ABSTRACT
Objectives
This study aims to investigate the role of excessive Protein Tyrosine Phosphatase Non-Receptor Type 21 (PTPN21) in the proliferation of Acute Lymphoblastic Leukemia (ALL) cells with EGF stimulation.
Methods
PTPN21 was overexpressed in ALL cell lines by lentiviral transfection. Apoptosis was assayed by Annexin V/7-AAD staining. The proliferation and cell cycle of EGF-treated ALL cells were assessed by MTT and Ki-67/7-AAD staining respectively. The phosphorylation of Src tyrosine kinase and mediators of distinct MAPK pathways were assessed by Western blot.
Results
Overexpression of PTPN21 had minimal effect on the apoptosis of ALL cells, but significantly promoted the proliferation and cell cycle progression of ALL cells stimulated with EGF. The activity of Src tyrosine kinase and the MAPK pathways was elevated. Inhibition of MAPK pathways by specific inhibitors mitigated this pro-proliferative effect of excessive PTPN21 on EGF-stimulated ALL cells.
Conclusion
PTPN21 may facilitate ALL progression by promoting cell proliferation via the Src/MAPK signaling pathways.
Introduction
Acute Lymphoblastic Leukemia (ALL) is an aggressive hematologic malignancy characterized by the abnormal proliferation of immature lymphocytes in bone marrow, peripheral blood, and other organs [Citation1]. While chemotherapeutic treatment has proven successful in curing over 85% of pediatric patients, its efficacy in adult patients is notably lower. Treatment failure and relapse are significant challenges in adult ALL patients, often resulting in poor outcomes [Citation2,Citation3]. Genome-wide analyses have identified numerous unfavorable genetic alterations closely associated with high-risk ALL [Citation4,Citation5]. The exploration of genetic alterations not only contributes to our understanding of ALL's pathogenesis but also aids in the identification of new therapeutic targets in ALL.
In the previous study, we were the first to reveal that protein tyrosine phosphatase non-receptor type 21 (PTPN21) mutations were implicated in the relapse of Philadelphia chromosome-negative ALL following allogeneic hematopoietic stem cell transplantation (allo-HSCT) [Citation6]. The PTPN21 gene encodes a member of the protein tyrosine phosphatase (PTP) family, which plays pivotal roles in the regulation of cellular phosphorylation and signal transduction [Citation7,Citation8]. Previous reports have uncovered that abnormal expression of PTPN21 is involved in the pathogenesis of various malignancies by promoting cell proliferation, abnormal differentiation, and resistance to apoptosis [Citation9–12]. For instance, PTPN21 expression significantly increases in bladder cancer and is positively correlated with the growth and invasiveness potential of cancer cells [Citation9]. High expression of PTPN21 has also been reported in B-cell non-Hodgkin's gastric lymphoma, where it enhances STAT5 activity, potentially leading to the development of hematologic-related human gastric carcinoma [Citation11]. Our previous study found that endogenous PTPN21 inhibits the sensitivity of ALL cells to natural killer (NK)-mediated lysis, providing partial insight into the role of PTPN21 in ALL pathogenesis through an immune escape mechanism [Citation13]. However, whether PTPN21 directly influences ALL cells has yet to be elucidated.
In this study, we aim to investigate the impact of excessive PTPN21 on proliferation and its underlying mechanism in ALL cells. Our results suggest that elevated PTPN21 expression may activate Src tyrosine kinase and the downstream MAPK signaling pathways, thereby promoting the proliferation of ALL cells. Furthermore, our findings indicate that the specific MAPK signal pathways involved may vary among different ALL cell lines.
Materials and methods
Cell culture
Human ALL cell lines Jurkat, NALM-6, and Reh were purchased from the Cell Bank of Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences (Shanghai, China). All ALL cells were cultured in Roswell Park Memorial Institute 1640 medium (Corning, NY, USA) supplemented with 10% fetal bovine serum (FBS, Gibco, Carlsbad, CA). HEK293 T cells were cultured in Dulbecco's modified Eagle's medium with 4.5 g/L glucose (Corning, NY, USA) supplemented with 10% FBS. All cell lines were cultured at 37°C in a humidified incubator with 5% CO2.
Chemicals and antibodies
Human epidermal growth factor (EGF) was purchased from Peprotech (London, UK). Cells were treated with 100 ng/ml EGF for stimulation. The inhibitors PD98059, SP600125, and SB203580 were purchased from Selleck Chemicals (Houston, TX, USA). PD98059 (20 μM), SP600125 (40 μM), and SB203580 (40 μM) were applied for the inhibition of MEK, JNK, and p38, respectively. The following antibodies were used: DYKDDDDK Tag (FLAG) antibody (ab205606, 1:1000) and PTPN21 (ab12550, 1:500) antibody were purchased from Abcam (Cambridge, UK); p-Src (Tyr527) antibody (#2105, 1:1000), Src antibody (#2108, 1:1000), p-ERK1/2 antibody (#9101, 1:1000), ERK1/2 antibody (#9102, 1:1000), p-p38 MAPK (Thr180/Tyr182) antibody (#9211, 1:1000), p38 MAPK antibody (#9212, 1:1000), p-SAPK/JNK antibody (#9251, 1:1000), SAPK/JNK antibody (#9252, 1:1000) were purchased from Cell Signaling Technology (Beverly, MA, USA); GAPDH antibody (AF0006, 1:1000) was purchased from Beyotime (China). Fluorescent IgG IRDye® 650 goat anti-mouse (#926-68020, 1:1000) and goat anti-rabbit secondary antibodies (#926-68021, 1:1000) were purchased from LI-COR (USA).
Plasmids and transfection
pHIV7/SFFV-GFP (a gift from Professor Jiing-kuan Yee, City of Hope National Medical Center, CA, USA) was digested by the BamHI enzyme (New England Biolabs, USA) and ligated with PTPN21-3×FLAG using T4 DNA ligase (New England Biolabs, USA). The resulting plasmid was transfected into HEK293 T cells with lentiviral packaging plasmids pMD2.G and psPAX2 (Thermo Fisher Scientific, USA) using Lipofectamine 2000 (Invitrogen, USA) according to the manufacturer’s guide. After 48 hours of culture, the virus-containing media were collected and filtered with a 0.45μm filter. Virus particles were concentrated by high-speed centrifugation. The target cells were infected with a virus titer at a multiplicity of infection (MOI) of 10. The MOI was determined by TaqMan real-time PCR on an ABI PRISM 7000 machine. The primers for plasmid construction are as follows: PTPN21-F: 5′-ACTGAGTCGCCCGGGTACCGAGGCCACCATGCCACTGCCATTTGGGTTGA-3′,
PTPN21-R: 5′-CACCGTCATGGTCTTTGTAGTCGATGAGCCTGGAGCTTTTC-3′;
53xFLAG-F: 5′-TGAAAAGCTCCAGGCTCATCGACTACAAAGACCATGACGGT-3′,
53xFLAG-R: 5′-GAAGATCTTCACTTGTCATCGTCATCCTTGTAATC-3′.
Immunofluorescence (IF) analysis
Cells were collected and rinsed with PBS twice, then smeared onto slides using cytocentrifugation. After fixation in 4% polyformaldehyde for 10 min and permeabilization with 0.5% Triton X-100 in PBS for 5 min, the slides were blocked with 1% BSA and 0.1% Tween-20 at room temperature (RT) for 1 h. Subsequently, the slides were incubated with 1:400 diluted Flag-tag primary antibody at 4°C overnight, followed by incubation with 1:100 diluted secondary antibody at RT for 1 h. Fluorescent images were captured by confocal microscopy and analyzed using LSM510 software 3D (Zeiss, Germany).
Apoptosis assay
Apoptosis was measured using the Apoptosis Detection kit (BD Pharmingen, San Diego, CA, USA). Briefly, cells were harvested and washed twice with cold PBS, then resuspended in Annexin V binding buffer, followed by incubation with the Annexin V-PE antibody and 7-aminoactinomycin D (7-AAD) solution at RT for 15 min. Finally, the cells were evaluated using an FC500 MCL flow cytometer (Beckman Coulter, USA). The FlowJo software (v 7.2.2) was utilized for automatic cell gating and subsequent analysis. Annexin V+ cells were considered apoptotic cells.
Proliferation assay
CCK-8 assay was employed to assess cell proliferation following the manufacturer’s instruction. In brief, the cells with the indicated treatment were incubated with CCK-8 solution (Keycell, China) for 1 hour at 37°C. Subsequently, the OD450 values were measured using a microplate reader (BIOBASE, China).
Cell cycle assay
Cells were deprived of FBS overnight and simulated with 100 ng/ml EGF for 30 min before being collected for cell cycle detection. The cells were permeabilized using the Foxp3/Transcription Factor Staining Buffer Set (eBioscience, San Diego, CA). Specifically, after incubation with Fixation/Permeabilization solution at 4°C for 45 min, cells were resuspended with 100μl Permeabilization Buffer and incubated with 5μl of Ki-67 antibody (Biolegend, San Diego, CA, USA) and 10μl of 7-AAD (BD Pharmingen, San Diego, CA, USA) at 4°C for 30 min. Finally, the stained cells were resuspended in 500 μl of PBS for flow cytometry analysis.
Western blot analysis
Cells were harvested and lysed in radioimmunoprecipitation assay (RIPA) lysis buffer (Boster Biological Technology, Wuhan, China) with protease inhibitor cocktail (Thermo Fisher Scientific, USA) and PhosSTOP phosphatase inhibitor (Roche Life Science, Basel, Switzerland) on ice for 1 h. The resulting cell lysates were centrifuged at 12,000 g at 4°C for 10 min. The supernatant was then mixed with loading buffer (Fu De Biological Technology, Hangzhou, China) and denatured at 100°C for 10 min. Protein concentrations were quantified using a BCA Kit (P0012S, Beyotime, Haimen, China) following the manufacturer’s instructions. Equal amounts of the protein samples were loaded onto SDS-PAGE gels (Fu De Biological Technology, Hangzhou, China), and separated by electrophoresis (110 V for 100 min), followed by transfer onto nitrocellulose membranes. Subsequently, the membranes were blocked with 5% non-fat milk (BD Pharmingen, San Diego, CA, USA) at RT for 2 h and then incubated with primary antibody solutions at 4°C overnight. After washing with TBST, the membranes were incubated with fluorescent secondary antibody solutions at RT for 1 h, followed by washing with TBST. Finally, the membranes were subject to visualization using an Odyssey Infrared Imaging System (LI-COR, NE, USA).
Statistical analysis
All experiments were repeated at least three times independently. Statistical analysis was performed using SPSS software (v.22, IBM, NY, USA). The Differences between mean values were evaluated using unpaired two-tailed Student’s t-test. Values are presented as the mean ± standard deviation and P < 0.05 was considered statistically significant.
Results
Overexpression of PTPN21 potentiated the proliferation and cell cycle progression of EGF-stimulated ALL cells
To explore the role of PTPN21 in ALL, we initially overexpressed PTPN21 in Jurkat, NALM-6, and Reh cells. Cells transfected with GFP-expressing plasmid (pHIV7-SFFV-GFP) were used as control. Next, we confirmed the overexpression of PTPN21 by Western blot and IF (a,b). Subsequently, we investigated apoptosis, proliferation, and cell cycle of control and PTPN21-overexpressing ALL cells. Annexin V/7-AAD assay results, which represent apoptosis [Citation14], revealed that overexpression of PTPN21 had no significant effect on the apoptosis of ALL cells (a,b).
Figure 1. PTPN21 was successfully overexpressed in Jurkat, NALM-6, and Reh cells. (a) Western blot data show that Flag-PTPN21 was overexpressed in Jurkat, NALM-6, and Reh cells. GAPDH was used as the internal control. (b) IF results reveal that Flag-PTPN21 could be detected in the transfected Jurkat, NALM-6, and Reh cells.
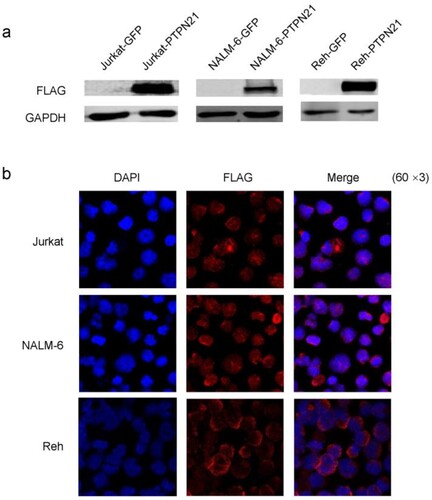
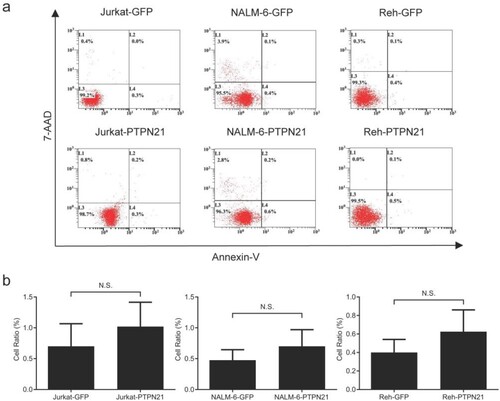
Figure 2. Excessive PTPN21 had no effects on apoptosis of Jurkat, NALM-6, or Reh cells. (a) Annexin V/7-AAD assay results reveal that overexpression of PTPN21 did not affect apoptosis of ALL cells. (b) Quantification of the Annexin V/7-AAD results. N.S., no significance.
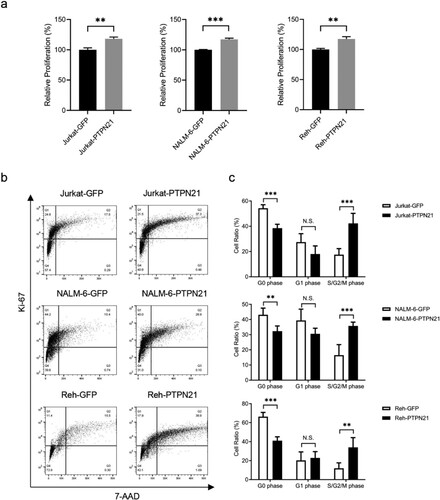
Since PTPN21 facilitates the EGF signal pathway and promotes cell proliferation in bladder cancer cells [Citation9], we then asked whether PTPN21 exerts pro-proliferative function in EGF-stimulated ALL cells. CCK-8 assay results uncovered that excessive PTPN21 substantially increased the proliferation of ALL cells upon EGF stimulation (a). Consistently, Ki-67/7-AAD staining outcomes, which indicate cell cycle [Citation15,Citation16], showed that overexpression of PTPN21 dramatically reduced the proportion of EGF-stimulated ALL cells in the G0 phase (resting phase) while substantially increasing the proportion of cells in the S/G2/M phase (b,c). These findings collectively demonstrate that excessive PTPN21 facilitates the proliferation and cell cycle progression of ALL cells with EGF stimulation.
Figure 3. Overexpression of PTPN21 facilitated proliferation and cell cycle progression in EGF-stimulated Jurkat, NALM-6, and Reh cells. (a) CCK-8 staining results show that overexpression of PTPN21 elevated cell proliferation in all three ALL cell types with EGF-stimulation. (b) Ki-67/7-AAD assay data uncover that the cell cycle progression is promoted by excessive PTPN21 in all three ALL cell lines. (c) Quantification of the Annexin V/7-AAD results. N.S., no significance. *P < 0.05, **P < 0.01, ***P < 0.001.
PTPN21 overexpression activated the Src/MAPK signal pathways
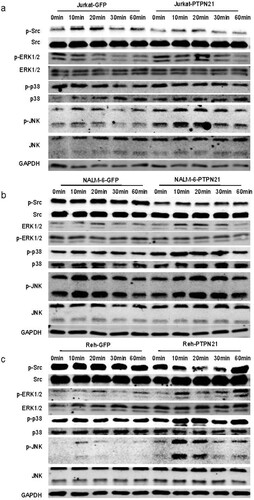
To delineate the mechanism by which PTPN21 fulfills the pro-proliferative function on ALL cells, we assessed the Src/MAPK pathways. Western blot data showed that the phosphorylation of Src at the Y527 residue (p-Src) was prominently decreased, indicating enhanced Src activity [Citation17]. By contrast, the phosphorylation of ERK (p-ERK) was elevated upon PTPN21 overexpression in all three ALL cell lines with EGF stimulation. Moreover, the p38 phosphorylation (p-p38) level was not altered in EGF-stimulated Jurkat cells but was upregulated in EGF-stimulated NALM-6 and Reh cells with PTPN21 overexpression. Additionally, excessive PTPN21 enhanced the phosphorylation of JNK (p-JNK) in EGF-stimulated Jurkat and Reh cells but had minimal effect on the p-JNK level in EGF-stimulated NALM-6 cells (a–c). These observations suggest that excessive PTPN21 can potentiate the Src and MAPK pathways in ALL cells.
Figure 4. Overexpression of PTPN21 activated Src and the MAPK signaling pathways in EGF-stimulated ALL cells. (a) Western blot data show that in Jurkat cells, PTPN21 overexpression reduced p-Src level and upregulated p-ERK1/2 and p-JNK levels, but had no impact on p-p38 level. (b) Western blot data reveal that in NALM-6 cells, PTPN21 overexpression led to a decreased level of p-Src and elevated levels of p-ERK1/2 and p-p38 but didn’t affect p-p38 level. (c) Western blot results uncover that in Reh cells, excessive PTPN21 downregulated p-Src level and elevated levels of p-ERK1/2, p-p38, and p-p38.
Inhibition of the MAPK pathways affected the pro-proliferative effect of PTPN21 on ALL cells
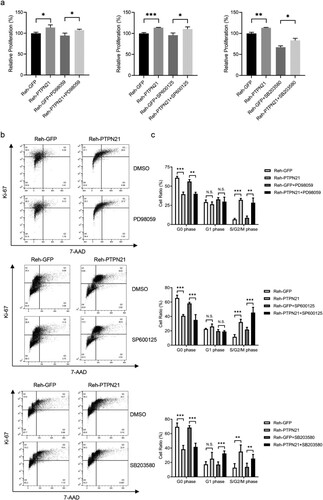
To determine whether inhibition of the MAPK pathways was sufficient to abolish the pro-proliferative effect of PTPN21 on EGF-stimulated ALL cells, we treated these cells with MEK inhibitor PD98059, JNK inhibitor SP600125 or p38 inhibitor SB203580, respectively. Because PTPN21 overexpression affected p-ERK and p-JNK levels in EGF-stimulated Jurkat cells, we treated these cells with PD98059 or SP600125. CCK-8 assay results indicated that excessive PTPN21 could increase the proliferation of EGF-stimulated Jurkat cells in the presence of PD98059 or SP600125 (a). However, while PD98059 treatment had minimal impact on the cell cycle of EGF-stimulated Jurkat cells with PTPN21 overexpression, SP600125 abolished the effect of excessive PTPN21 on the cell cycle of these cells, as indicated by the Ki-67/7-AAD staining assay data (b,c). Similarly, we treated control and PTPN21-overexpressing NALM-6 cells with PD98059 or SB203580 and assayed their proliferation and cell cycle. The results revealed that SB203580, but not PD98059, abolished the promotion effect of PTPN21 on the proliferation and cell cycle of EGF-stimulated NALM-6 cells (a–c). Furthermore, we treated Reh cells with or without PTPN21 overexpression with PD98059, SP600125, or SB203580 as excessive PTPN21 influenced all three MAPK pathways. CCK-8 staining assay outcomes showed that none of the inhibitors attenuated the pro-proliferative function of PTPN21 in Reh cells (a). However, Ki-67/7-AAD data uncovered that while PD98059 or SP600125 had a minimal impact on the cell cycle of these cells, SB203580 partially reversed the effect of PTPN21 overexpression on their cell cycle (b,c). These data further underscore the importance of the MAPK pathways in mediating the effect of PTPN21 in promoting the proliferation of ALL cells.
Figure 5. JNK inhibitor mitigated the pro-proliferative effect of PTPN21 on EGF-stimulated Jurkat cells. (a) CCK-8 staining results reveal that MEK inhibitor PD90859 and JNK inhibitor SP600125 have little impact on PTPN21’s pro-proliferative function in EGF-stimulated Jurkat cells. (b) Ki-67/7-AAD assay data show that the cell cycle progression promoted by excessive PTPN21 in Jurkat cells was not affected by PD90859 but was mitigated by SP600125. (b) Quantification of the Annexin V/7-AAD results. N.S., no significance. *P < 0.05, **P < 0.01, ***P < 0.001.
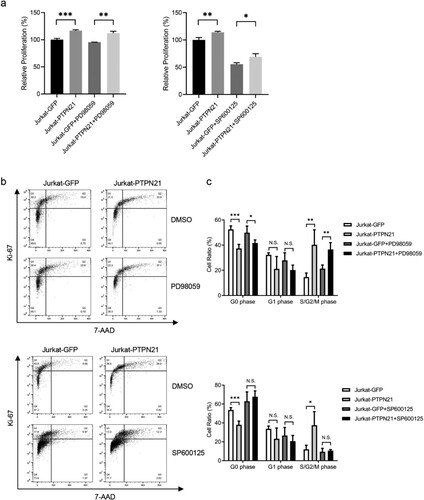
Figure 6. p38 pathway mediated the pro-proliferative function of PTPN21 in EGF-stimulated NALM-6 cells. (a and b) CCK-8 staining (a) and Ki-67/7-AAD assay data (b) show that the proliferation and cell cycle progression facilitated by PTPN21 overexpression in NALM-6 cells was not influenced by MEK inhibitor PD90859 but was affected by p38 inhibitor SB203580. (c) Quantification of the Annexin V/7-AAD results. N.S., no significance. *P < 0.05, **P < 0.01, ***P < 0.001.
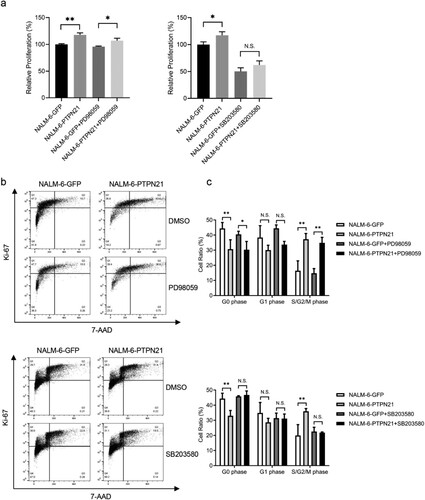
Figure 7. Inhibition of p38 partially attenuated the proliferation of EGF-stimulated Reh cells driven by PTPN21 overexpression. (a) CCK-8 staining assay data show that none of the inhibitors can abolish the proliferation-promoting effect of PTPN21 on Reh cells. (b) Ki-67/7-AAD assay results illustrate that MEK inhibitor PD90859 and JNK inhibitor SP600125 had minimal impact on the cell cycle of EGF-stimulated Reh cells while p38 inhibitor SB203580 significantly increased the proportion of Reh cells at the G1 phase with PTPN21 overexpression. (c) Quantification of the Annexin V/7-AAD results. N.S., no significant difference. *P < 0.05, **P < 0.01, ***P < 0.001.
Discussion
PTPN21, a member of the non-receptor tyrosine phosphatase family, was initially identified in a human skeletal muscle cDNA library [Citation18]. As a classical protein tyrosine phosphatase, PTPN21 plays pivotal roles in the regulation of cellular phosphorylation and signal transduction [Citation19,Citation20]. It has been demonstrated to be associated with the pathologic process in a variety of solid neoplasms [Citation10,Citation21,Citation22].
In our previous study, we conducted longitudinal whole-exome sequencing analysis on samples from Ph-B-cell ALL patients who received allo-HSCT. We compared genomic characteristics between patients with different prognoses. PTPN21 was specifically found to be mutated in relapsed cases but not in any of the non-relapsed cases, suggesting that PTPN21 mutation is associated with relapse. We hypothesize that PTPN21-mutated leukemia cells might acquire additional survival advantage, thus contributing to the pathogenesis of ALL relapse post-HSCT [Citation6]. However, the mechanism by which PTPN21 promotes leukemia is poorly understood.
To delineate the role of PTPN21 in ALL, we overexpressed PTPN21 in three representative ALL cell lines of different origins, including Jurkat cells (T-ALL derived), NALM-6 cells (B-ALL derived), and Reh cells (non-B/non-T-ALL derived). PTPN21 overexpression did not induce apoptosis in any of these three cell lines. We then assessed cell proliferation status by comparing the ratio of cells in different cell cycle phases. Our results revealed that PTPN21 overexpression led to a significant decrease in the number of cells in the G0 phase and a remarkable increase in the number of cells in the S/G2/M phases upon EGF stimulation, indicating that PTPN21 could substantially potentiate ALL cell proliferation. Given that abnormal cell proliferation is one of the most important mechanisms of tumorigenesis [Citation23], we speculate that PTPN21 may facilitate ALL progression by inducing aberrant activation of cell proliferation.
Previous studies have demonstrated that Src tyrosine kinase is a direct substrate of PTPN21 [Citation7]. PTPN21 increases the magnitude and duration of EGF signaling by activating Src tyrosine kinase, which requires dephosphorylation of its Y527 residue [Citation7]. In this study, our results showed that after EGF stimulation, overexpression of PTPN21 significantly promoted the dephosphorylation of Src at Y527 residue in ALL cells. Src kinase activation is considered an important oncogenic pathway that plays a vital role in the tumorigenesis of various malignancies [Citation24]. Exacerbated Src family kinase activation has been reported to predict a poor prognosis of acute leukemia [Citation25]. Thus, Src kinase may be a promising druggable target of leukemia carrying PTPN21 mutations.
As downstream pathways of Src kinase, the MAPK cascade pathways play pivotal roles in oncogenic processes, including cell proliferation, differentiation, genetic expression, and apoptosis [Citation26–28]. MAPK signaling activation has also been reported to efficiently promote progenitor proliferation and be associated with poor prognosis in ALL [Citation29,Citation30]. The MAPK signaling comprises three pathways: JNK, ERK, and p38 pathways [Citation31]. We examined the effect of PTPN21 on these MAPK pathways in ALL cell lines of different origins. The results revealed that overexpression of PTPN21 significantly increased the phosphorylation of ERK1/2 and JNK in Jurkat cells, ERK1/2 and p38 in NALM-6 cells, and ERK1/2, JNK and p38 in Reh cells, respectively.
We further identified the role of the MAPK pathways in PTPN21-mediated regulation of ALL cell proliferation using small molecule inhibitors for each MAPK pathway. Although ERK1/2 was activated in all ALL cell lines, its specific inhibitor PD98059 failed to eliminate the PTPN21-mediated proliferation promotion, implying that ERK1/2 is not the key regulator. The pro-proliferative effect of PTPN21 is partially attenuated by JNK inhibitor SP600125 in Jurkat cells and efficiently disrupted by p38 inhibitor SB203580 in NALM-6 cells, demonstrating that JNK and p38 contributed to the PTPN21-induced cell proliferation in Jurkat and NALM-6 cells, respectively. As to Reh cells, while PD98059 and SP600125 exerted no influence, SB203580 partially restrained their cell cycle progression, indicating that P38 may participate in this regulation process. The poor efficiency of individual inhibitors in mitigating the proliferation-promoting effect of PTPN21 on EGF-stimulated Reh cells may be attributed to the multiple MAPK pathways that PTPN21 employs to fulfill its function, and inhibitors blocked all these pathways are likely more effective in abolishing PTPN21’s impact. These findings also suggest that the MAPK pathway by which PTPN21 fulfills its pro-proliferative effect on ALL cells is cell type-dependent. However, the precise mechanisms underlying the pro-proliferative effect of PTPN21, particularly on Reh cells, need further investigation.
In summary, our data suggest that PTPN21 promotes the proliferation of ALL cells via activating the Src kinase-mediated MAPK pathways and that PTPN21 and its downstream pathways may potentially serve as novel therapeutical targets for ALL treatment.
Authors' contributions
Ni Zhu and Jieping Wei made equal contributions in research, data analysis and manuscript writing; He Huang and Haowen Xiao designed the experiment and ensured correct data analysis; Li-Mengmeng Wang assisted in research and supervised data collection; Haowen Xiao participated in the revision and review of the article.
Ethics statement
Not applicable since this article does not contain any studies with human or animal subjects.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Additional information
Funding
References
- Terwilliger T, Abdul-Hay M. Acute lymphoblastic leukemia: a comprehensive review and 2017 update. Blood Cancer J. 2017;7(6):e577. doi:10.1038/bcj.2017.53
- Gokbuget N, Dombret H, Ribera JM, et al. International reference analysis of outcomes in adults with B-precursor Ph-negative relapsed/refractory acute lymphoblastic leukemia. Haematologica. 2016;101(12):1524–1533. doi:10.3324/haematol.2016.144311
- Toft N, Birgens H, Abrahamsson J, et al. Results of NOPHO ALL2008 treatment for patients aged 1-45 years with acute lymphoblastic leukemia. Leukemia. 2018;32(3):606–615. doi:10.1038/leu.2017.265
- Mullighan CG, Goorha S, Radtke I, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446(7137):758–764. doi:10.1038/nature05690
- Roberts KG, Morin RD, Zhang J, et al. Genetic alterations activating kinase and cytokine receptor signaling in high-risk acute lymphoblastic leukemia. Cancer Cell. 2012;22(2):153–166. doi:10.1016/j.ccr.2012.06.005
- Xiao H, Wang LM, Luo Y, et al. Mutations in epigenetic regulators are involved in acute lymphoblastic leukemia relapse following allogeneic hematopoietic stem cell transplantation. Oncotarget. 2016;7(3):2696–2708. doi:10.18632/oncotarget.6259
- Cardone L, Carlucci A, Affaitati A, et al. Mitochondrial AKAP121 binds and targets protein tyrosine phosphatase D1, a novel positive regulator of Src signaling. Mol Cell Biol. 2004;24(11):4613–4626. doi:10.1128/MCB.24.11.4613-4626.2004
- Carlucci A, Gedressi C, Lignitto L, et al. Protein-tyrosine phosphatase PTPD1 regulates focal adhesion kinase autophosphorylation and cell migration. J Biol Chem. 2008;283(16):10919–10929. doi:10.1074/jbc.M707248200
- Carlucci A, Porpora M, Garbi C, et al. PTPD1 supports receptor stability and mitogenic signaling in bladder cancer cells. J Biol Chem. 2010;285(50):39260–39270. doi:10.1074/jbc.M110.174706
- Korff S, Woerner SM, Yuan YP, et al. Frameshift mutations in coding repeats of protein tyrosine phosphatase genes in colorectal tumors with microsatellite instability. BMC Cancer. 2008;8:329. doi:10.1186/1471-2407-8-329
- Plani-Lam JH, Chow TC, Fan YH, et al. High expression of PTPN21 in B-cell non-Hodgkin's gastric lymphoma, a positive mediator of STAT5 activity. Blood Cancer J. 2016;6(1):e388. doi:10.1038/bcj.2015.107
- Wu ZZ, Lu HP, Chao CC. Identification and functional analysis of genes which confer resistance to cisplatin in tumor cells. Biochem Pharmacol. 2010;80(2):262–276. doi:10.1016/j.bcp.2010.03.029
- Wang H, Zhu N, Ye X, et al. PTPN21-CDS(long) isoform inhibits the response of acute lymphoblastic leukemia cells to NK-mediated lysis via the KIR/HLA-I axis. J Cell Biochem. 2020;121(5-6):3298–3312. doi:10.1002/jcb.29601
- Vermes I, Haanen C, Steffens-Nakken H, et al. A novel assay for apoptosis. Flow cytometric detection of phosphatidylserine expression on early apoptotic cells using fluorescein labelled Annexin V. J Immunol Methods. 1995;184(1):39–51. doi:10.1016/0022-1759(95)00072-I
- Kim KH, Sederstrom JM. Assaying cell cycle status using flow cytometry. Curr Protoc Mol Biol. 2015;111:2861–2611. doi:10.1002/0471142727.mb2806s111
- Rahme R. Assaying cell cycle status using flow cytometry. Methods Mol Biol. 2021;2267:165–179. doi:10.1007/978-1-0716-1217-0_11
- Roskoski Jr. R. Src kinase regulation by phosphorylation and dephosphorylation. Biochem Biophys Res Commun. 2005;331(1):1–14. doi:10.1016/j.bbrc.2005.03.012
- Moller NP, Moller KB, Lammers R, et al. Src kinase associates with a member of a distinct subfamily of protein-tyrosine phosphatases containing an Ezrin-like domain. Proc Natl Acad Sci U S A. 1994;91(16):7477–7481. doi:10.1073/pnas.91.16.7477
- Sacco F, Boldt K, Calderone A, et al. Combining affinity proteomics and network context to identify new phosphatase substrates and adapters in growth pathways. Front Genet. 2014;5:115. doi:10.3389/fgene.2014.00115
- Cho YC, Kim BR, Cho S. Protein tyrosine phosphatase PTPN21 acts as a negative regulator of ICAM-1 by dephosphorylating IKKbeta in TNF-alpha-stimulated human keratinocytes. BMB Rep. 2017;50(11):584–589. doi:10.5483/BMBRep.2017.50.11.169
- Li XQ, Liu BC, Jiang XB, et al. Inhibition of PTPN21 has antitumor effects in glioma by restraining the EGFR/PI3 K/AKT pathway. Toxicol Appl Pharmacol. 2022;451:116180. doi:10.1016/j.taap.2022.116180
- Chen L, Qian Z, Zheng Y, et al. Structural analysis of PTPN21 reveals a dominant-negative effect of the FERM domain on its phosphatase activity. Sci Adv. 2024;10(9):eadi7404. doi:10.1126/sciadv.adi7404
- Sulic S, Panic L, Dikic I, et al. Deregulation of cell growth and malignant transformation. Croat Med J. 2005;46(4):622–638.
- Dehm SM, Bonham K. Src gene expression in human cancer: the role of transcriptional activation. Biochem Cell Biol. 2004;82(2):263–274. doi:10.1139/o03-077
- De Grandis M, Bardin F, Fauriat C, et al. JAM-C Identifies Src Family Kinase-Activated Leukemia-Initiating Cells and Predicts Poor Prognosis in Acute Myeloid Leukemia. Cancer Res. 2017;77(23):6627–6640. doi:10.1158/0008-5472.CAN-17-1223
- Fei X, Dou YN, Sun K, et al. TRIM22 promotes the proliferation of glioblastoma cells by activating MAPK signaling and accelerating the degradation of Raf-1. Exp Mol Med. 2023;55(6):1203–1217. doi:10.1038/s12276-023-01007-y
- Gao Z, Zhao GS, Lv Y, et al. Anoikis-resistant human osteosarcoma cells display significant angiogenesis by activating the Src kinase-mediated MAPK pathway. Oncol Rep. 2019;41(1):235–245. doi:10.3892/or.2018.6827
- Santarpia L, Lippman SM, El-Naggar AK. Targeting the MAPK-RAS-RAF signaling pathway in cancer therapy. Expert Opin Ther Targets. 2012;16(1):103–119. doi:10.1517/14728222.2011.645805
- Huang Y, Thoms JA, Tursky ML, et al. MAPK/ERK2 phosphorylates ERG at serine 283 in leukemic cells and promotes stem cell signatures and cell proliferation. Leukemia. 2016;30(7):1552–1561. doi:10.1038/leu.2016.55
- van der Zwet JCG, Buijs-Gladdines J, Cordo V, et al. MAPK-ERK is a central pathway in T-cell acute lymphoblastic leukemia that drives steroid resistance. Leukemia. 2021;35(12):3394–3405. doi:10.1038/s41375-021-01291-5
- Morrison DK. MAP kinase pathways. Cold Spring Harb Perspect Biol. 2012;4(11):a011254. doi:10.1101/cshperspect.a011254
