ABSTRACT
Background:
The BCL2 interacting protein 3-like (BNIP3L) protein is involved in multiple myeloma (MM) development and progression. This study aims to explore the connection between BNIP3L single-nucleotide polymorphisms (SNPs) and MM.
Methods:
SNaPshot was used to examine six SNP loci of the BNIP3L gene in enrolled subjects. The relationship between these loci and MM susceptibility and prognosis was explored. Survival analysis was used to evaluate the impact of different factors on patient survival.
Results:
The rs2874670 AA genotype and A allele were associated with increased MM risk (P < 0.05). The CCACAC haplotype had a higher frequency in MM, while CCGCAC had a higher frequency in normal patients (all P < 0.05). Patients with R-ISS stage I and II had higher survival rates than those with stage III (P < 0.05). Patients, who received chemotherapy followed by autologous stem cell transplantation, had longer survival time than those who only received chemotherapy (P < 0.05). Low levels of LDH and β2-MG were associated with better survival rates (P < 0.05). Cox regression identified that LDH levels, β2-MG levels, and R-ISS staging were the risk factors for the death of MM. Mann-Whitney U test found a significant difference in survival time between MM patients with different BNIP3L rs2874670 genotypes after BD chemotherapy (P < 0.05).
Conclusion:
To our knowledge, this is the first study to find that BNIP3L rs2874670 could increase MM susceptibility in China. Different BNIP3L rs2874670 genotypes may affect the prognosis of MM patients receiving BD chemotherapy.
Background
Multiple myeloma (MM) is known as a cell dyscrasia characterized by the proliferation of intramedullary clonal plasma cells along with an increase of monoclonal immunoglobin present in serum and urine [Citation1], usually considered to be a pivotal contributor to end-organ dysfunction, which include renal insufficiency, bone disease, hypercalcemia, anemia, and severe infections. While the advancement of drug development and biomarker identification has enhanced patient diagnosis and outcomes, there is still insufficient data to use biomarkers to cure multiple myeloma patients completely or to target a patient's unique biology in a predictive manner [Citation2,Citation3]. The treatment of MM has evolved with strategies focusing on achieving deep and durable responses, prolonging survival, and improving quality of life. Standard treatment involves chemotherapy, immunomodulatory drugs, proteasome inhibitors, steroids, and stem cell transplantation. Novel agents like monoclonal antibodies [Citation4] and targeted therapies have also emerged as effective options. Supportive care [Citation5] is crucial in managing MM-related complications. Recent advancements include minimal residual disease assessment and CAR-T cell therapy [Citation6]. Ongoing research aims to identify new targets and treatment modalities to improve outcomes for patients.
BNIP3L and BNIP3 share similarities with the BH3-only subgroup of the BCL2 family. They have sequence homology in the BH3 domain, reside in the mitochondrial outer membrane, and can interact with BCL2 and BCL-XL [Citation7]. Our previous study [Citation8] revealed that protein expression of BNIP3L in the bone marrow of patients with MM was significantly lower than that in the bone marrow of healthy donors. Then, based on the clinical parameters of 559 patients with MM in the prognosis-related microarrays of the GSE24080 cohort indicated that BNIP3L expression was negatively correlated with β2 microglobulin and creatinine levels, while it was positively correlated with albumin and hemoglobin levels in patients with MM. The above finding has revealed that BNIP3L can play an important role in the occurrence and development of MM, and it may be an independent indicator for predicting survival in MM [Citation8]. To date, no studies have investigated the association between single-nucleotide polymorphisms (SNPs) of BNIP3L, some clinical parameters and MM. Therefore, we conducted this study to explore their relationship.
In our study, we examined the distribution of six SNPs (rs17310286, rs3758098, rs2874670, rs10503786, rs73217800, and rs1042992) of BNIP3L gene and explored the relationship between six SNPs loci and the susceptibility and prognosis of MM in China, and analyzed the correlation between BNIP3L SNPs genotype, clinical indicators, treatment protocol and survival time in MM patients for providing more information for the diagnosis, treatment, and prognosis of MM.
Methods
Study subjects
This study comprised 185 subjects, 94 MM patients who visited the Hematology Department between Jan 2022 and Oct 2022, and 91 healthy controls. Blood samples were collected from the above 94 MM patients. All subjects included in this experiment met the National Comprehensive Cancer Network guidelines. Any patient with one of the following conditions would be excluded from our issues: (1) hepatitis B virus, hepatitis C virus, HIV, or other viral infections; (2) neoplastic disease; and (3) other hematological diseases. The stage of MM patients was differentiated according to the Revised International Staging System (R-ISS) and Durie-Sallon Staging System (DS). We recruited 91 healthy individuals as a control group with no previous anemia, inflammation, or tumor history. The clinical characteristics of all the individuals involved in the experiment are presented in .
Table 1. Clinical features of patients with MM and healthy control patient.
The study was approved by the ethics committee (NO.2023-E160-01). The subjects were briefed about the purpose, steps, benefits, risks, inconveniences, and interests of the study, and they all agreed to participate in the study and signed a written informed consent.
Measurement of serum-related clinical parameters
The hemoglobin levels were measured with a Beckman Coulter LH780 blood analyzer (Beckman Coulter, Brea, CA). The serum biochemical indices were analyzed with a Hitachi 7600 automatic biochemical analyzer (Hitachi, Tokyo, Japan). The following biochemical indicators were measured: β2-microglobulin (β2-MG), albumin (Alb), calcium (Ca), serum creatinine (Cr), and LDH. The erythrocyte sedimentation rate (ESR) was measured with an Automated erythrocyte sedimentation (Sysmex, Kobe, Japan). The immunoglobulin subtype was differentiated using the Abbott Axsym System (Abbott Laboratories, Chicago, IL). Bone marrow smears were stained with Wright stain solution (Baso, Zhuhai, China), and the ratio of plasma cells was calculated.
Selection of BNIP3L SNPs
In this study, SNPs information on the BNIP3L gene was selected by data from the PubMed and NCBI-SNP databases and results about the relationship between BNIP3L gene polymorphisms and susceptibility [Citation9]. All SNPs were established with the minor allele frequencies (MAF) > 5% to prevent false negative results. In this way, six SNPs were selected:rs17310286, rs3758098, rs2874670, rs10503786, rs73217800, and rs1042992.
DNA extraction and genotyping
Genomic DNA was collected from peripheral blood samples according to the instructions of the UE Blood genomic DNA preparation kit (Jiangsu Kang Wei Century Biotechnology Co, LTD). The primer sequences for the BNIP3L gene loci were summarized in . The Snapshot polymerase chain reaction was consistent with previous work [Citation10].
Table 2. Primer sequences used for detecting BNIP3L SNPs.
Follow-up of the enrolled patients
We collected the overall survival (OS) time of the patients from their initial diagnosis to the cut-off time for follow-up, November 6, 2023, in this study. Patients were excluded if they: (1) were smokers; (2) were taking any medications potentially affecting glucose levels for at least three months before recruitment (we did not exclude patients taking folic acid, iron, vitamin D or iodine supplements); (3) had a history of cardiovascular disease; (4) pregestational diabetes mellitus, or (5) had multiple pregnancies. Finally, we got the complete clinical and survival data of 70 patients for analysis. Survival analysis was conducted by the Kaplan-Meier method and Cox regression analysis. Clinical parameters are grouped according to the Guidelines for the diagnosis and management of multiple myeloma in China (2022 revision) [Citation11].
Treatment
We retrospectively reviewed the medical records of 70 MM patients, of which 33 patients(47.1%) received chemotherapy and autologous stem cell transplantation, 37 patients (52.9%) only received chemotherapy because some patients failed to collect sufficient stem cells before transplantation. Patients in this study chose their chemotherapy treatments based on their individual conditions, including five main protocols: BD(Bortezomib + Dexamethasone), VRD(bortezomib + lenalidomide + dexamethasone), VCD (bortezomib + cyclophosphamide + dexamethasone), IRD(Icazomib + Cyclophosphamide + Dexamethasone), PAD(Bortezomib + Azithromycin + Dexamethasone). The VRD protocol is preferred in primary care, accounting for 54.8% of all patients who received chemotherapy. Additionally, 13.9% of patients received the BD protocol, 8.6% received the VCD protocol, 3.2% received the IRD protocol, and 6.5% received the PAD protocol.
Statistical analysis
The IBM SPSS software package version 26 (IBM, Armonk, NY) was used to perform the statistical analyses. A Kolmogorov–Smirnov test was used to make a judgment on the distribution type of the data. The Hardy-Weinberg equilibrium (HWE) of polymorphisms for genome types in healthy and case groups was evaluated using the goodness-of-fit chi-square. The chi-square test or Fisher’s exact probability method was applied to carry out a comparison of genotype and allele frequencies of rs17310286, rs3758098, rs2874670, rs10503786, rs73217800 and rs1042992 of the MM and control groups. The chi-square test for categorical variables was chosen to compare the clinical parameters of MM patients with BNIP3L polymorphisms. To assess the relative risk of a given allele and genotype, binary logistic regression was used to calculate odds ratios (ORs) and 95% confidence intervals (CIs) after adjusting for age and gender. Kaplan-Meier method and Cox proportional hazards regression model were used to compare the survival time of different BNIP3L polymorphisms and clinical parameters. Mann–Whitney test was used to calculate the difference in survival time of MM patients with different alleles after receiving different chemotherapy protocols. The SHEsis online software [Citation12] (https://analysis.bio-x.cn) was used to conduct linked disequilibrium (LD) and haplotype analyses. P < 0.05 were considered statistically significant.
Results
Basic clinical features of MM patients
The clinical features of all the subjects in our experiment are listed in . In total, 94 patients and 91 healthy controls were enrolled. There was no difference in gender or age between the two different groups (all P > 0.05). IgG immunoglobulin subtypes among the patients we included were the most common, accounting for 45.7%. It was followed by IgA subtypes, accounting for 21.3%. The proportion of patients with MM in DS stage III is the most significant, accounting for 83.0%, while the proportion of patients in R-ISS stage II and III is similar, accounting for 40.9% and 38.7% respectively.
Correlation between BNIP3L polymorphisms and the risk of MM
Frequencies of genotypes and alleles of the six SNPs (rs17310286, rs3758098, rs2874670, rs10503786, rs73217800, and rs1042992) polymorphisms of the BNIP3L gene in MM and healthy control groups were showed in . The distribution of genotypes of the six SNPs in both patient and control groups fits with Hardy-Weinberg equilibrium (all P > 0.05). The genotype distribution and the allele frequency of rs2874670 were different between MM patients and healthy control patients (P < 0.05). After analyzing the BNIP3L rs2874670 polymorphisms, we found that the presence of the AA genotype exhibited a visibly raised risk of MM compared to patients with the GG genotype (OR = 3.053, 95%CI 1.264-7.374, P = 0.013). Likewise, patients with the BNIP3L rs2874670 A allele were 1.651 times higher (OR = 1.651, 95%CI 1.095-2.490, P = 0.017) than those with the G allele. A binary logistic regression analysis after adjusting for age and gender demonstrated that there existed no significant difference between the BNIP3L rs17310286, rs3758098, rs10503786, rs73217800, and rs1042992 polymorphisms and the risk of MM (all P > 0.05).
Table 3. Genotype and Allele frequencies of BNIP3L polymorphisms for MM and healthy control groups.
Association of BNIP3L polymorphisms with clinical parameters
BNIP3L SNPs was analyzed to explore their correlations with different clinical parameters, including β2-microglobulin (β2-MG), albumin (Alb), calcium (Ca), serum creatinine (Cr) and LDH, erythrocyte sedimentation rate (ESR) and immunoglobulin subtype. The analytical results were summarized in (rs17310286, rs3758098, and rs2874670) and (rs10503786, rs73217800, and rs1042992). Unfortunately, we failed to find anything significantly different between the BNIP3L polymorphisms and the clinical parameters of MM patients.
Table 4. Associations of BNIP3L polymorphisms with clinical parameters of patients with MM.
Table 5. Associations of BNIP3L polymorphisms with clinical parameters of patients with MM.
LD analysis
Seraligned alleles on the same SNPs (such as SNPs within the same gene) are usually inherited together, a phenomenon called linkage disequilibrium (LD) [Citation13]. In an association study, risk variants can usually be detected through direct or indirect assay of those associated markers in linkage disequilibrium (LD) containing risk variants [Citation14]. We performed an LD analysis to explore the possible relationship between the polymorphisms of these six SNPs and the risk of MM. Detection of the LD intensity between each unknown marker and a certain test marker can predict the ability of this group of SNPs to detect risk variants indirectly. The observed D’ value and r2 were chosen to represent the degree of association. For patients with MM, as the SNPs of BNIP3L gene, rs17310286 and rs3758098, rs17310286 and rs2874670, rs17310286 and rs10503786, rs17310286 and rs73217800, rs17310286 and rs1042992, rs3758098 and rs2874670, rs3758098 and rs10503786, rs3758098 and rs73217800, rs3758098 and rs1042992, rs2874670 and rs10503786, rs2874670 and rs73217800, rs2874670 and rs1042992, rs10503786 and rs73217800, rs10503786 and rs1042992, rs73217800 and rs1042992 showed strong LD (all D’ > 0.8, ). However, the other SNP did not show a strong LD between them.
Table 6. LD analysis the relationships between six SNPs of BNIP3L gene and risk of MM.
Haplotype analysis
The inheritance of a new mutation will be inherited along with the allele around the mutation site, which is all in LD with each other. These contiguous sets of alleles in which co-inheritance occurs are called ‘haplotypes’ [Citation13]. Haplotype analyses for all patients and healthy control groups were performed with the SHEsis software. Frequencies of four major haplotypes of the six SNPs are listed in . All those frequencies under 0.03 will be ignored in the analysis. Findings revealed that CCACAC showed a higher frequency in patients with MM. CCACAC showed a link with a significantly raised risk of MM (OR, 1.754; 95% CI, 1.109–2.775; P = 0.016). Instead, CCGCAC in the control group showed a higher frequency, and our findings suggested a protective effect against the development of the disease (OR, 0.478; 95% CI, 0.262–0.869; P = 0.014). Our results showed no correlation between CCATGC and TAGCAT haplotypes and MM (all P > 0.05).
Table 7. Major haplotype frequencies of BNIP3L gene in MM and control patients.
Survival analysis
Seventy MM patients completed follow-up records in this study. The median follow-up period was 21.9 months (range, 3.3–94.7 months), and the median OS was 26.4 months (95%CI 22.4-30.1). Although there was a tendency towards lower survival rates in some groups, we found no significant difference in OS between different groups of BNIP3L polymorphisms (A–F). Further research was performed to analyze the clinical parameters affecting the OS of MM patients. Our results showed that the survival rates of R-ISS stage I and II MM patients were higher than those of R-ISS stage III patients (G, log-rank P < 0.001), and the survival rates of MM patients with lower levels of LDH and β2-MG were higher than those of higher level of LDH and β2-MG(H,I, all log-rank P < 0.05). In addition, MM patients who received chemotherapy followed by autologous stem cell transplantation had significantly longer survival time than those who only received chemotherapy (J, log-rank P < 0.05).
Figure 1. Kaplan-Meier survival curves of MM patients with different BNIP3L polymorphisms and clinical parameters by log-rank test (A–F: rs17310286, rs3758098, rs2874670, rs10503786, rs73217800, rs1042992, all P > 0.05; G–J: R-ISS stage, LDH, β2-MG, Treatment, all *P < 0.05; K–P: Age, Gender, Hb, Cr, and Plasma cells, Chemotherapy regimens, all P > 0.05).
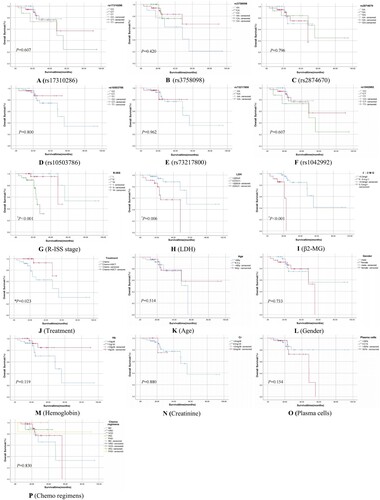
Cox regression analysis of risk factors of mortality in MM patients was presented in . MM patients with low levels of LDH and β2-MG had better survival rates than those with high levels (HR = 8.707, 5.474, respectively; all P < 0.05), and with R-ISS stage I and II had higher survival rates than those with stage III (HR = 15.427,132.84, respectively; all P < 0.05). The difference in survival time between the BNIP3L SNP rs2874670 genotype of MM patients who received different treatment protocols is shown in Supplementary Table 1. The results showed that there was a significant difference in survival time between MM patients with BNIP3L rs2874670 AA genotype and those with GG + GA genotype after receiving BD chemotherapy protocol (Mann–Whitney U test, Z = −1.974, P < 0.05).
Table 8. Cox regression analysis of risk factors for mortality in MM patients.
Discussion
BNIP3L, a member of the BCL-2 family, has been identified for over two decades [Citation15]. Mitophagy induced by BNIP3L has since been found in neurons, retinal ganglion cells, renal cells, and numerous tumor cells [Citation16–19]. BNIP3 and BNIP3L are considered crucial receptors of mitophagy and respond to the hypoxic microenvironment and induce mitophagy-mediated suppression of several tumor developments. BNIP3L plays a crucial role in mitophagy by either promoting the formation of autophagosomes or aiding in their recruitment to target mitochondria [Citation20]. Mitophagy improves tumor stem cell plasticity through metabolic remodeling to help tumor cells adapt better to the tumor microenvironment [Citation9,Citation21]. As a result, the early studies of these proteins were centered around their involvement in the process of cell death. It was reported that BNIP3L is associated with an imbalance in the process of autophagy in mitochondria [Citation22,Citation23]. Mitochondrial dysregulation is closely associated with various human diseases and has been documented to play a contradictory role in tumorigenesis [Citation24–26]. Previous research has revealed that BNIP3L is associated with various malignant tumors, such as acute myeloid leukemia (AML) and myelodysplastic syndromes (MDS) [Citation27], glioblastoma [Citation28], pancreatic cancer [Citation29], and lung cancer [Citation30]. In 2016, Lazarini et al. suggested that reduced BNIP3L expression in terminal MDS and AML may be involved in apoptosis imbalance, and also proposed that BNIP3L could serve as an independent prognostic factor for MDS [Citation27]. Additionally, Zhou et al. indicated a significant association between BNIP3L genetic polymorphisms (rs147389989, rs1042992, and rs17310286) and schizophrenia, and the BNIP3L gene had potential pathogenic mutations and functions [Citation9]. Research recently implied that BNIP3L could recruit TR3 to mitochondria to inactivate melanoma cells [Citation31]. Another study showed that Ewing sarcoma cells can survive by degrading the endogenous BNIP3L [Citation32]. Besides, silencing of BNIP3L in AML cells showed an obvious connection with the increased sensitivity to mitochondria, indicating a possible correlation between BNIP3L-induced mitophagy and tumor cell survival [Citation33]. Previous research [Citation11] also proved that the downregulation of BNIP3L may play an important role in the occurrence and development of MM, and the promoting cancer capacity may be related to the pathway of the adipocytokine signaling pathway.
In this study, the research results showed that there was an obvious correlation between BNIP3L rs2874670 polymorphisms and MM patients. After a binary logistic regression statistical analysis adjusted by age and gender, we found that the presence of the rs2874670 AA genotype, as well as the rs2874670 A allele, exhibited a 3.053 times higher and 1.651 times higher risk of MM than the average, respectively (all P < 0.05). Among the MM patients with BNIP3L rs2874670 in our study, 29(30.85%) patients have the AA genotype and 75(79.79%) patients have the A allele. However, the genotype distributions or allele frequencies of rs17310286, rs3758098, rs10503786, rs73217800, and rs1042992 showed no significant difference between MM and healthy patients. Therefore, our results indicated that BNIP3L rs2874670 polymorphism is involved with susceptibility to MM in China.
As we know, gender, aging, DS stage, R-ISS stage, Hb, Cr, Alb, β2-MG, LDH, ESR, and Plasma cells are the important clinical indicators for the occurrence and development of MM. Therefore, we further analyzed the correlations of BNIP3L SNP polymorphisms with these clinical parameters for MM patients. Nevertheless, our analysis has not found any significant discrepancy between the BNIP3L SNP polymorphisms and these clinical parameters in MM patients based on current data. Therefore, these parameters may be not confounding factors for BNIP3L SNP polymorphism susceptibility to MM patients.
In addition, as the BNIP3L gene SNPs, an LD analysis determined that fifteen SNPs combinations all showed strong LD (all D’>0.8). Through a haplotype analysis for the diseased and healthy groups, we found that one BNIP3L haplotype, CCGCAC, was linked with MM in the Chinese population and showed a protective effect against MM. The CCACA haplotypes had an association with the raised risk of MM.
Finally, we conducted a survival analysis to analyze the differences between BNIP3L SNPs genotype, clinical indicators, treatment protocol and survival time in MM patients. Our research has revealed that MM patients with low levels of LDH and β2-MG had better survival rates than those with high levels (log-rank P < 0.05). This finding was consistent with our results that patients with R-ISS stage I and II had higher survival rates than those with stage III. (log-rank P < 0.05). Due to R-ISS incorporates LDH and cytogenetic abnormalities in addition to ISS parameters (defined by β2-MG and albumin), it is necessary to pay attention to the LDH and β2-MG levels of MM patients and manage MM patients' R-ISS stages. In addition, patients in our study who received chemotherapy followed by autologous stem cell transplantation had significantly longer survival time than those who only received chemotherapy (log-rank P < 0.05). This demonstrates the crucial role of autologous stem cell transplantation in providing complete relief from MM and even achieving complete remission. However, our survival analysis failed to find any difference in survival analysis between different genotypes of six SNP loci of BNIP3L (P > 0.05). It may be due to effective treatment options available presently, which can alleviate MM patients’ conditions and prolong their survival time. Surprisingly, we found a significant difference in survival time between MM patients with BNIP3L rs2874670 AA genotype and those with GG + GA genotype after receiving BD chemotherapy protocol (P < 0.05). However, we cannot confidently conclude the influence of the BNIP3L rs2874670 AA genotype on the efficacy of MM patients receiving different chemotherapy protocols due to the small sample size. Cox regression analysis showed the same results that high levels of LDH and β2-MG, and R-ISS stage II and III increased the risk of mortality in MM patients(P < 0.05). The other researchers have also found a correlation between clinical indicators and the survival time of patients. Liu et al. found that the death of patients had significantly higher levels of serum IgG, κ light chain, λ light chain, C3, and C4 compared to surviving patients [Citation34]. The positive expression of Bcl-2 and c-Myc affects the progression of MM, as found by Guo et al. [Citation35].
Above all, our research has revealed that BNIP3L SNPs rs2874670 polymorphism had some specific correlation with the genetic susceptibility to MM in Chinese populations. The risk of MM was increased in subjects with the rs2874670 A allele and the rs2874670 AA genotype of the BNIP3L gene. Although we have not found the difference in survival time between different genotypes of BNIP3L SNPs, the results indicated that MM patients with lower levels of LDH and β2-MG would improve their quality of life. It is important to closely monitor the levels of LDH and β2-MG in MM patients, and to consider the BNIP3L gene genotype and RISS stage for treatment of MM patients.
Of course, several potential limitations existed in our study. Firstly, the sample size for the study was not large enough, and the sample size in each subgroup was too small, so the evidence for different effects on the part of genotype and allele on MM risk was only suggestive but not conclusive. Secondly, this research was based on data from individual subjects, and only six SNPs of the BNIP3L gene were selected, which restricted interpretations for gene-to-gene interactions. Finally, the follow-up time for the enrolled MM patients should be longer, there will be a more accurate evaluate the impact on survival time by BNIP3L SNPs genotypes, clinical indicators, and treatment protocols. Of course, the mechanisms underlying these differences also need further study. I believe it would be interesting to explore more validation cohorts from more SNPs in the BNIP3L gene and extend follow-up observation time, thus investigating their associations with MM risk and the quality of life.
Conclusion
To the best of our knowledge, it was firstly found that the BNIP3L SNPs rs2874670 could increase susceptibility to MM, and MM patients with lower levels of LDH and β2-MG would improve the quality of life in the Chinese population. Moreover, we also found that fifteen SNPs pairs of the BNIP3L gene displayed strong LD. The CCGCAC haplotype suggested a protective effect against the development of the disease. The CCACA haplotype was correlated with a significantly high risk of MM. There is a significant difference in survival time between MM patients with BNIP3L rs2874670 AA genotype and those with GG + GA genotype after receiving BD chemotherapy protocol. Due to the limitations of sample size, SNPs loci, and follow-up time, these results based on current evidence are relative and suggestive, further observational studies and functional analyses are warranted to confirm these findings in the future.
Author contributions
Ruolin Li and Zhian Ling drafted the overall design of this paper. Yu Wang, Hongping Qin, and Chunni Huang collected resources of blood samples. Qicai Wang, Jing Wu, and Zhongqing Li conducted experiments. Qicai Wang and Xing Xie conducted data curation and analyzed the data using Software. Qicai Wang wrote the original draft article. Ruolin Li and Zhian Ling reviewed and edited the initial draft.
Ethics approval and consent to participate
This research program was approved by the Ethics Committee of First Affiliated Hospital of Guangxi Medical University (NO: 2023-E160-01). All participants signed informed consent forms.
Supplemental Material
Download MS Word (23.9 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Rollig C, Knop S, Bornhauser M. Multiple myeloma. Lancet. 2015;385:2197–2208. doi:10.1016/S0140-6736(14)60493-1
- Sonneveld P, Avet-Loiseau H, Lonial S, et al. Treatment of multiple myeloma with high-risk cytogenetics: a consensus of the International Myeloma Working Group. Blood. 2016;127:2955–2962. doi:10.1182/blood-2016-01-631200
- Wallington-Beddoe CT, Mynott RL. Prognostic and predictive biomarker developments in multiple myeloma. J Hematol Oncol. 2021;14:151, doi:10.1186/s13045-021-01162-7
- Gozzetti A, Ciofini S, Simoncelli M, et al. Anti CD38 monoclonal antibodies for multiple myeloma treatment. Hum Vaccin Immunother. 2022;18:2052658, doi:10.1080/21645515.2022.2052658
- Guzdar A, Costello C. Supportive care in multiple myeloma. Curr Hematol Malig Rep. 2020;15:56–61. doi:10.1007/s11899-020-00570-9
- Mailankody S, Devlin SM, Landa J, et al. GPRC5D-Targeted CAR T cells for myeloma. N Engl J Med. 2022;387:1196–1206. doi:10.1056/NEJMoa2209900
- Zhang J, Ney PA. Role of BNIP3 and NIX in cell death, autophagy, and mitophagy. Cell Death Differ. 2009;16:939–946. doi:10.1038/cdd.2009.16
- Ruolin L, Chen G, Dang Y, et al. Expression and clinical significance of BCL2 interacting protein 3 like in multiple myeloma. Technol Cancer Res Treat. 2021;20:15330338211024551.
- Zhou J, Ma C, Wang K, et al. Identification of rare and common variants in BNIP3L: a schizophrenia susceptibility gene. Hum Genomics. 2020;14:16, doi:10.1186/s40246-020-00266-4
- Li R, Hu Z, Chen H, et al. Association of growth differentiation factor-15 polymorphisms and growth differentiation factor-15 serum levels with susceptibility to multiple myeloma in a Chinese population. Clin Lab. 2021;6(1). doi:10.7754/Clin.Lab.2020.200341
- [Guidelines for the diagnosis and management of multiple myeloma in China (2022 revision)]. Zhonghua Nei Ke Za Zhi. 2022;61:480–487.
- Shi YY, He L. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res. 2005;15:97–98. doi:10.1038/sj.cr.7290272
- Manunta P, Tripodi G. Haplotype analysis in human hypertension. J Hypertens. 2005;23:711–712. doi:10.1097/01.hjh.0000163135.69643.e4
- Collins FS, Guyer MS, Charkravarti A. Variations on a theme: cataloging human DNA sequence variation. Science. 1997;278:1580–1581. doi:10.1126/science.278.5343.1580
- Imazu T, Shimizu S, Tagami S, et al. Bcl-2/E1B 19 kDa-interacting protein 3-like protein (Bnip3L) interacts with Bcl-2/Bcl-xL and induces apoptosis by altering mitochondrial membrane permeability. Oncogene. 1999;18:4523–4529. doi:10.1038/sj.onc.1202722
- Yuan Y, Zheng Y, Zhang X, et al. BNIP3L/NIX-mediated mitophagy protects against ischemic brain injury independent of PARK2. Autophagy. 2017;13:1754–1766. doi:10.1080/15548627.2017.1357792.
- O'Sullivan TE, Johnson LR, Kang HH, et al. BNIP3- and BNIP3L-mediated mitophagy promotes the generation of natural killer cell memory. Immunity. 2015;43:331–342. doi:10.1016/j.immuni.2015.07.012
- Xu D, Chen P, Wang B, et al. NIX-mediated mitophagy protects against proteinuria-induced tubular cell apoptosis and renal injury. Am J Physiol Renal Physiol. 2019;316:F382–F395. doi:10.1152/ajprenal.00360.2018.
- da Silva Rosa SC, Martens MD, Field JT, et al. BNIP3L/Nix-induced mitochondrial fission, mitophagy, and impaired myocyte glucose uptake are abrogated by PRKA/PKA phosphorylation. Autophagy. 2021;17:2257–2272. doi:10.1080/15548627.2020.1821548
- Li Y, Zheng W, Lu Y, et al. BNIP3L/NIX-mediated mitophagy: molecular mechanisms and implications for human disease. Cell Death Dis. 2022;13:14. doi:10.1038/s41419-021-04469-y
- Panigrahi DP, Praharaj PP, Bhol CS, et al. The emerging, multifaceted role of mitophagy in cancer and cancer therapeutics. Semin Cancer Biol. 2020;66:45–58. doi:10.1016/j.semcancer.2019.07.015
- Schweers RL, Zhang J, Randall MS, et al. NIX is required for programmed mitochondrial clearance during reticulocyte maturation. Proc Natl Acad Sci USA. 2007;104:19500–19505. doi:10.1073/pnas.0708818104
- Sandoval H, Thiagarajan P, Dasgupta SK, et al. Essential role for Nix in autophagic maturation of erythroid cells. Nature. 2008;454:232–235. doi:10.1038/nature07006
- Chourasia AH, Boland ML, Macleod KF. Mitophagy and cancer. Cancer Metab. 2015;3:4, doi:10.1186/s40170-015-0130-8
- Drake LE, Springer MZ, Poole LP, et al. Expanding perspectives on the significance of mitophagy in cancer. Semin Cancer Biol. 2017;47:110–124. doi:10.1016/j.semcancer.2017.04.008
- Nunnari J, Suomalainen A. Mitochondria: in sickness and in health. Cell. 2012;148:1145–1159. doi:10.1016/j.cell.2012.02.035
- Lazarini M, Machado-Neto JA, Duarte AD, et al. BNIP3L in myelodysplastic syndromes and acute myeloid leukemia: impact on disease outcome and cellular response to decitabine. Haematologica. 2016;101:e445–e448. doi:10.3324/haematol.2016.142521
- Jung J, Zhang Y, Celiku O, et al. Mitochondrial NIX promotes tumor survival in the hypoxic niche of glioblastoma. Cancer Res. 2019;79:5218–5232. doi:10.1158/0008-5472.CAN-19-0198
- Humpton TJ, Alagesan B, DeNicola GM, et al. Oncogenic KRAS induces NIX-mediated mitophagy to promote pancreatic cancer. Cancer Discov. 2019;9:1268–1287. doi:10.1158/2159-8290.CD-18-1409
- Sun JL, He XS, Yu YH, et al. Expression and structure of BNIP3L in lung cancer. Ai Zheng. 2004;23:8–14.
- Wang WJ, Wang Y, Chen HZ, et al. Orphan nuclear receptor TR3 acts in autophagic cell death via mitochondrial signaling pathway. Nat Chem Biol. 2014;10:133–140. doi:10.1038/nchembio.1406
- Gallegos ZR, Taus P, Gibbs ZA, et al. EWSR1-FLI1 activation of the cancer/testis antigen FATE1 promotes ewing sarcoma survival. Mol Cell Biol. 2019:39(14):e00138-19. doi:10.1128/MCB.00138-19
- Rodrigo R, Mendis N, Ibrahim M, et al. Knockdown of BNIP3L or SQSTM1 alters cellular response to mitochondria target drugs. Autophagy. 2019;15:900–907. doi:10.1080/15548627.2018.1558002
- Liu X, Zhou Z, Sun D. Values of immunoglobulin and complements for evaluating treatment outcomes of patients with multiple myeloma. Clin Lab. 2023:69(11). doi:10.7754/Clin.Lab.2023.230210
- Guo S, Zhi Y, Yang H, et al. Bcl-2 expression is associated with poor prognosis of solitary plasmacytoma of bone. Ann Hematol. 2014;93:471–477. doi:10.1007/s00277-013-1897-8
