Abstract
The health pandemic (covid-19) caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has infected, to date, over 233 million people and over 4.77 million deaths worldwide. Therefore, it is important to offer a wide understanding of the ability of potential antiviral biomolecules to combat this virus, which begins to develop new variants, potentially more transmissible and infectious. Sulphated polysaccharides (SPs) from seaweeds have been reported for their antiviral activity and their potential as vaccine adjuvants. However, their effect against SARS-CoV-2 remains very limited. In this study, seventeen ligands of SPs were screened for effective interaction with the domain (RBD) spike protein using a computational approach. The obtained results indicated that two PS ligands (Xylan sulphate and Heparin tetrasaccharide N-sulphated) effectively bind to the RBD protein and could efficiently inhibit viral entry. Overall, data support the potential importance of SPs against SARS-CoV-2 as entry inhibitors and vaccine adjuvants.
Highlights
Seventeen ligands of sulphated polysaccharides (SPs) were subjected to a molecular docking to assess their interaction with SARS-CoV-2 RBD spike protein;
Two SP ligands showed an effective binding affinity to the RBD protein and could efficiently inhibit viral entry;
One effective SP ligand (Xylan Sulphate) was predicted using ADMETox evaluation as good inhibitor;
Effective candidates can have an importance against SARS-CoV-2 as entry inhibitors and vaccine adjuvants.
1. Introduction
The novel SARS-CoV-2 is the infectious agent responsible for the global COVID-19 (coronavirus disease 2019) outbreak [Citation1]. To date, the epidemic has spread around the world and has infected over 233 million people and over 4.77 million deaths [Citation2].
SARS-CoV-2 is an enveloped positive-sense RNA virus that belongs to a group of β-coronavirus [Citation3]. From attachment; and through the stages of the viral cycle (penetration, transcription, translation, replication of the genome), the SARS-CoV-2 virus interacts with various targets [Citation4]. During interaction, therapeutic protein targets of SARS-CoV-2, known to be essential at different stages of the viral cycle including 3CLpro (main protease), PLpro (papain-like protease), SGp-RBD (spike glycoprotein-receptor binding domain), RdRp (RNA dependent RNA polymerase) and ACE2 (angiotensin-converting enzyme 2) are involved and therefore constitute a good targets of drug research [Citation5]. The mechanism of blocking viral attachment through hindering the binding of the virus to the cell surface receptor is very essential in early stopping the action of the virus [Citation6].
The initial step of viral invasion into host cells start by the binding of viral particles to the host cell surface via electrostatic interaction, and the subsequent process is then achieved by transforming the unstable reversible electrostatic binding into stable irreversible binding [Citation6]. Coronaviruses use the homotrimeric spike glycoprotein (comprising a S1 subunit and S2 subunit in each spike monomer) on the envelope to bind to their cellular receptors [Citation7].
The CoV spike (S) glycoprotein is a key target for vaccines, therapeutic antibodies, and diagnostics [Citation8]. Glycosylated S proteins cover the surface of SARS-CoV-2 which the RBD (receptor binding domain) of the S1 subunit mediates binding by recognizing host cell receptors, angiotensin-converting enzyme 2 (ACE2) leading to cell attachment, and fusion during viral infection [Citation5,Citation9]. SARS-CoV-2 RBD binds to ACE2 with an affinity in the low nanomolar range, indicating that the RBD is a key functional component within the S1 subunit that is responsible for binding of SARS-CoV-2 by ACE2 [Citation5,Citation10]. Thereafter, the RdRp facilitates the viral genome replication [Citation27]. The 3CLpro and PLpro act as proteases in the process of proteolysis of the viral polyprotein into functional units [Citation5,Citation10].
Therefore, the search of natural biomolecules with antiviral efficacy is currently a challenge for the scientific community for the discovery of efficient SARS-CoV-2 antiviral agents [Citation11,Citation12], especially with the emergence and spread of new SARS-CoV-2 variants [Citation13,Citation14]. The choice of an effective antiviral agent depends on its potential compatibility to occupy and block the site of attachment, which renders the virus unable to complete the subsequent infection process [Citation15,Citation16].
Seaweeds provide a great variety of bioactive compounds with antimicrobial activity. The main components of them are usually polysaccharides [Citation17]. Among these natural and safe bioactive substances, sulphated polysaccharides (SPs) are recognized to possess various biological activities including anticoagulant, antiviral, and immuno-inflammatory activities [Citation16]. The major SPs are galactans, fucans, ulvans and their derivatives synthesized by the three major groups of marine algae, brown algae (Phaeophyta), red algae (Rhodophyta), and green algae (Chlorophyta), respectively [Citation18]. These bioactive substances don’t have equivalents in terrestrial plants and resemble the chemical and biological properties of mammalian glycosaminoglycans [Citation19].
SPs are considered as efficiency candidates in antiviral activities against several viruses, among them the coronavirus family [Citation20], which inhibit the viral infection in different phases of the viral life cycle and/or by improving the host antiviral immune response [Citation21]. Their efficiencies appear to be based on their abilities to interfere with the entry process by blocking the pathogenic virus receptors [Citation21,Citation22,Citation23]. Furthermore, SPs show other potentialities, which make it possible to promote antiviral efficacy via an immunity action. Several experimental studies demonstrate that SPs from seaweeds have potential vaccine adjuvant properties, with immunostimulatory activity on macrophages, dendritic cells and other types of immune cells [Citation24,Citation25]. In a recent study, Kim et al [Citation26] found that both monomeric and trimeric SARS-CoV-2 spike bind more tightly to immobilized heparin with remarkably high affinity and it prefers long, heavily sulphated structures. Despite their antiviral potential, SPs effect against SARS-CoV-2 remain very limited.
In this study, seventeen SPs from seaweeds were selected on the basis of their potential antiviral activities and screened for their effective interaction with the SARS-CoV2 RBD spike protein using a computational approach.
2. Material and methods
2.1. Ligand preparation
We selected seventeen compounds from literature representing different ranges of SPs to screen their potential inhibitory action against SARS-CoV-2. The repeating units as a basic structure of each SPs were considered as ligands; and were prepared for molecular docking with AutoDock Tools [Citation27]. Discovery studio 2020 was used to visualize the interactions between ligands and their target protein [Citation28]. The 2D and 3D structures of ligands conjugated to protein are presented in supplementary I. All the parameters were set to default.
2.2. Preparation of protein
The structure of SARS-CoV-2 RBD was obtained from the Research Collaboratory for Structural Bioinformatics, the Protein Data Bank RCSB PDB (PDB ID: 6VW1) [Citation29] with 2.1 Å as a resolution. Then, it was prepared for molecular docking using AutoDock Tools [Citation27] by removing defects such as missing hydrogen atoms, water, charge states, incorrect bond order assignments, orientations of various groups, and missing side chains.
2.3. Molecular docking
AutoDock Vina [Citation30] was used to predict binding affinities (kcal/mol) between RBD protein of SARS-CoV-2 and 17 ligands of SPs using a Lamarckian genetic algorithm [Citation27]. The grid box size was set at 120 Å for x, y and z, and the grid centre was set to −24.98, 14.11, and 57.25 for x, y and z respectively. The standard docking protocol was applied which is based on the population size of 150 randomly placed individuals, a maximum number of 250000 energy evaluations, a mutation rate of 0.02, a crossover rate of 0.80 and an elitism value of 1. Twenty-five independent docking runs were carried out for each inhibitor and cluster tolerance was kept at 1.0 Å.
2.4. ADMETox profiling
The term ADME/Tox refers to the absorption, distribution, metabolism, excretion, and toxicity of medicines. The in silico ADME/Tox profile is a valuable technique for predicting the pharmacological and toxicological characteristics of drug candidates, particularly during the pre-clinical stage. In silico models have been used to enhance ADME/Tox predictions. The application of these models has notably contributed to medication optimization and preventing late-stage failures, which is essential since such failures result in significant wasteful investment of time and money [Citation31].
ADMETox properties were studies for the best-docked compound for further analysis and evaluation to give more insight about our most activated compound (Xylan sulphate).
3. Results and discussion
3.1. Molecular docking
Molecular docking results of the seventeen candidates are presented in Table . Heparin was used as a positive control for the screening of the interaction of all candidates with SARS-CoV-2 RBD protein, which the binding energy is −7.0 kcal/mol. The interactions between SARS-CoV-2 RBD protein and all candidates show various types of molecular contacts (Table ). The completely docking results were included in the Supplementary Table 2. The results showed the potentialities of candidates, especially those with high binding affinities to SARS-CoV-2 RBD protein using several interactions and types of bonds. Candidates with a docking score between −8.3 and −7.0 kcal/mol are considered as interesting compounds that can be proposed as potential compounds to inhibit the SARS-CoV-2. Among them, two candidates, Xylan sulphate and Heparin tetrasaccharide N-sulphated showed the highest binding affinities, and demonstrated lower binding energies compared to the positive control, with −8.3, −7.3 kcal/mol, respectively (Table ).
Table 1. Docking score results for SARS–CoV-2 receptors binding with the potential inhibitors.
Ligands interact with sites on the protein surface, which several amino acids were involved in the binding process. Globally, five kinds of bonds belonging to the broad category of non-covalent interactions were dominated in interactions (Hydrogen, Carbon–Hydrogen, π-Sulphure, π–π-shaped, π – Alkyl bonds). It should be noted that the sulphate groups of ligands were involved in the majority of bonds, not only via the π-Sulphure bonds, but also in the other bonds, in particular by involving the oxygen atoms to binding with amino acid residues. Lodish et al. [Citation32] reported that although certain bonds are weak, the coexistence of multiple noncovalent bonds produces highly stable and specific associations between different macromolecules.
3.2. Entry inhibitor action
The number and diversity of bonds, involving the sulphate group of ligand and the amino acid residues may be one of the binding efficiency and stability of the ligand–protein complex. Indeed, the interaction results of Xylan sulphate and SARS-CoV-2 RBD protein (Supplementary Table 2) show Hydrogen Bond with ASP 428, and MET 430, π – π T-shaped with PHE 464, π – Sulphur with ARG 355, ASP 428, PHE 464, GLU 516 residues. Meanwhile, Heparin tetrasaccharide, N-sulphated interacts with ARG 355, LYS 356, LYS 462 residues, forming hydrogen bonds, and π – Alkyl bond with LYS 356 residue (Supplementary Table 2). Notably, it is observed that unlike the bonds recognized in all candidates, Xylan and Heparin tetrasaccharide N-sulphated, in their interactions with SARS-CoV-2 RBD protein, specifically develop two new types of bonds (π – π T-shaped, π – Alkyl). These last, although also qualified as non-covalent, they involve the aromatic cycle of amino acids; hence probably their high affinities in binding to the protein; in addition to the hydrophilic interaction of the two ligands with the hydrophilic sites of the active region of the RBD protein that favoured by the presence of sulphonic groups (Figure ).
Figure 1. The hydrophobic surface of protein receptor interactions with Haparin (A) and Xylan sulphate (B), from blue for hydrophilic to brown for hydrophobic.
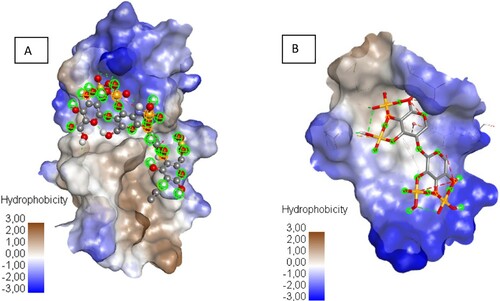
Apart from these two candidates, none of the screened candidates were more active than heparin, an anticoagulant used as the positive control [Citation33]. Indeed, in vitro studies, heparin was recently reported to exhibit antiviral activity, and was used as antiviral drugs [Citation26,Citation33]. Other studies report that soluble unfractionated heparin and its derivatives inhibit host cell entry of SARSCoV2 in vitro [Citation34,Citation35].
Nebulized heparin is now being studied [Citation36], and a separate clinical investigation shows a relation between heparin and decreased mortality in COVID-19 patients [Citation37]. Heparin could be a promising option for repurposing as a prophylacticCOVID-19 therapy with more randomized controlled studies in various therapeutic regimes [Citation37]. However, Xylan sulphate who demonstrated the more affinity than Heparins to bind SARS-CoV-2 RBD protein, has not been involved in any study to demonstrate anti SARS-CoV-2 potential. This result appears very important in the search for a new drug combating SARS-CoV-2.
Recently, in vitro studies using the SP fucoidans from brown algae and iota-carrageenan from red algae were demonstrated significant antiviral activities against SARS-CoV-2 [Citation23]. At the cellular mechanism, the efficiencies of SPs may be due to their interference with the entry process at the initial attachment by blocking the positive charge of the pathogen surface receptors, to prevent them from binding to the surface of the host cell [Citation24,Citation25]. Despite their antiviral activities, the implication of SPs against SARS-CoV-2 remains very limited [Citation23].
From the obtained results, and in terms of seaweeds origin, we can deduce that the SPs from red algae are more effective, than brown and green ones, with a range of energy, (−8.2 to −5), (−6.9 to −5.1), (−5.7 to −5.3) kcal/mol, respectively.
3.3. Potential use of SPs as vaccine adjuvants
Based on our current study, we can hypothesize that small molecules may be formulated as derivatives from SPs. They can be designed and tested to be efficient as entry inhibitors and vaccine adjuvants. Individual sites of the sulphated chains of SPs can act as mimetic natural ligands and interact with the protein [Citation38]
SPs alone or after their attachment to the antigen substrate induce an immune reaction [Citation39]. Several experimental studies demonstrate that SPs from seaweeds have potential vaccine adjuvant properties, with low toxicity, high biocompatibility, safety, as well as their mechanisms of immunomodulatory activity [Citation40,Citation41]. Various observations were reported for a variety of SPs. Fucoidan from brown algae was shown to have stimulatory effects on macrophages, dendritic cells, and other types of immune cells [Citation24,Citation25]. The sulphated galactans from green algae increase the production of inflammatory mediators by macrophages, which enhance both adaptive and innate immunity [Citation40,Citation42]. Carrageenans from red seaweeds stimulate macrophages, lymphocytes, phagocytosis, natural killer cells, antibody-dependent cell cytotoxicity; and the production of inflammatory cytokines [Citation41,Citation43,Citation44].
3.4. ADMET and drug-likeness prediction
Water solubility, lipophilicity, and the percentage of intestinal human absorption (HIA) characteristics were used to predict absorption. SwissADME’s Silicos IT LogSw descriptor was used to forecast water solubility. Our compound’s LogSw values were anticipated to be 7.58. Compounds having scores less than (more negative than) 6 on the SwissADME LogSw scale are deemed weakly soluble.
Lipophilicity was determined by calculating the logarithm of the n-octanol/water partition coefficient, which was predicted using SwissADME’s Consensus LogPo/w descriptor. LogPo/w is intimately associated with transport mechanisms such as membrane permeability and distribution to various tissues and organs [Citation45]. A moderate logP (0 log P 3) is a typical guideline for excellent oral bioavailability (good permeability and solubility) [Citation46]. The expected value of logPo/w for our compound 15 (Xylan sulphate) is 2.14.
The logSw and logPo/w predictions revealed a link between solubility and lipophilicity. The proportion of compound 15 that would be absorbed via the human intestine (% HIA) was estimated to be 19.66 percent using pkCSM-pharmacokinetics.
The glycoprotein P (P-gp) substrate, blood–brain barrier (BBB) permeability, and percentage-unbound descriptors were used to predict distribution. pkCSM-pharmacokinetics was used to predict descriptors. Pgp is an ATP-dependent drug-extracting pump identified in a variety of human tissues. The chemical 15 was anticipated to be P-gp substrates [Citation47]. Our compound’s BBB permeability was anticipated to be −5.193.
SwissADME was used to estimate metabolism by inhibiting the major cytochromes (CYP) of the P450 superfamily, notably CYP2C19, CYP1A2, CYP2C9, CYP3A4, and CYP2D6. Inhibiting CYP enzymes, a key mechanism for metabolism-based drug–drug interactions, often entails competing with another drug for the same enzyme binding site [Citation48]. Compound 15 was expected to inhibit CYP1A2 but not CYP2C19, CYP2C9, CYP2D6, or CYP3A4.
Excretion occurs largely as a result of a mix of hepatic and renal clearance, is related to bioavailability, and is critical in calculating dosage rates to reach steady-state concentrations [Citation49]. Using the total clearance (CLtot) descriptor of pkCSM-pharmacokinetics, excretion values were expected to be −0.109 ml/min/kg.
The compound 15’s toxicity levels were anticipated utilizing pkCSM-pharmacokinetics to predict hepatotoxicity and oral rat acute toxicity LD50 values[Citation46]. 2.489 mol/kg is the predicted LD50 value.
4. Conclusion
This study underlines the great potential of SP ligands as entry inhibitors and vaccine adjuvants that may offer promising use as potential agents in treating COVID-19. The molecular docking results validated the two SP ligands (Xylan sulphate and Heparin tetrasaccharide N-sulphated) that have a higher capability to interact with the target SARS-CoV-2 RBD protein. However, Xylane sulphate was predicted to be a potential spike protein of SARS-COV-2 inhibitor using ADMETox evaluation. The Admetox of our inhibitor is good, and it deserves to be studied in the laboratory. Hence, they will be effective as promising entry inhibitors that neutralize the attachment site; and therefore inhibit binding at a very early stage of the virus cycle.
In addition to this capacity, their use as vaccine adjuvants can be a real alternative in the research of the effectiveness of vaccines. From the obtained prediction results, SP ligands can be formulated, as derivatives, and tested to obtain good and safe drugs and/or vaccine adjuvants. Future researches, in vitro, will be necessary to validate the antiviral potentialities of SP ligands against SARS-CoV-2.
Supplemental Material
Download PDF (1.2 MB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Ceraolo C, Giorgi FM. Genomic variance of the 2019-nCoV coronavirus. J Med Virol. 2020;92(5):522–528. https://doi.org/10.1002/jmv.25700.
- Coronavirus disease (COVID-19) pandemic, emergencies & diseases, coronavirus disease (COVID-19) data updated on 01 October 2021. https://www.who.int/emergencies/diseases/novel-coronavirus-2019.
- Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 2020;382:727–733.
- Zaporozhets TS, Besednova NN. Biologically active compounds from marine organisms in the strategies for combating coronaviruses. AIMS Microbiol. 2020;6(4):470–494. DOI:https://doi.org/10.3934/microbiol.2020028.
- Vardhan S, Sahoo S. In silico ADMET and molecular docking study on searching potential inhibitors from limonoids and triterpenoids for COVID-19. Comput Biol Med. 2020;124; DOI:https://doi.org/10.1016/j.compbiomed.2020.103936.
- Shi Q, Wang A, Lu Z, et al. Overview on the antiviral activities and mechanisms of marine polysaccharides from seaweeds. Carbohydr Res. 2017;453–454:1–9.
- Lan J, Ge J, Yu J, et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi:https://doi.org/10.1038/s41586-020-2180-5.
- Wrapp D, Wang N, Corbett KS, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. DOI:https://doi.org/10.1126/science.abb2507.
- Huang Y, Yang C, Xu X. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol Sin. 2020;41:1141–1149. DOI:https://doi.org/10.1038/s41401-020-0485-4.
- Vardhan S, Sahoo SK. Virtual screening by targeting proteolytic sites of furin and TMPRSS2 to propose potential compounds obstructing the entry of SARS-CoV-2 virus into human host cells. J Tradit Complement Med. 2021. DOI:https://doi.org/10.1016/j.jtcme.2021.04.001
- Adejoro IA, Babatunde DD, Tolufashe GF. Molecular docking and dynamic simulations of some medicinal plants compounds against SARS-CoV-2: an in silico study. J Taibah Univ Sci. 2020;14(1):1563–1570. DOI:https://doi.org/10.1080/16583655.2020.1848049.
- Soumia M, Hanane Z, Benaissa M, et al. Towards potential inhibitors of COVID-19 main protease Mpro by virtual screening and molecular docking study. J Taibah Univ Sci. 2020;14(1):1626–1636. DOI:https://doi.org/10.1080/16583655.2020.1850002.
- Hodcroft EB, Zuber M, Nadeau S, et al. Emergence and spread of a SARS-CoV-2 variant through Europe in the summer of 2020. medRxiv. 2020. DOI:https://doi.org/10.1101/2020.10.25.20219063
- Volz EM, Hill V, McCrone JT, et al. Evaluating the effects of SARS-CoV-2 spike mutation D614G on transmissibility and pathogenicity. Cell. 2021. DOI:https://doi.org/10.1016/j.cell.2020.11.020
- Damonte EB, Matulewicz MC, Cerezo AS. Sulfated seaweed polysaccharides as antiviral agents. Curr Med Chem. 2004;11:2399–2419. DOI:https://doi.org/10.2174/0929867043364504.
- Jiao G, Yu G, Zhang J, et al. Chemical structures and bioactivities of sulfated polysaccharides from marine algae. Mar Drugs. 2011;9:196–223.
- Ahmadi A, Zorofchian Moghadamtousi S, Abubakar S, et al. Antiviral potential of algae polysaccharides isolated from marine sources: a review. Biomed Res Int. 2015;825203:10. DOI:https://doi.org/10.1155/2015/825203.
- Ngo DH, Kim SK. Sulfated polysaccharides as bioactive agents from marine algae. Int J Biol Macromol. 2013;62:70–75.
- Silva TH, Alves A, Popa EG, et al. Marine algae sulfated polysaccharides for tissue engineering and drug delivery approaches. Biomatter. 2012;2:278–289.
- Kwon S, et al. Sulfated polysaccharides effectively inhibit SARS-CoV-2 in vitro. Cell Discov. 2020;6:50. DOI:https://doi.org/10.1038/s41421-020-00192-8.
- Chen X, Han W, Wang G. Application prospect of polysaccharides in the development of anti-novel coronavirus drugs and vaccines. Int J Biol Macromol. 2020;164:331–343. DOI:https://doi.org/10.1016/j.ijbiomac.2020.07.106.
- Laurienzo P. Marine polysaccharides in pharmaceutical applications: an overview. Mar Drugs. 2010;8(9):2435–2465.
- Kato D, Era S, Watanabe I, et al. Antiviral activity of chondroitin sulphate E targeting dengue virus envelope protein. Antiviral Res. 2010;88:236–243.
- Kim SY, Joo HG. Evaluation of adjuvant e_ects of fucoidan for improving vaccine e_cacy. J Vet Sci. 2015;16:145–150.
- Kuznetsova TA, Ivanushko LA, Persiyanova EV, et al. Evaluation of adjuvant effects of fucoidane from brown seaweed Fucus evanescens and its structural analogues for the strengthening vaccines effectiveness. Biomed Khim. 2017;63:553–558.
- Kim SY, Jin W, Sood A, et al. Characterization of heparin and severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) spike glycoprotein binding interactions. Antiviral Res. 2020;181:104873. DOI:https://doi.org/10.1016/j.antiviral.2020.104873.
- Morris GM, Huey R, Lindstrom W, et al. Autodock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem. 2009;30:2785–2791.
- Dassault Systèmes BIOVIA. BIOVIA Discovery Studio. Dassault Systèmes, 2020.
- Shang J, Ye G, Shi K, et al. Structural basis of receptor recognition by SARS-CoV-2.The protein data bank. Nature. 2020;581:221–224.
- Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–461.
- Durán-Iturbide NA, Díaz-Eufracio BI, Medina-Franco JL. In silico ADME/Tox profiling of natural products: a focus on BIOFACQUIM. ACS Omega. 2020;5(26):16076–16084. DOI:https://doi.org/10.1021/acsomega.0c01581.
- Lodish H, Berk A, Zipursky SL, et al. Molecular cell biology. New York: W. H. Freeman; 2000.
- Liu J, Li J, Arnold K, et al. Using heparin molecules to manage COVID-2019. Res Pract Thromb Haemost. 2020;4:518–523. DOI:https://doi.org/10.1002/rth2.12353.
- Mycroft-West CJ, Su D, Pagani I, et al. Heparin inhibits cellular invasion by SARS-CoV-2: structural dependence of the interaction of the spike S1 receptor-binding domain with Heparin. Thromb Haemost. 2020;120(12):1700–1715. DOI:https://doi.org/10.1055/s-0040-1721319.
- Tandon R, Sharp JS, Zhang F, et al. Effective inhibition of SARS-CoV-2 entry by heparin and enoxaparin derivatives. J Virol. 2020;95(3):e01987–20. DOI:https://doi.org/10.1128/JVI.01987-20.
- Dixon B, Smith RJ, Artigas A, et al. Can nebuilsed heparin reduce time to extubation in SARS-CoV-2 (CHARTER study) - protocol. medRxiv. 2020. DOI:https://doi.org/10.1101/2020.04.28.20082552
- Ayerbe L, Risco C, Ayis S. The association between treatment with heparin and survival in patients with covid-19. J Thromb Thrombolysis. 2020. DOI:https://doi.org/10.1007/s11239-020-02162-z
- Kuznetsova TA, Persiyanova EV, Ermakova SP, et al. The sulfated polysaccharides of brown algae and products of their enzymatic transformation as potential vaccine adjuvants. Nat Prod Commun. August 2018. DOI:https://doi.org/10.1177/1934578X1801300837
- Liang Z, Zhu H, Wang X, et al. Adjuvants for coronavirus vaccines. Front Immunol. 2020;11:589833. DOI:https://doi.org/10.3389/fimmu.2020.589833.
- Barbosa JS, Costa MSSP, Melo LFM, et al. In vitro immunostimulating activity of sulfated polysaccharides from Caulerpa cupressoides var. flabellata. Mar Drugs. 2019;17:105.
- Sanina, N. Vaccine adjuvants derived from marine organisms. Biomolecules 2019, 9 (8), 340. DOI:https://doi.org/10.3390/biom9080340.
- Lee JB, Ohta Y, Hayashi K, et al. Immunostimulating e_ects of a sulfated galactan from Codium fragile. Carbohydr Res. 2010;345:1452–1454.
- Barth CR, Funchal GA, Luft C, et al. Carrageenan-induced inflammation promotes ROS generation and neutrophil extracellular trap formation in a mouse model of peritonitis. Eur J Immunol. 2016;46:964–970.
- Bondu S, Deslandes E, Fabre MS, et al. Carrageenan fromSolieria chordalis (Gigartinales): Structural analysis and immunological activities of the low molecular weight fractions. Carbohydr Polym. 2010;81:448.
- Kennewell P. Global Perspective, In Comprehensive Medicinal Chemistry II (Taylor, J.B; Triggle, D. J., Eds) Vol. 1, 2006.
- Zerroug A, Belaidi S, BenBrahim I, et al. Virtual screening in drug-likeness and structure/activity relationship of pyridazine derivatives as anti-Alzheimer drugs. J King Saud Univ. 2019;31(4):595–601. DOI:https://doi.org/10.1016/j.jksus.2018.03.024.
- Małkiewicz MA, Szarmach A, Sabisz A, et al. Blood-brain barrier permeability and physical exercise. J Neuroinflammation. 2019;16(1):1–16. DOI:https://doi.org/10.1186/s12974-019-1403-x.
- Manikandan P, Nagini S. Cytochrome P450 structure, function and clinical significance: a review? Curr Drug Targets. 2017;19(1):38–54. DOI:https://doi.org/10.2174/1389450118666170125144557.
- Pires DEV, Blundell TL, Ascher DB. pkCSM: predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. J Med Chem. 2015;58(9):4066–4072. DOI:https://doi.org/10.1021/acs.jmedchem.5b00104.
