Abstract
The present study was focused on the potentiality of agro-based residues for the production of pectinase to meet the growing market demand by improving the yield with low cost of production. Among the agro-based residues used for the production of pectinase, apple pomace was able to produce the maximum of 1366.30 ± 36.71 U/ml using Aspergillus parvisclerotigenus KX928754 in liquid static surface fermentation, followed by sugarcane bagasse (973.12 ± 22.43 U/ml) and used tea (686.7 ± 45.06 U/ml). The process parameters optimization using a single variable at a time affirmed that pH 7.0, incubation period of 168 h, 30°C temperature, sucrose 2% as carbon source and peptone 3% as nitrogen source was found to be optimum for better production. The crude filtrate was purified by precipitation, dialyzed, eluted on Sephadex G-100 column followed by lyophilization and stored at −20°C. A. parvisclerotigenus KX928754 pectinase was purified to 2.10-fold, 2.91% of yield rate and having a specific activity of 1081.66 U/mg. Moreover, the electrophoretic analysis through sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) revealed 37.4 kDa of protein from the purified pectinase. Thus, the use of apple pomace as a substrate for scaling up pectinase with efficient recovery could reduce the price of the enzyme and increase its avenue for different industrial exploitation.
1. Introduction
The ecosphere, part of the universe habitable by living organisms is a closed system for energy, raw material and accumulation of pollutants or waste. Industrialization has caused serious consequences directly and indirectly on the ecosphere, adversely affecting life and its environment. Therefore, controlled exploitation of natural resources and the use of cleanup technology to establish a sustainable environment has been the focus in the business arena. The processes involved in agricultural production such as preparation, transformation and preservation of agricultural products for intermediary or final consumption have led to the generation of a large quantity of agro-processing residues. These residues comprise a high concentration of organic matter rich in nutrients and bioactive compounds [Citation1]. The availability of these nutrients (proteins, sugars and minerals) in such residue offers a suitable environment for the growth of the microbes and hence can be considered as raw material which can be used for the synthesis of microbial-based products such as organic acids, vitamins, hormones, cellular proteins, secondary metabolites, fermentable sugars and various enzymes [Citation2].
Enzymes are natural catalysts produced by living organisms to activate or enhance the array of various biochemical reactions for manifesting and sustaining life. Commercial exploitation of these enzymes over the years for performing some particular chemical transformation has led to their increased uses in industrial processes [Citation3]. The agro-industrial residues mainly constitute cellulose, hemicellulose and pectic substances as the major component of their primary cell wall. The cellulose and hemicellulose concentrations are higher in the vegetative part of the plant, while the pectin is found in large quantities in fruits and vegetables. Homogalacturonan, rhamnogalacturonan I and rhamnogalacturonan II are the three major pectic polysaccharides found in plants [Citation4]. These pectic substrates are degraded by the process called depolymerization (hydrolases and lyases) and de-esterification (esterase). The various enzymes that are suitable for carrying out these processes are grouped as protopectinases (solubilize protopectin), esterase (pectin methylesterase that removes the methoxyl residues from pectin, pectin acetyl esterase that removes the acetyl residues from pectin), depolymerase (polygalacturonase that hydrolyzes the α-1,4-glycosidic linkages in polygalacturonic acid, polymethylgalacturonase targets highly esterified pectin for cleaving α-1,4-glycosidic bonds present in the backbone of pectin by hydrolysis process), lyases (pectate lyase cleaves glycosidic linkages of pectate and forms oligosaccharides, pectin lyase cleaves pectin in a random manner and forms unsaturated methyloligogalacturonates) [Citation4, Citation5]. Different microorganisms are known to produce pectinase enzymes however, their optimization and characterization properties are varied with respect to bacteria [Citation6, Citation7], yeast [Citation8], fungi [Citation2, Citation9] and actinomycetes [Citation10, Citation11]. Pectinases producing several fungal species have been documented until now; however, the pectinases from the genus Aspergillus are well reported. More particularly, Aspergillus sojae [Citation12], Aspergillus oryzae [Citation13], Aspergillus candidus and Aspergillus flavus [Citation14], Aspergillus tamari [Citation15], Aspergillus niger [Citation16] and A. fumigatus [Citation17] contributed lots for efficient production of pectinase however, no such report of production from the tested species is till now available.
A primary constraint associated with new enzyme source commercialization is a higher cost incurred during enzyme production, which can be reduced by the use of optimized fermentation conditions, economically viable raw materials and proficient strains [Citation18]. Many studies have focused to characterize various microbial pectic enzymes with their biochemical properties, mode of action and process parameters (optimized pH, temperature, incubation duration, carbon source and nitrogen source) with the aim for increasing enzyme production and diminishing the production cost [Citation2, Citation6, Citation19]. Various agro-processing residues like mosambi peel, pineapple, wheat bran, papaya, orange peel [Citation20], apple, lemon peel, grape skin, tamarind kernel [Citation21], cocoa beans [Citation22], banana peel [Citation2] and rice bran [Citation23] have been used for the production of pectinase enzymes. Jayani et al. [Citation24] proclaimed microbial pectinase, which accounts for 25% of the global food enzyme sales. It is becoming more evident that the enzymes are the major barriers because of their production cost. Therefore, the present work aims at screening various agro-processing residues, evaluation and optimization of cultural conditions for the enhanced production of pectinase from A. parvisclerotigenus KX928754 followed by enzyme purification so as to bring down the cost of the enzyme production and assessing the suitability of the strain as a promising candidature for the production of industrially pertinent pectinase.
2. Materials and methods
2.1. Substrates and chemicals
Agro-processing residues (used as substrates) such as raw tea (RT) and used tea (UT), citrus peel (CP), orange peel (OP), banana peel (BP), apple pomace (AP), sugarcane bagasse (SB) and coffee husk (CH) were collected from the local markets of Bhubaneswar, Odisha, India. The collected residues were cut into tiny pieces, dried at 60°C in a hot air oven for 48 h. Dried samples were ground to a coarse powder in a blender and kept inside sterile containers for further use. The chemicals used in this study were of analytical grade and procured from Sigma Chemicals Co. (USA), Sisco Research Laboratories Pvt. Ltd. (India), Merck India Ltd. (India) and Hi-Media Laboratories Pvt. Ltd. (India).
2.2. Preparation of fungal inoculum for fermentation
The fungal strain was isolated from the soil sample rich with decomposed fruits and vegetable waste collected from the vegetable market of Bhubaneswar, Odisha, India and was identified as Aspergillus parvisclerotigenus SSB9 (NCBI accession no. KX928754). The inoculum was streaked onto potato dextrose agar (PDA) and incubated for 168 h at 30 ± 2°C to form colonies. A single isolated colony was picked and mixed in 1 ml of sterile deionized water. The mixture was vortexed for a minute (for spores to get released) and filtered through sterile cotton, followed by spreading (0.1 ml) onto pre-prepared fermentation PDA agar medium containing 200 g/l potato infusion, 20 g/l dextrose. The plate was incubated at 30 ± 2°C for 168 h and allowed to form a lawn of spores. The plate was scraped using a sterile spatula and the spore content was mixed in sterile water to form a uniform spore suspension. The estimated total spore count was 5.0 × 108 spores ml–1 and it was stored for further use. The suspension (spore density of 1 × 107 spores ml–1) was utilized as inoculum for process optimization and production of pectinase enzyme as per the method of Sethi et al. [Citation25].
2.3. Screening of suitable substrate for enzyme production
Screening for various agro-processing residues for the biosynthesis of pectinase was performed by the process of liquid static surface fermentation (LSSF) in a 150 ml Erlenmeyer flask, with a fermentation medium volume of 50 ml containing various agro-based residues (10 gm/100 ml) inoculated with 1 × 107 spores ml–1 and incubated at 30 ± 2°C for 96 h. For control, the samples were allowed to grow in a Czapek-Dox medium containing 1% pectin but devoid of sucrose. At the end of incubation, the culture broth was filtered to recover crude extracellular pectinase and the filtrate (crude enzyme) was stored at −20°C for further studies. Biomass dry-weight was diligently estimated after passing it through a hot air oven at an optimum temperature of 80°C up to a time span of 24 h.
2.4. Optimization of process parameters
Various physical and nutritional parameters required for enhancing pectinase biosynthesis were standardized. The strategy that was taken up for this study was the optimization of one variable at a time (OVAT) and keeping others as constant. The parameters were optimized against pH of the media [Citation3–9, with constant temperature (30°C) and incubation period (144 h)], incubation period [24–192 h, with constant temperature (30°C) and pH (7.0)] and temperature [24–45°C, with constant pH (7.0) and incubation period (168 h)]. After that, various sources of carbon (maltose, pectin, mannitol, starch, sucrose, glucose and lactose; 1%, w/v) and additional sources of nitrogen (peptone, yeast extract, beef extract, urea, ammonium nitrate, ammonium sulphate, ammonium chloride and sodium nitrate; 1%, w/v) were applied for optimization of pectinase production. At the end of incubation (168 h except for incubation period), the key samples were processed for the evaluation of the catalytic activity of pectinase by DNS method [Citation26] and quantification of total protein [Citation27].
2.5. Purification of pectinase enzyme
The enzyme in culture filtrate (crude) was precipitated by the gradual addition of different concentrations (40%, 50%, 60%, 70%, 80%, 90%, 100%) of ammonium sulfate with constant shaking with the help of a magnetic stirrer at 4°C for 24 h, followed by centrifugation at 10,000 rpm for 15 mins at 4°C to separate the protein precipitate from the solution. The resulting pellets were carefully dissolved in 0.1 M sodium citrate buffer (pH 5.0) at an established 0.1 g/ml ratio for obtaining 10X concentrated solution [Citation28]. The concentrated enzyme solution was dialyzed for 24 h at 4°C with intermittent stirring for eliminating lesser molecular weight substances and ammonium sulfate from the dialysate. The enzyme solution was purified as per Sethi et al. [Citation2] by eluting the dialysate with 0.1 M sodium citrate buffer (pH 5.0) on Sephadex G-100 column (Sigma Aldrich, USA) with the flow rate of 1 ml/ min. 2 ml of each collected sample was meticulously analysed for its total protein content following the protocol of Lowry et al. [Citation27]. Fractions having absorption maxima at 750 nm were pooled for evaluation of the catalytic activity of the pectinase by the protocol enunciated by Vasanthi and Meenakshisundaram [Citation29]. The fractions with maximal protein concentration were pooled, lyophilized and preserved at −20°C for further characterization [Citation2].
2.6. Protein profiling and determination of molecular weight by SDS-PAGE
The samples (crude and purified pectinase) thus obtained were carefully separated by using 10% SDS-PAGE as per the method of Laemmli [Citation30]. A molecular weight protein marker ranging from 14.4–97.4 kDa (Bio-Rad Laboratories India Pvt. Ltd., India) was applied as a standard for obtaining protein bands. The Coomassie Brilliant Blue staining method was adopted for the visualization of separated proteins [Citation31]. The relative band positions were analysed by using a Bio-Rad Gel documentation system (Bio-Rad, CA).
2.7. Statistical analysis
The results observed are the mean of three independent experimental replicates (n = 3) and values are represented as the mean ± standard error.
3. Results and discussion
3.1. Selection of suitable substrate for enhanced pectinase production
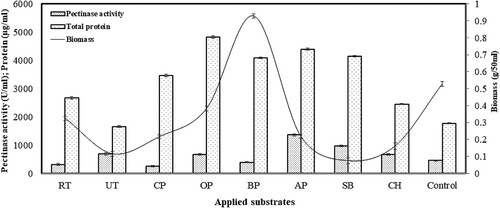
The study was focused initially on optimization of the fermentation process using various economic substrates. The use of RT, UT, CP, OP, BP, AP, SB and CH as principal media component or substrate for the production of pectinase revealed that AP was proved to be a viable substrate for enhanced pectinase production (1366.30 ± 36.71 U/ml), followed by SB (973.12 ± 22.43 U/ml), UT (686.70 ± 45.06 U/ml) and CH (673.64 ± 28.89 U/ml) (Figure ). Various fungal species have been screened for the production of pectinase earlier [Citation2, Citation32, Citation33] however, A. parvisclerotigenus KX928754 was found to produce more pectinase in comparison to other species studied. A combination of apple bagasse and wheat bran was found to be effective for the production of polygalacturonase [Citation34] but, observation related to pectinase was not reported. However, the availability of pectin components from the AP was studied [Citation35, Citation36]. The better pectinase production from AP can be attributed due to the higher concentration of pectin and also the presence of other nutrients (protein, carbohydrates, fat and minerals), that are most essential for the growth of the A. parvisclerotigenus [Citation37, Citation38]. AP is also known as the best source of higher concentration of antioxidant polyphenols [Citation39] that could be an additional reason for the enhanced production of pectinase in the present investigation. Moreover, AP is a versatile substrate for the production of numerous metabolites like enzyme, aroma compound, nutritional enrichment, bio-polymer, polysaccharide, citric acid, bio-inoculant, ethanol, linolenic acid, and pigment from the different kind of microbes including bacterial, fungus and yeast [Citation40]. Various studies have reported that liquid static surface fermentation can produce a higher amount of enzyme and can be standardized for industrial application [Citation41]. The addition of agricultural residues such as apple residue, citrus pectin, banana peel, wheat bran, orange peel, rice bran, pomegranate peel, coffee pulp, cassava flake, sugarcane bagasse, pomelo peel, and pineapple peel have shown to assist in the production of pectinase. Citrus peel and apple pomace are known to be ideal sources of pectin and have been used for the extraction of pectin commercially. Further, the uses of these substrates are considered as the excellent source of production of pectinase however, its concentration varies with the application of various fungi and particularly Aspergillus spp. [Citation36, Citation41–44].
Figure 1. Effect of various substrates on the pectinase production, protein concentration and mycelia growth in liquid static surface ferementation (LSSF) by Aspergillus parvisclerotigenus KX928754. Where RT: raw tea; UT: used tea; CP: citrus peel; OP: orange peel; BP: banana peel; AP: apple pomace; SB: sugarcane bagasse; CH: coffee husk; Control: Czapek-Dox medium containing 1% pectin but without sucrose. The data represents mean ± standard error of replicates (n = 3).
3.2. Process parameters optimization for enhanced pectinase biosynthesis
3.2.1. Effect of initial pH (medium) on the pectinase production
Various parameters have to be standardized for augmenting the pectinase production using A. parvisclerotigenus KX928754. Initially, the pH was optimized with an incubation period of 144 h and a temperature of 30°C. It was observed that the production of pectinase was maximum (1361.52 ± 7.30 U/ml) at pH of 7.0 and then decreased to 1329.43 ± 4.36 U/ml at pH 6.0 whereas above the pH 7.0, a sudden fall of activity was noticed. The overall pectinase production was found to be more in basic pH as compared to all acidic range of pH which reflects acidic pectinase. Moreover, the generation of growth biomass showed a similar kind of trend with respect to pH which was maximum (0.114 ± 0.003 g/50 ml) at pH 7.0 (Figure a). The results are in concordance with the findings of Barkavi et al. [Citation45] and Ketipally et al. [Citation46] who have used A. niger and A. nomius, respectively. Various microbes are also known to produce pectinase that is active at higher pH and is termed alkaline pectinase. However, the alkalization or acidification of the medium depends upon the utilization of substrate by the applied organism, synthesized enzyme and its enzymatic activity [Citation47].
Figure 2. Effect of pH (a), incubation period (b) and incubation temperature (c) on the pectinase production and mycelia growth by Aspergillus parvisclerotigenus KX928754. pH optimization at 30°C and 144 h of incubation; incubation period optimization at 30°C and pH 7; temperature optimization at pH 7 and 168 h of incubation. The data represents mean ± standard error of replicates (n = 3).
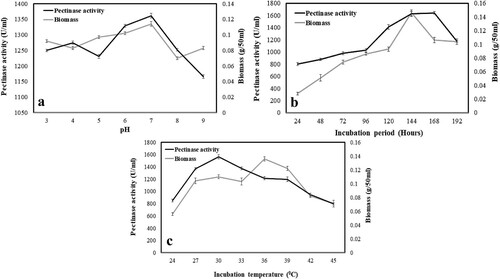
3.2.2. Effect of incubation time on the pectinase production
Under constant pH of 7.0 and temperature of 30°C, the LSSF was performed with varying incubation time and was observed that 168 h was optimal for the better production of pectinase (1642.21 ± 19.57 U/ml) followed by 144 h (1623.46 ± 42.89 U/ml). The production of pectinase increased drastically till 144 log h and reached its peak (1642.21 ± 19.56 U/ml) at 168 log h and then suddenly fallen to 1172.09 ± 15.1 U/ml at 192 h. Such a pattern of pectinase production may be due to depletion of the nutrients after a specific duration, which was confirmed through biomass production (Figure b). Few organisms are known to produce pectinase within a short period of incubation time because of its fast-metabolic activity and/ or simple nutrients in the media, while some are also slow and that depend on the complex nutrients supplied (agro-residues). In the present investigation, the organism was cultured with AP, where the culture has to produce the enzyme required for degrading the substrate, followed by utilization of the same for growth that explains the slow growth and production of pectinase which strongly support with findings of Barkavi et al. [Citation45], Abbasi et al. [Citation34] and Ketipally et al. [Citation46]. A few strains of Aspergillus are also known to produce pectinase in a short period of time [Citation41, Citation46, Citation48]. The time required for fermentation depends upon certain factors such as the amount of nutrients in the medium, nature of the medium, physiological parameters and fermentative microorganisms [Citation49, Citation50].
3.2.3. Effect of temperature on the production of pectinase
The media was cultured from 24°C to 45°C with constant pH (pH 7.0) and incubation time (168 h). It was observed that 30°C was the optimal temperature for the best production of pectinase (1562.3 ± 38.88 U/ml) however, the maximum (0.136 ± 0.004 g/50 ml) biomass generation was achieved at 36°C. After that, a decreasing trend of both pectinase production and biomass generation was noticed under increasing temperature conditions (Figure c). Hours et al. [Citation42] observed that media supplemented with AP and cultured with Aspergillus foetidus was able to produce maximum pectinase at 30°C, but the concentration of pectinase was lower than the present study and this may be attributed to the nature of selected microorganism. Similar kind of results showing 30°C as the optimum temperature for pectinase production were also reported in A. terreus [Citation2] and A. niger [Citation23, Citation33, Citation41, Citation51]. The production of enzyme was enhanced with an increase in temperature up to 30°C and then decreased. The increase in temperature leads to exuberant growth of fungi, but only up to a certain extent and after that, a dwindling tendency in the growth profile has been observed.
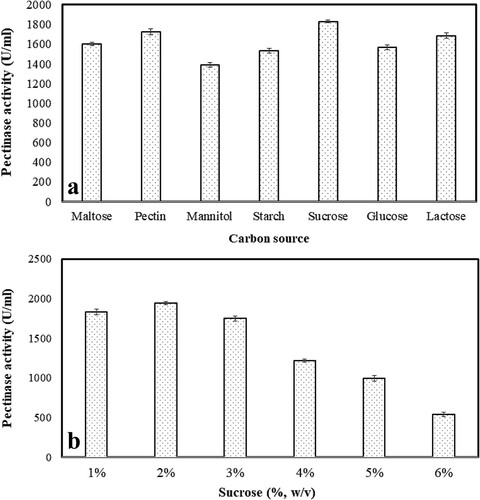
3.2.4. Effect of additional carbon source along with its concentration on the pectinase production
Carbon source has a great impact on the initial growth of microorganisms, thus supplementing a suitable carbon source helps in biomass buildup, which in turn can support for enzyme production [Citation34]. A. parvisclerotigenus KX928754 was able to produce 1829.27 ± 14.01 U/ml of pectinase at constant pH (7.0), incubation temperature (30°C) and incubation period for 168 h, when supplemented with sucrose, followed by pectin (1726.88 ± 30.24 U/ml) and lactose (1686.42 ± 26.10 U/ml) [Figure a]. However, 2% sucrose was found to be more suitable for the better synthesis of pectinase (1943.51 ± 21.70 U/ml) in comparison to other concentrations [Figure b]. Various studies have shown that the addition of different carbon sources into media has increased the rate of enzyme production. The organisms were able to utilize the supplemented carbon sources and produce pectinase which is sensitive to implemented carbon sources [Citation41, Citation44, Citation46, Citation52, Citation53]. The lowest pectinase was produced by using media supplemented with mannitol (1389.21 ± 25.83 U/ml), which was about 31% of less production in comparison to sucrose (1829.27 ± 14.01 U/ml). The reason behind using sucrose as a superior carbon source by fungi is due to the extracellularly bound enzyme invertase that hydrolyze sucrose to glucose and fructose which is then absorbed immediately into the cell for its metabolic activity [Citation54, Citation55]. A. parvisclerotigenus KX928754 can utilize multiple carbon sources as compared to A. terreus [Citation2]. Sucrose at various concentrations was also supplemented in the media and it was observed that 2% sucrose was able to produce 1943.51 ± 21.70 U/ml of the enzyme (Figure b). The production of the enzyme increased up to 2% of sucrose and then declined as its concentration increased. A similar effect of sucrose has also been observed by Peksel and Kubicek [Citation56] and Teixeira et al. [Citation57] on A. niger and A. japonicus, respectively.
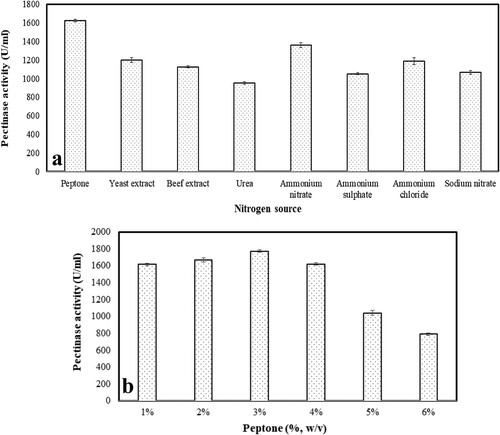
3.2.5. Effect of additional nitrogen source along with its concentration on the pectinase production
Adequate nitrogen supply is highly critical not only for achieving optimal growth of the microorganisms but also for their metabolic rate. To find a suitable nitrogen source, media was supplemented with different inorganic and organic nitrogen sources and was observed that the presence of peptone exhibited maximum (1623.51 ± 13.93 U/ml) pectinase production in comparison to others. Ammonium nitrate resulted in 1356.42 ± 26.80 U/ml of pectinase which was considered as the second-best results whereas, no such significant differentiation occurred in the case of urea, ammonium nitrate, ammonium sulphate, ammonium chloride and sodium nitrate (Figure a). When the increasing concentration of peptone was individually tried, an immense effect on the rate of pectinase production was observed (1773.70 ± 12.56 U/ml) at 3% of concentration (Figure b). Juwon et al. [Citation58] reported that media supplemented with different nitrogen sources has an impressive effect on fungal growth and enzyme production. Peptone being an organic nitrogen source had a positive effect on the various productivity [Citation46, Citation59]. The inorganic nitrogen source was found to be promising and it depends on the organism used for the production but critically controlling of concentration of a suitable nitrogen source in the media is essential for the superlative enzyme production [Citation60, Citation61].
3.3. Purification of pectinase
The purification of the enzyme was conducted in order to inhibit the loss of product viability by implementing a series of steps. Each step not only enhances the purity of the enzyme starting from the crude extract but also emphasizes the minimal loss throughout the process. With the help of gel filtration chromatography, extracts were efficiently obtained with a single absorption peak having 2.10-fold purification and 2.91% yield, where the specific activity was increased from 514.07 to 1081.66 U/mg (Table ). Various purification steps are available for the extraction of pectinase from the crude sample, but the extraction has to be concentrated on the enzyme property, insolubility and viability [Citation62]. Anand et al. [Citation63] purified pectinase from A. niger by implementing acetone precipitation followed by CM-cellulose column chromatography and obtained an exo-polygalacturonase which was about 15.28-fold with 1.2% yield and specific activity of 33.47 U/mg protein. Similarly, Siddiqui et al. [Citation64] also purified polygalacturonase from Rhizomucor pusillus by Sephadex G-200 followed by Sephacryl S-100 column chromatography and attained purified polygalacturonase of 12.34-fold purification with a 27.06% yield.
Table 1. Summary of purification and activity of pectinase obtained from Aspergillus parvisclerotigenus KX928754.
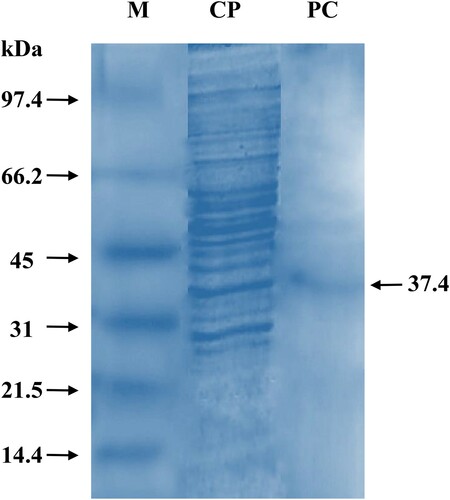
3.4. Determination of molecular weight by SDS-PAGE
The crude extracts when subjected to SDS-PAGE, produced various prominent bands ranging from 14.4 kDa to 97.4 kDa and above. The purified pectinase had a molecular weight of approximately 37.4 kDa (Figure ) which indicates monomeric nature. However, the in-gel activity on native gel electrophoresis was not conducted. A similar kind of molecular weight determination of purified pectinase was performed earlier but no reports are available in A. parvisclerotigenus. The purified polygalacturonase having 124 kDa of molecular mass [Citation63] and various pectinases ranging from 34 kDa to 42 kDa [Citation65] were characterized in A. niger.
4. Conclusion
Through the present study, it can be concluded that the indigenous strain A. parvisclerotigenus KX928754 has incredible potential for the production of an appreciable amount of pectinase. This study enlightens the possibility of utilization of agro-processing residues (apple pomace) efficiently in the LSSF process, which proves an effective substrate for producing industrially pertinent enzymes. The parameters affecting enzyme production rate were studied and the optimal level required for maximal production was also identified. Implementation of agro-processing or agro-industrial residues in fungal enzyme production is not only efficient but also offers several process advantages because of its nutritional constituents and thus helps in effective management of agro-waste residues. Hence, it can be proclaimed that the present investigation paves the path for a high yield process that is economically viable for enhancing pectinase production.
Acknowledgements
One of the authors (JRR) wishes to thank the authorities of AIPH University for providing encouragement and support.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Sadh KP, Duhan S, Duhan JS. Agro-industrial wastes and their utilization using solid state fermentation: a review. Bioresour Bioprocess. 2018;5:1.
- Sethi BK, Nanda PK, Sahoo S. Enhanced production of pectinase by Aspergillus terreus NCFT 4269.10 using banana peels as substrate. 3 Biotech. 2016;6:36.
- Shuang L, Yang X, Yang S, et al. Technology prospecting on enzymes: application, marketing and engineering. Comput Struct Biotechnol J. 2012;2(3):e201209017.
- Dinu D, Nechifor MT, Stoian G, et al. Enzymes with new biochemical properties in the pectinolytic complex produced by Aspergillus niger MIUG 16. J Biotechnol. 2007;131(2):128–137.
- Satapathy S, Rout JR, Kerry RG, et al. Biochemical prospects of various microbial pectinase and pectin: an approachable concept in pharmaceutical bioprocessing. Front Nutr. 2020;7(117):1–17.
- Kashyap DR, Vohra PK, Chopra S, et al. Applications of pectinases in the commercial sector: a review. Bioresour Technol. 2001;77:215–227.
- Oumer OJ, Abate D. Characterization of pectinase from Bacillus subtilis strain Btk 27 and its potential application in removal of mucilage from coffee beans. Enz Res. 2017;2017:7686904.
- Carrasco M, Rozas JM, Alcaino J, et al. Pectinase secreted by psychrotolerant fungi: identification, molecular characterization and heterologous expression of a cold-active polygalacturonase from Tetracladium sp. Microb Cell Fact. 2019;18:45.
- Patidar MK, Nighojkar S, Kumar A, et al. Pectinolytic enzymes–solid state fermentation, assay methods and applications in fruit juice industries: a review. 3 Biotech. 2018;8:199.
- Beg QK, Bhushan B, Kapoor M, et al. Production and characterization of thermostable xylanase and pectinase from Streptomyces sp. QG-11-3. J Ind Microbiol Biot. 2000;24:396–402.
- Salehghamari E, Nasrollahzadeh Z, Tahmaseb M, et al. Pectinase enzyme from Streptomyces coelicoflavus GIAL86 isolated from Meyghan salt lake, Arak. Iran. Int J Aquat Biol. 2019;7(2):106–111.
- Demir H, Tari C. Valorization of wheat bran for the production of polygalacturonase in SSF of Aspergillus sojae. Ind Crop Prod. 2014;54:302–309.
- Dange VU, Harke S. Production and purification of pectinase by fungal strain in solid-state fermentation using agro-industrial bioproduct. Int J Life Sci Res. 2018;6(4):85–93.
- Naseer O, Khan JA, Omer MO, et al. Exogenous factors affecting growth of Aspergillus species. Indian J Anim Res B. 2018;820:1–6.
- Munir M, Abdullah R, Haq IU, et al. Isolation and identification of multi stress tolerant polygalacturonase producing fungi from various fruits. J Anim Plant Sci. 2019;29(4):2019.
- Khalil M, Khattak S, Ali Q, et al. Isolation and characterization of pectinase producing Aspergillus niger from orange. Int J Bot Studies. 2020;5(2):45–48.
- Zehra M, Syed MN, Sohail M. Banana peels: a promising substrate for the coproduction of pectinase and xylanase from Aspergillus fumigatus MS16. Pol J Microbiol. 2020;69(1):19–26.
- Murad HA, Azzaz HH. Microbial pectinases and ruminant nutrition. Res J Microbiol. 2011;3:246–269.
- Gummadi SN, Panda T. Purification and biochemical properties of microbial pectinases-a review. Proc Biochem. 2003;38:987–996.
- Priya V, Sashi V. Pectinase enzyme production by using agrowastes. Int J Eng Sci Res Technol. 2014;3(4):8041–8046.
- Martinez-Trujillo A, Arreguin-Rangel L, Garcia-Rivero M, et al. Use of fruit residues for pectinase production by Aspergillus flavipes FP-500 and Aspergillus terreus FP-370. Lett Appl Microbiol. 2011;53:202–209.
- Akintobi AO, Oluitiola PO, Olawale AK, et al. Production of pectinase enzymes system in culture filtrates of Penicillium variabile Sopp. Nat Sci. 2012;10(7):99–109.
- Abdullah R, Jafer A, Nisar K, et al. Process optimization for pectinase production by locally isolated fungal strain using SmF. Biosci J. 2018b;34(4):1025–1032.
- Jayani RS, Saxena S, Gupta R. Microbial pectinolytic enzymes: a review. Process Biochem. 2005;40:2931–2944.
- Sethi BK, Nanda PK, Sahoo SL. Isolation, identification and conservation of potent hydrolase producer from different soils of Odisha, India. Int J Pharma Bio Sci. 2013;4(2):B89–B100.
- Miller GL. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem. 1959;31:426–428.
- Lowry OH, Rosebrough NJ, Farr AL, et al. Protein measurements with the folin phenol reagent. J Biol Chem. 1951;193:265–275.
- Jana A, Maity C, Halder SK, et al. Structural characterization of thermostable, solvent tolerant, cytosafe tannase from Bacillus subtilis PAB2. Biochem Eng J. 2013;77:161–170.
- Vasanthi M. Optimization of pectinase enzyme production by using sour orange peel as substrate in solid state fermentation. Asian J Biochem Pharm Res. 2012;2(1):16–26.
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685.
- Blakesley RW, Boezi JA. A new staining technique for proteins in polyacrylamide gels using coomassie brilliant blue G250. Anal Biochem. 1977;82(2):580–582.
- Patil NP, Patil KP, Chaudhari BL, et al. Production, purification of exo-polygalacturonase from soil isolate paecilomyces variotii NFCCI 1769 and its application. Indian J Microbiol. 2012;52:240–246.
- Maller A, Da Silva TM, Damasio ARL, et al. Functional properties of a manganese-activated exo-polygalacturonase produced by a thermotolerant fungus Aspergillus niveus. Folia Microbiol. 2013;58:615–621.
- Abbasi H, Seyed RM, Mehrdad S. Polygalacturonase (PG) production by fungal strains using agro-industrial bioproduct in solid state fermentation. Chem Eng Res Bull. 2011;15:1–5.
- Muller-Maatsch J, Bencivenni M, Caligiani A, et al. Pectin content and composition from different food waste stream. Food Chem. 2016;201:37–45.
- May CD. Industrial pectins: sources, production and applications. Carbohydr Polym. 1990;12:79–99.
- Kruczek M, Gumul D, Kacaniova M, et al. Industrial apple pomace by-products as a potential source of pro-health compounds in functional food. J Microbiol Biotechnol Food Sci. 2017;7(1):22–26.
- Skinner RC, Gigliotti JC, Ku KM, et al. A comprehensive analysis of the composition, health benefits, and safety of apple pomace. Nutr Rev. 2018;76(12):893–909.
- Fernandes PAR, Ferreira SS, Bastos R, et al. Apple pomace extract as a sustainable food ingredient. Antioxidants. 2019;8(6):189.
- Vendruscolo F, Albuquerque PM, Streit F, et al. Apple pomace: a versatile substrate for biotechnological applications. Crit Rev Biotechnol. 2008;28(1):1–12.
- Kamalambigeswari R, Yadav SA, Sivaswamy N, et al. Isolation, identification, screening and optimization of pectinase producing soil fungi (Aspergillus niger). Int J Res Pharm Sci. 2018;9(3):762–768.
- Hours RA, Voget CE, Ertola RJ. Some factors affecting pectinase production from apple pomace in solid-state cultures. Biol Wastes. 1988;24(2):147–157.
- Joshi VK, Parmar M, Rana N. Purification and characterization of pectinase produced from apple pomace and evaluation of its efficacy in fruit extraction and clarification. Indian J Nat Prod Resour. 2011;2(2):189–197.
- Jalis H, Ahmad A, Khan SA, et al. Utilization of apple peels for the production of plant cell-wall degrading enzymes by Aspergillus fumigatus MS16. J Anim Plant Sci. 2014;24(2):664–667.
- Barkavi D, Siva Sankari P. Optimization of solid-state fermentation conditions for the production of pectinases by Aspergillus niger. J Pharm BioSci. 2014;2:50–57.
- Ketipally R, Karnthi KG, Raghu RM. Polygalacturonase production by Aspergillus nomius MR103 in solid state fermentation using agro-industrial wastes. J Appl Nat Sci. 2019;11(2):305–310.
- Zeni J, Cence K, Grando CE, et al. Screening of pectinase-producing microorganisms with polygalacturonase activity. Appl Biochem Biotechnol. 2011;163(3):383–392.
- Abdullah R, Farooq I, Kaleem A, et al. Pectinase production from Aspergillus niger IBT-7 using solid state fermentation. Bangladesh J Bot. 2018a;47:473–478.
- Patil SR, Dayanand A. Optimization of the process for the production of fungal pectinases from deseeded sunflower head in submerged and solid-state conditions. Bioresour Technol. 2006;97:2340–2344.
- Adeleke AJ, Odunfa SA, Olannbiwonninu A, et al. Production of cellulose and pectinase from orange peels by fungi. Nat Sci. 2012;10(5):107–112.
- Dhembare AJ, Kakad SL, Rajani R. Effect of pH, temperature and kinetics of pectinase enzyme using Aspergillus niger by solid-state of fermentation. Der Pharmacia Sinica. 2015;6(8):1–5.
- Khan A, Sahay S, Rai N. Production and optimization of pectinase enzyme using Aspergillus Niger strains in solid state fermentation. Res Biotechnol. 2012;3(3):19–25.
- Singh S, Mandal SK. Optimization of processing parameters for production of pectinolytic enzymes from fermented pineapple residue of mixed Aspergillus species. Jordan J Biol Sci. 2012;5(4):307–314.
- Kubicek-Pranz EM, Mozelt M, Rohr M, et al. Changes in the concentration of fructose 2,6-bisphosphate in Aspergillus niger during stimulation of acidogenesis by elevated sucrose concentration. Biochim Biophys Acta. 1990;1033(3):250–255.
- Raju AI-CH, Pulipati K, Jetti A. Production of invertase by Aspergillus niger under solid state fermentation using orange fruit peel as substrate. Adv Crop Sci Tech. 2016;4(6):247.
- Peksel A, Kubicek C. Effects of sucrose concentration during citric acid accumulation by Aspergillus niger. Turk J Chem. 2003;27(5):581–590.
- Teixeira MFS, Filho JLL, Duran N. Carbon source effect on pectinase production from Aspergillus japonicus 586. Braz J Microbiol. 2000;31(4):286–290.
- Juwon AD, Emmanuel OF. Experimental investigations on the effects of carbon and nitrogen sources on concomitant amylase and polygalacturonase production by Trichodermaviride BITRS-1001 in submerged fermentation. Biotechnol Res Int. 2012;2012:904763.
- Akhter N, Morshed A, Uddin A, et al. Production of pectinase by Aspergillus niger cultured in solid state media. Int J Biosci. 2011;1:33–42.
- Ibrahim D, Salikin NH, Hong LS, et al. Pomelo peels as alternative substrate for extracellular pectinase production by Aspergillus niger HFM-8. Malays J Microbiol. 2013;9:308–316.
- Doughari JH, Onyebarachi GC. Production, purification and characterization of polygalacturonase from aspergillus flavus grown on orange peel. Appl Microbiol Open Access. 2019;4:155.
- Lee SY, Khoiroh I, Ling TC, et al. Enhanced recovery of lipase derived from Burkholderia cepacia from fermentation broth using recyclable ionic liquid/polymer-based aqueous two-phase systems. Sep Purif Technol. 2017;179:152–160.
- Anand G, Yadav S, Yadav D. Production, purification and biochemical characterization of an exo-polygalacturonase from Aspergillus niger MTCC 478 suitable for clarification of orange juice. 3 Biotech. 2017;7:122.
- Siddiqui MA, Pande V, Arif M. Production, purification and characterization of polygalacturonase from Rhizomucor pusillus isolated from decomposting orange peels. Enz Res. 2012;2012:138634.
- Barman S, Sit N, Badwaik LS, et al. Pectinase production by Aspergillus niger using banana (Musa balbisiana) peel as substrate and its effect on clarification of banana juice. J Food Sci Technol. 2015;52(6):3579–3589.