinib-resistant T315I-mutant cells, as well as drug efflux activity through regulation of multidrug resistance 1 (MDR1) gene [Citation17]. MiR-146a has been found to be abnormally expressed in both adults and children with acute leukemia, and circulating miR-146a level is much lower in CML patients than in healthy individuals. However, the circulating miR-146a level displays a significant rise after 14 days of imatinib treatment and a negative correlation with BCR-ABL expression level at 90 days. It is logical to speculate that circulating miR-146a has a prognostic role in the prediction of early response to imatinib treatment in CML patients [Citation14]. According to the study of Chen et al., human umbilical cord mesenchymal stem cell (hucMSC) exosomes, via miR-146a-5p, promoted imatinib-induced apoptosis by suppressing glutaminase-1 (GLS1) ubiquitination to increase GLS protein while weakening ubiquitin-specific protease 6 (USP6) regulation of CML cells resistant to imatinib [Citation1]. MiR-155 is highly expressed in CD34+ CML stem cells and promotes their proliferation, which is expected to be a therapeutic target for CML [Citation18] Overexpression of miR-146a inhibits the NF-κB pathway, which leads to apoptosis in CML [Citation19]. Therefore, miR-146a has been proved to be a predictor in CML [Citation14]. MiR-106a, a target of lncRNA-BGL3, is closely related to the expression of Bcr-Abl, a regulatory gene of CML [Citation20]. And miR-155 was found to be associated with tumor angiogenesis [Citation21]. Collectively, the four miRNAs may possess potential clinical value as prognostic biomarkers and therapeutic targets in the field of hematologic oncology.
In this study, patients with CML who received TKI treatment were selected as the study subjects [Citation22], and the association of CML resistance with MVD and the four miRNAs was analyzed.
Materials and methods
Study subjects and data collection
Totally 234 patients with CML who received TKI treatment in the Department of Hematology of Taihe Hospital, Hubei University of Medicine from December 2022 to January 2023 participated in this study. They were divided into resistance group (n = 59) and non-resistance group (n = 175) according to the results of the 5-year follow-up. Inclusion criteria: patients (i) diagnosed as CML; (ii) TKI treatment: Patients over 60 years of age were given imatinib (400 mg/time, orally, once a day); Patients younger than 60 years of age were given one of three drugs: Flumatinib (600 mg/time orally once a day), dasatinib (100 mg/time orally once a day), and nilotinib (300 mg/time orally twice a day). Exclusion criteria: patients (i) combined with malignancy; (ii) combined with dysfunction of important organs like heart, liver, and kidney; (iii) with incomplete clinical history and follow-up data [Citation23]. This study was approved by the Ethics Committee of Taihe Hospital, Hubei University of Medicine (KY2308).
Basic information of patients was collected, including gender, age, white blood cells (WBC, ×109/L), platelets (PLT, ×109/L), histopathological type (central nervous system, lymph node, skin, etc.), (pre-TKI treatment) disease stage (chronic, accelerated, acute), (pre-TKI treatment) risk level (low, intermediate, high), EUTOS long-term survival (ELTS) score (low risk, medium risk, high risk), therapeutic drugs (imatinib, nilotinib, dasatinib), from diagnosis to TKI reception.
Microvessel count
Bone marrow biopsy specimens were formalin-fixed, decalcified with EDTA/HCL and paraffin-embedded. Then they were sectioned at a thickness of 4 μm per section for microvessel counting. Endothelial cells were labeled using polyclonal rabbit anti-human von Willebrand Factor (vWF) antibody (Beijing Zhongshan Biotechnology Co., Ltd.) [Citation6]. The anti-rabbit IgG-HRP multimer (Beijing Zhongshan Biotechnology Co., Ltd.) was used as the secondary antibody. The obtained sections were subjected to conventional color development, hematoxylin re-staining, dehydration, transparency, and sealing.
The images were first analyzed at low magnification (×40) to identify the areas with the greatest number of blood vessels, the so-called ‘hot spots’. These hot spots were then observed at a high magnification (×400). The microvessels in the three hot spots were counted, and the results were averaged to express MVD. MVD is the total number of microvessels (×400) per 0.5 mm2 [Citation24].
qPCR assay
The total RNA of bone marrow biopsy specimens of patients with CML was extracted using RNA extraction kits. cDNA was synthesized from the obtained RNA with a cDNA kit and then received PCR amplification with SYBR Mix. The primer sequences are shown in , and the 2−ΔΔCt method was used to calculate the relative expression of miR-106a, miR-340, miR-155, and miR-146a.
Table 1. qRT-PCR primer sequences.
Statistical analysis
SPSS software was used for data analysis. The measurement data conforming to normal distribution were expressed as (mean ± standard deviation), and an independent sample t-test was used for comparison between two groups. Pearson correlation coefficient was applied to analyze the correlation between MVD and miRNA levels. The Kaplan-Meier method was employed for survival analysis. Multivariate logistic regression was used for the analysis of factors associated with CML resistance and non-resistance after TKI treatment. Time-dependent Receiver Operator Characteristic (ROC) curves were plotted to analyze the accuracy of MVD, miR-106a, miR-340, miR-155, miR-146a levels in predicting disease resistance. P < 0.05 indicates significantly different.
Results
Baseline characteristics of patients in the resistance group and the non-resistance group
The basic information of patients in the resistance group and the non-resistance group in this study is summarized in . The resistance group consisted of 16 men and 43 women, with a mean age of 57.54 ± 8.57 years. There were 10 cases in the chronic stage, 19 in the accelerated stage, and 30 in the acute stage in the resistance group. In regard to the risk level of patients in the resistance group, 9 cases were at low risk, 18 at intermediate risk, and 32 at high risk. As for the non-resistance group, there were 62 men and 113 women, with a mean age of 57.88 ± 8.57 years; 82 cases in the chronic stage, 55 cases in the accelerated stage, and 38 cases in the acute stage; 77 cases at low risk, 62 at intermediate risk, and 36 at high risk. There were no statistically significant differences in age, gender, WBC, PLT, ELTS score and therapeutic drugs between the resistance and non-resistance groups (P > 0.05). However, the differences between the two groups were statistically significant in terms of disease stage and risk level (P < 0.001).
Table 2. Baseline characteristics.
Patients with CML respond to TKI drug therapy
Subsequently, the therapeutic response to TKI was evaluated. There were 172 cases in the Optimum response and 62 cases in the Warning/Failure group. And the results were shown in . There were no significant differences in age, gender, PLT (×109/L), risk level (%), ELTS score and therapeutic drugs between CML patients in the Optimum response group and the Warning/Failure group (P > 0.05). There were significant differences in WBC (×109/L) and disease stage between the two groups (P < 0.05).
Table 3. Comparison of clinicopathologic features between groups.
Differences in microvessel density and levels of 4 miRNAs between the resistance group and non-resistance group
As shown in A–E, MVD, miR-106a, miR-155 and miR-146a levels were significantly higher in the resistance group than those in the non-resistance group, while miR-340 level was much lower. And the difference was statistically significant (P < 0.05). According to A–D, the MVD in bone marrow biopsy specimens were correlated with miR-155 (B) and miR-340 (D) levels (P < 0.05), but not correlated with miR-106a (A) and miR-146a (C) levels (P > 0.05). In and , the results of ROC curve analysis showed that MVD, miR-155 and miR-340 levels had good accuracy in predicting CML resistance after allo-HSCT, with optimal cut-off values of 32.5, 1.252 and 0.823, respectively. The area under the curve (AUC) of miR-106a and miR-146a were 0.594 and 0.574, suggesting that they were not associated with prognosis of CML resistance.
Figure 1. Differences in MVD and miRNA levels between the resistance group and the non-resistance group. The differences in MVD (A), miR-106a level (B), miR-155 level (C), miR-146a level (D), miR-340 level (E) between the resistance group and the non-resistance group. *P < 0.05; **P < 0.01.
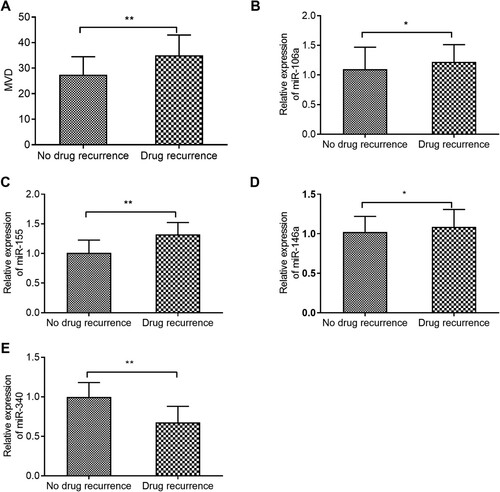
Figure 2. Correlation analysis of MVD with the levels of the 4 miRNAs. A: Correlation analysis of MVD with miR-106a. B: Correlation analysis of MVD with miR-155. C: Correlation analysis of MVD with miR-146a. D: Correlation analysis of MVD with miR-340.
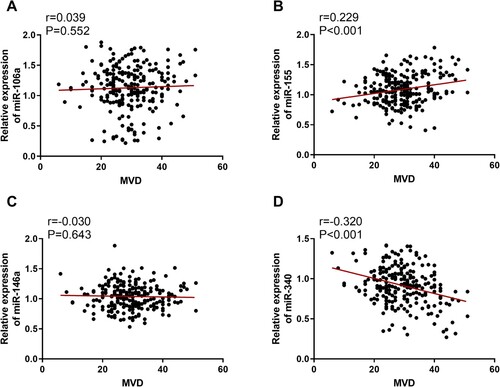
Figure 3. ROC curves of MVD, miR-106a, miR-340, miR-155, and miR-146a for predicting CML resistance after TKI treatment.
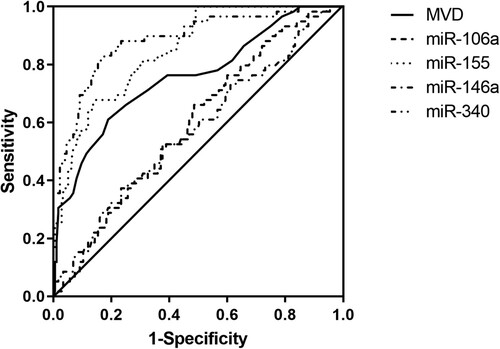
Table 4. Value of MVD and miRNA expression in predicting CML resistance after TKI treatment.
Survival analysis of patients without resistance to chronic myelogenous leukemia with different microvessel density and miRNA expression levels
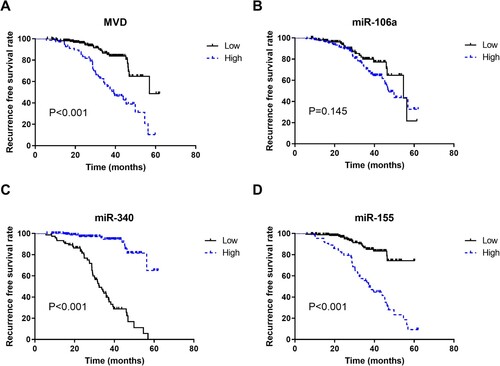
Kaplan-Meier survival curves were established to analyze the correlation between relapse-free prognosis after 5 years of follow-up and MVD, miR-106a, miR-340, miR-155 and miR-146a expression levels. The results are shown in . The 5-year resistance-free survival rate was higher in patients with low MVD (48.7%) than in patients with high MVD (10.4%) (P < 0.001) (A); the 5-year resistance-free survival rate of patients was not statistically different between the miR-106a low expression group (21.6%) and the miR-106a high expression group (32.7%) (P = 0.145) (B); the 5-year resistance-free survival rate of patients in the miR-340 low expression group (5.6%) was lower than that in the miR-340 high expression group (65.1%) (P < 0.001) (C); as for miR-155, the low expression group (74.3%) presented a much higher 5-year resistance-free survival rate than the high group (9.3%) (P < 0.001) (D).
Multivariate logistic regression analysis for prognosis of patients with chronic myelogenous leukemia
As shown in , the results of multivariate logistic regression analysis showed that disease stage, risk level (high risk), high MVD, low miR-340 expression and high miR-155 expression were all independent risk factors affecting CML resistance after TKI treatment (P < 0.05). Notably, WBC, miR-106a and miR-146a expression levels did not correlate with prognostic CML resistance (P > 0.05).
Table 5. Multivariate logistic regression analysis of prognosis in CML patients.
Discussion
The incidence of CML, the most common type of myeloproliterative neoplasm, keeps growing in the world [Citation1]. CML patients will develop resistance after TKI therapy such as imatinib mesylate. Imatinib resistance has been reported to be brought on by mutations in the BCR-ABL kinase domain (KD), but no mutations in BCR-ABL-KD are detected in many imatinib-resistant patients [Citation25]. MiRNAs are short non-coding RNAs involved in controlling gene expression [Citation25]. In order to investigate the role of miRNAs in TKI resistance in CML patients, we discussed the correlation of miR-106a, miR-340, miR-155 and miR-146a with TKI resistance in CML patients as well as their prediction in CML resistance. We finally revealed the predictive value of MVD, miR-155 and miR340 for CML resistance after TKI treatment.
In this study, 234 patients with CML who received TKI treatment were statistically analyzed, of which 59 cases were in the resistance group and 175 ones in the non-resistance group. There were substantial differences between the two groups in regards disease stage and risk level. The prognosis of patients with different disease stages is different. The therapeutic response to TKI was evaluated, there were 172 cases in the Optimum response and 62 cases in the Warning/Failure group. There were no significant differences in age, gender, PLT, risk level, ELTS score and therapeutic drugs between CML patients in the Optimum response group and the Warning/Failure group. There were significant differences in WBC and disease stage between the Optimum response and the Warning/Failure group. For example, TKIs are safe and well-tolerated for patients with advanced CML before and after allogeneic hematopoietic stem cell transplantation (HSCT), and they may improve the prognosis of this aggressive disease [Citation26]. Although most CML patients have normal quality of life and survival time after TKI treatment, some patients progress to the accelerated phase (AP) and blast phase (BP) with poor outcomes [Citation27]. Although the prognosis of patients present with de novo CML-AP is almost similar to that of CML-CP patients, patients who have developed from CML-CP and experienced relevant treatment should receive second- or third-generation TKIs and strongly consider allogeneic HSCT [Citation27]. Similarly, patients transformed from CML-BP have a particularly poor prognosis, with median survival typically less than one year; some of them can obtain long-term survival if the combination of potent TKIs (such as ponatinib) and allogeneic HSCT is applied [Citation27]. These discoveries of new drugs and new combination therapies are collected as an Arsenal for the treatment of advanced CML. The results of this study suggest that patients receiving TKI treatment in the chronic stage are less likely to experience CML resistance. The likelihood of CML resistance will therefore be decreased in individuals with chronic CML who are low risk. Angiogenesis plays a key role in hematologic malignancies. MVD is considered an indicator of malignant potential of the tumors and possesses the ability to predict tumor resistance [Citation28]. For instance, MVD was useful in predicting drug resistance in patients with squamous cell cervical cancer stage II who were younger than 40 years old, and a higher MVD is associated with disease resistance [Citation29]. The relationship between MVD and CML resistance hasn't been examined in any studies, though. Our study identified a significant difference in MVD between the resistance and non-resistance groups, and high MVD was strongly connected to postoperative CML resistance. The AUC of MVD for prediction of CML resistance was 0.757, indicating that MVD has some predictive value for prognostic CML resistance.
Research on the relationship between miRNAs and CML resistance is lacking. In this case, we conducted such research and discovered that there was a significant difference in the expression levels of miR-106a, miR-340, miR-155, and miR-146a between the resistance and non-resistance groups. The correlation analysis showed that MVD was positively correlated with miR-155 level, negatively correlated with miR-340 level, and not correlated with the other two miRNAs. For the ROC curve, miR-340 and miR-155 had AUCs of 0.886 and 0.849, respectively. More importantly, the AUC of miR-340 is closer to 1, suggesting its better predictive accuracy.
MiR-340 is aberrantly expressed in a variety of human tumors, such as breast cancer [Citation30], colorectal cancer [Citation31], and gastric cancer [Citation32]. It can also serve as a potential therapeutic target for gliomas [Citation33]. In non-small cell lung cancer tissues, MiR-340 is markedly underexpressed, and this underexpression is strongly correlated with a poor prognosis [Citation34]. Specifically, a survival analysis of 136 patients with non-small cell lung cancer showed that low miR-340 expression was one of the contributors to poor 5-year overall survival [Citation34]. In recent studies, knockdown of Circ-CHI3L1.2 has been discovered to modulate cisplatin resistance of osteosarcoma cells via the miR-340-5p-LPAATβ axis [Citation35]; SOX2 mediates cisplatin resistance in small cell lung cancer by down-regulating the expression of hsa-miR-340-5p [Citation36]. To date, no studies have been done on the correlation between miR-340 and CML resistance. Our study disclosed that patients with low miR-340 expression were more likely to have CML resistance after TKI treatment than those with high miR-340 expression, which may be attributed to the antitumor efficacy of miR-340.
MiR-155 is implicated in the regulation of key genes related to myelopoiesis [Citation37] and is up-regulated in several lymphomas. In the study of Rokah et al. [Citation38], miR-155 was found to be abnormally down-regulated in CML patients compared to healthy controls. Patients who have recently been diagnosed with CML have lower levels of miR-155 expression in their peripheral blood cells [Citation39]. It follows that low miR-155 expression may be linked to the development of CML. However, in our findings, high miR-155 expression was a risk factor for CML resistance. This opposite result may be related to the disease process. The role of miR-155 in CML still needs further validation. Overall, it also implies that miRNAs have a variety of roles in CML. In addition, our study uncovered that MVD, miR-340 and miR-155 levels were strongly correlated with the 5-year resistance-free survival of patients. Concretely speaking, low miR-340 expression, high miR-155 expression and high MVD may lead to CML resistance after TKI treatment, which finally lowered the survival rates of patients.
Although this study demonstrated the predictive value of high MVD, low miR-340 expression and high miR-155 expression in CML resistance after TKI treatment, it still has certain limitations due to its small sample size and single center. Large sample sizes may be required for future analysis. Additionally, the specific mechanisms of action of MVD, miR-340 and miR-155, which may serve as therapeutic targets for CML, need to be further validated in animal models.
Conclusion
MVD, miR-340 and miR-155 are independent risk factors affecting CML resistance after TKI treatment, which have predictive value for postoperative CML resistance. Patients with high MVD, low miR-340 expression and high miR-155 expression are more likely to experience CML resistance after TKI treatment. In order to optimize TKI treatment outcomes, it is necessary to identify patients with the above conditions earlier before disease progression.
Authors’ contributions
Yi-gang Guo and Xue-lian Feng designed the study. Lu-lu Zhang, Ping Hu, Zhang-zhi Li and Rui-bo Zhang collated the data, carried out data analyses and produced the initial draft of the manuscript. Xi Lv and Qiong Yi contributed to drafting the manuscript. All authors have read and approved the final submitted manuscript.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data used to support the findings of this study are included within the article.
Additional information
Funding
References
- Chen X, Chen Y, Zhang M, et al. HucMSC exosomes promoted imatinib-induced apoptosis in K562-R cells via a miR-145a-5p/USP6/GLS1 axis. Cell Death Dis. 2022;13(1):92. doi:10.1038/s41419-022-04531-3
- Habib EM, Nosiar NA, Eid MA, et al. MiR-150 expression in chronic myeloid leukemia: relation to imatinib response. Lab Med. 2022;53(1):58–64. doi:10.1093/labmed/lmab040
- Jabbour E, Kantarjian H. Chronic myeloid leukemia: 2022 update on diagnosis, therapy, and monitoring. Am J Hematol. 2022;97(9):1236–1256.
- Ciftciler R, Haznedaroglu IC. Tailored tyrosine kinase inhibitor (TKI) treatment of chronic myeloid leukemia (CML) based on current evidence. Eur Rev Med Pharmacol Sci. 2021;25(24):7787–7798.
- Jiang X, Wang J, Deng X, et al. The role of microenvironment in tumor angiogenesis. J Exp Clin Cancer Res: CR. 2020;39(1):204. doi:10.1186/s13046-020-01709-5
- Antic D, Jovanovic MP, Fekete MD, et al. Assessment of bone marrow microvessel density in chronic lymphocytic leukemia. Appl Immunohistochem Mol Morphol. 2010;18(4):353–356. doi:10.1097/PAI.0b013e3181d18ae2
- Kvasnicka HM, Thiele J. Bone marrow angiogenesis: methods of quantification and changes evolving in chronic myeloproliferative disorders. Histol Histopathol. 2004;19(4):1245–1260.
- Cojbasic I, Macukanovic-Golubovic L, Mihailovic D, et al. The significance of angiogenesis for predicting optimal therapeutic response in chronic myeloid leukaemia patients. Pol J Pathol. 2017;68(3):241–251. doi:10.5114/pjp.2017.71532
- Yeh CH, Moles R, Nicot C. Clinical significance of microRNAs in chronic and acute human leukemia. Mol Cancer. 2016;15(1):37. doi:10.1186/s12943-016-0518-2
- Ufkin ML, Peterson S, Yang X, et al. miR-125a regulates cell cycle, proliferation, and apoptosis by targeting the ErbB pathway in acute myeloid leukemia. Leuk Res. 2014;38(3):402–410. doi:10.1016/j.leukres.2013.12.021
- Martins JRB, Moraes LN, Cury SS, et al. MiR-125a-3p and MiR-320b differentially expressed in patients with chronic myeloid leukemia treated with allogeneic hematopoietic stem cell transplantation and imatinib mesylate. Int J Mol Sci. 2021;22(19).
- Shen X, Si Y, Yang Z, et al. MicroRNA-542-3p suppresses cell growth of gastric cancer cells via targeting oncogene astrocyte-elevated gene-1. Med Oncol. 2015;32(1):361. doi:10.1007/s12032-014-0361-5
- Zhao J, Lu Q, Zhu J, et al. Prognostic value of miR-96 in patients with acute myeloid leukemia. Diagn Pathol. 2014;9:76.
- Habib EM, Nosiar NA, Eid MA, et al. Circulating miR-146a expression predicts early treatment response to imatinib in adult chronic myeloid leukemia. J Investig Med. 2021;69(2):333–337. doi:10.1136/jim-2020-001563
- Cao LL, Lu H, Soutto M, et al. Multivalent tyrosine kinase inhibition promotes T cell recruitment to immune-desert gastric cancers by restricting epithelial-mesenchymal transition via tumour-intrinsic IFN-gamma signalling. Gut. 2023.
- Litwinska Z, Machalinski B. miRNAs in chronic myeloid leukemia: small molecules, essential function. Leuk Lymphoma. 2017;58(6):1297–1305. doi:10.1080/10428194.2016.1243676
- Rittavee Y, Artus J, Desterke C, et al. miR-495-3p sensitizes BCR-ABL1-expressing leukemic cells to tyrosine kinase inhibitors by targeting multidrug resistance 1 gene in T315I mutated cells. Exp Hematol. 2023;118:40–52. doi:10.1016/j.exphem.2022.12.003
- Mahdloo T, Sahami P, Ramezani R, et al. Up-regulation of miR-155 potentiates CD34+ CML stem/progenitor cells to escape from the growth-inhibitory effects of TGF-ss1 and BMP signaling. EXCLI J. 2021;20:748–763.
- Liu W, He J, Yang Y, et al. Upregulating miR-146a by physcion reverses multidrug resistance in human chronic myelogenous leukemia K562/ADM cells. Am J Cancer Res. 2016;6(11):2547–2560.
- Guo G, Kang Q, Zhu X, et al. A long noncoding RNA critically regulates Bcr-Abl-mediated cellular transformation by acting as a competitive endogenous RNA. Oncogene. 2015;34(14):1768–1779. doi:10.1038/onc.2014.131
- Golestaneh AF, Lecker LSM, Schlegel J, et al. Large scale in vivo micro-RNA loss of function screen identified miR-29a, miR-100 and miR-155 as modulators of radioresistance and tumor-stroma communication. Int J Cancer. 2019;144(11):2774–2781. doi:10.1002/ijc.32019
- Ocheni S, Iwanski GB, Schafhausen P, et al. Characterisation of extramedullary relapse in patients with chronic myeloid leukemia in advanced disease after allogeneic stem cell transplantation. Leuk Lymphoma. 2009;50(4):551–558. doi:10.1080/10428190902755513
- Hochhaus A, Saussele S, Rosti G, et al. Chronic myeloid leukaemia: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2017;28(suppl_4):iv41–iv51. doi:10.1093/annonc/mdx219
- Yuri P, Hendri AZ, Danarto R. Association between tumor-associated macrophages and microvessel density on prostate cancer progression. Prostate Int. 2015;3(3):93–98. doi:10.1016/j.prnil.2015.06.002
- Yap E, Norziha ZA, Simbun A, et al. MicroRNAs that affect the Fanconi Anemia/BRCA pathway are downregulated in imatinib-resistant chronic myeloid leukemia patients without detectable BCR-ABL kinase domain mutations. Leuk Res. 2017;59:32–40. doi:10.1016/j.leukres.2017.05.015
- Shulman DS, Lee MA, Lehmann LE, et al. Outcomes following bone marrow transplantation in children with accelerated phase or blast crisis chronic myelogenous leukemia in the era of tyrosine kinase inhibitors. J Pediatr Hematol Oncol. 2016;38(8):610–614. doi:10.1097/MPH.0000000000000636
- Senapati J, Jabbour E, Kantarjian H, et al. Pathogenesis and management of accelerated and blast phases of chronic myeloid leukemia. Leukemia. 2023;37(1):5–17. doi:10.1038/s41375-022-01736-5
- Martinovic Z, Kovac D, Martinovic C. Recurrences in stage II rectal carcinoma after curative resection alone: from the viewpoint of angiogenesis. World J Surg Oncol. 2016;14:122. doi:10.1186/s12957-016-0877-6
- Cantu De Leon D, Lopez-Graniel C, Frias Mendivil M, et al. Significance of microvascular density (MVD) in cervical cancer recurrence. Int J Gynecol Cancer. 2003;13(6):856–862. doi:10.1136/ijgc-00009577-200311000-00018
- Shi Z, Li Y, Qian X, et al. MiR-340 inhibits triple-negative breast cancer progression by reversing EZH2 mediated miRNAs dysregulated expressions. J Cancer. 2017;8(15):3037–3048. doi:10.7150/jca.19315
- Flatmark K, Borgen E, Nesland JM, et al. Disseminated tumour cells as a prognostic biomarker in colorectal cancer. Br J Cancer. 2011;104(9):1434–1439. doi:10.1038/bjc.2011.97
- Xiao C, Hong H, Yu H, et al. MiR-340 affects gastric cancer cell proliferation, cycle, and apoptosis through regulating SOCS3/JAK-STAT signaling pathway. Immunopharmacol Immunotoxicol. 2018;40(4):278–283. doi:10.1080/08923973.2018.1455208
- Huang Z, Xu Y, Wan M, et al. miR-340: a multifunctional role in human malignant diseases. Int J Biol Sci. 2021;17(1):236–246. doi:10.7150/ijbs.51123
- Qin Y, Zhou X, Huang C, et al. Lower miR-340 expression predicts poor prognosis of non-small cell lung cancer and promotes cell proliferation by targeting CDK4. Gene. 2018;675:278–284. doi:10.1016/j.gene.2018.06.062
- Zhang Z, Zhou Q, Luo F, et al. Circular RNA circ-CHI3L1.2 modulates cisplatin resistance of osteosarcoma cells via the miR-340-5p/LPAATbeta axis. Hum Cell. 2021;34(5):1558–1568. doi:10.1007/s13577-021-00564-6
- Cui F, Hao ZX, Li J, et al. SOX2 mediates cisplatin resistance in small-cell lung cancer with downregulated expression of hsa-miR-340-5p. Mol Genet Genomic Med. 2020;8(5):e1195. doi:10.1002/mgg3.1195
- Soltani I, Bahia W, Farrah A, et al. Potential functions of hsa-miR-155-5p and core genes in chronic myeloid leukemia and emerging role in human cancer: a joint bioinformatics analysis. Genomics. 2021;113(4):1647–1658. doi:10.1016/j.ygeno.2021.04.014
- Rokah OH, Granot G, Ovcharenko A, et al. Downregulation of miR-31, miR-155, and miR-564 in chronic myeloid leukemia cells. PLoS One. 2012;7(4):e35501. doi:10.1371/journal.pone.0035501
- Fallah P, Amirizadeh N, Poopak B, et al. Expression pattern of key microRNAs in patients with newly diagnosed chronic myeloid leukemia in chronic phase. Int J Lab Hematol. 2015;37(4):560–568. doi:10.1111/ijlh.12351