Abstract
MicroRNA-128 (miRNA-128) exhibits tumour regulatory role in several human cancers including the colon cancer. However, regulatory mechanics of miRNA-128 through Rho family GTPase-3 (RND3) targeting is elusive in colon cancer and was the objective of present study. The results showed that miRNA-128 has significant (P < 0.05) level of down-regulation in colon cancer. Over-expression of miRNA-128 significantly (P < 0.05) decreased the viability and colony forming ability of colon cancer cells through induction of apoptosis. The percentage of apoptosis was 6.01% and 37.02% for miR-NC and miR-128 mimics transfected HT-29 cancer cells, respectively. Induction of apoptosis was associated with up-regulation of Bax and down-regulation of Bcl-2. Rho family GTPase-3 (RND3) to be target of miRNA-128 and mediated the regulatory control of miRNA-128 in colon cancer. Finally, miR-128 was found to inhibit the growth of xenografted tumours in mice. Thus, miRNA-128 exerts tumour-suppressive role in human colon cancer and may serve in colon cancer management.
1. Introduction
The human colon cancer or colorectal cancer is the third most commonly diagnosed human cancer with more than twelve lakh cases reported per year [Citation1]. In terms of the disease severity, colon cancer is ranked as the second most lethal cancerous disorder for both male and female population throughout the world [Citation2]. The application of the advanced diagnostic and therapeutic procedures though has led to considerable decline in the overall mortality rate of colon cancer during recent times but for the adults younger than fifty years the clinical outcomes are comparatively less soothing [Citation3]. Presently, the scientific community in actively involved in exploring the pathogenesis of colon cancer at a molecular level with prime focus on the regulatory molecules controlling its growth and proliferation. The micro-RNAs (miRNAs) are being particularly investigated in this regard for several members falling in this endogenous class of non-coding RNAs have been found to profoundly affect the tumourigenesis of various human cancers [Citation4,Citation5]. The miRNAs are having key involvement in crucial human biological and physiological processes [Citation6]. The miRNAs repress the expression of protein coding eukaryotic genes at translational level through their interaction with the un-translated regions of gene transcripts [Citation7]. The vital biological processes like cell differentiation, cell cycle, cell death, and so on are regulated by miRNAs [Citation8]. Many human cancers were shown to exhibit the altered expression profiles of miRNAs making it suitable to employ the usage of such miRNA molecules for cancer prognosis [Citation9]. Moreover, researchers have suggested using miRNAs as therapeutic targets against human cancers as alternative treatment strategy [Citation10]. The microRNA-128 (miRNA-128) has been deduced to exert pivotal role on the molecular mechanics of many human cancers including the colon cancer [Citation11]. The miR-128 has been shown to suppress the growth of breast cancer by modulating the expression of LINK1 [Citation12]. It has been reported to target ZEB1 to regulate the cisplatin resistance in gastric cancer and ovarian cancer [Citation13,Citation14]. Similarly, miR-128 has been shown to regulate the proliferation and metastasis of bladder cancer by targeting VEGF-C [Citation15]. Moreover, miRNA-128 was shown to interact with specific molecular targets which also include the members of Rho GTPase family to regulate the progression of human cancers [Citation16]. However, the interactional assessment of miRNA-128 with Rho GTPse-3 (RND3) has not been worked out in human colorectal cancer. The present study explored the role miR/RND3 molecular axis in controlling the growth and progression of colon cancer.
2. Materials and methods
2.1. Human tissues and cell lines
A total of 15 paired colon cancer and the corresponding normal adjacent tissues (30 in total) were obtained from the cancer patients of age group 30–75 years from the Shanghai East Hospital, Tongji University School of Medicine, Shanghai, China. The clinical histories of all the patients were noted at the time of tissue collection, and the patients were informed in advance along with the consent signing. Only those patients were included who had not gone any chemo or radiotherapy. The tissues were transported in liquid nitrogen and stored in freezing conditions at −80°C till their experimental usage. The experimental protocols and the tissue study procedures were approved by the ethics committee of the Shanghai East Hospital, Tongji University School of Medicine, Shanghai, China (Approval number: TU/EH/09/HP/0455).
All the colon cell lines, cancerous (HT-29, RKO, SW480 and SW948) and normal epithelial (CCD-18Co) were provided by the American Type Collection Center, USA. The culture medium RPMI-1640 (Hyclone), containing 10% fetal bovine serum (FBS) (Gibco) and 1% penicillin/streptomycin (Thermo Fisher Scientific), was used for cell culturing at 37°C with 5% CO2 in humidified incubator. The transfection of the cell lines was performed using Lipofectamine-2000 (Thermo Fisher Scientific) at 80% cellular confluence. The over-expression transfection construct of miRNA-128 (miRNA-128 mimics) and silencing construct of Rho family GTPase-3 (si-RND3) together with the respective controls (miRNA-NC and si-NC) were synthesized by Guangzhou RiboBio Co. Ltd., China. The mammalian over-expression vector pcDNA3.1 was used for RND3 over-expression.
2.2. Determination of relative gene expression
Total RNA was extracted from the tissues and cell lines through TRIzol reagent (Thermo Fisher Scientific) based method. The RNA isolated was given DNase I (Thermo Fisher Scientific) treatment and was then quantified using NanoDrop spectrometer. Around 2 µg RNA was used for the synthesis of complementary DNA with the help of iScriptTM cDNA synthesis kit (Bio-rad) as per the manufacturer protocol. For the gene expression studies quantitative real-time PCR (qRT-PCR) was performed on QuantStudio 5.0 RT-PCR system (Thermo Fisher Scientific) using the SYBR Green PCR reaction mix (Thermo Fisher Scientific). The relative expression levels were deuced through 2-ddCt cycle threshold method. Human GADPH gene was used as internal control.
2.3. Cell viability
The viabilities of transfected colon cancer cells were assessed through 3-(4,5-dimethylthiazol-2-yl)-2, 5-diphenyl tetrazolium bromide (MTT) assay. 2 × 105 cells were seeded into each well of 96-well plate. MTT reagent (5 mg/L) was added to each well after 0, 12, 24, 48 and 96 h incubation at 37°C. The cell cultures were again incubated for 4 h at 37°C. Afterwards, the cell cultures were added with 250 µL dimethyl sulfoxide for the solubilization of formazan crystals. Finally, spectrophotometer was used for recording the optical density (OD) measurements at 570 nm wavelength for determining the cell viabilities.
2.4. Colony formation assay
The colony formation of transfected colon cancer cells was analysed using clonogenic assay. The cells were plated into 6-well plate with 200 cells per well. The cell culturing was performed for 14 days at 37°C with 5% CO2 in humidified incubator. The colonies formed were PBS washed, ethanol fixed and stained with 0.2% crystal violet solution. The colonies were visualized under light microscope, and manual cell counting was used for determining the percent colony formation.
2.5. Examination of cancer cell apoptosis
Following their plating into the 12-well plates with initial density of 105 cells per well, the transfected colon cancer cells were cultured for 48 h at 37°C. The cells were then harvested and washed carefully with PBS buffer. Paraformaldehyde (4%) was used for fixing the cells. For the assessment of their nuclear viabilities, the cells were stained with either 4′,6-diamidino-2-phenylindole (DAPI) or acridine orange (AO)/ethidium bromide (EB) solutions and examined under the fluorescent microscope. Dual annexin V-FITC/PI staining was also performed followed by the flow cytometric examination for the analysis of cell apoptosis.
2.6. miRNA-128 target analysis
The online bioinformatics was performed using TargetScan Human 7.2 database (http://www.targetscan.org/vert_72/) for specifically predicting the miRNA-128 target in colon cancer. To validate the prediction, dual luciferase reporter assay was used. The cancer cells were co-transformed with miR-128 mimics and luciferase construct of 3′-UTR of RND3 gene (either native, WT or mutated, MUT). After 24 h incubation at 37°C, the luciferase activity of cancer cells was analysed using Dual-luciferase Reporter assay system (Promega) following the manufacturer guidelines. The luciferase activity of Renilla luciferase gene was used for normalization of the luciferase activity measurements.
2.7. In vivo mice tumourigenesis study and immuno-histochemical fluorescence assay
Around 6–8 weeks old healthy female NOD/SCID mice were used in the present study. The maintenance conditions, experimental handling and in vivo study procedures were approved by the institutional animal ethics committee of Shanghai East Hospital, Tongji University School of Medicine, Shanghai, China. The HT-29 cancer cells were stably transfected with miRNA-128 over-expression constructs (miR-128 mimics) or normal control, miR-NC for 48 h. The transfected cells were re-suspended in matrigel in a 1:1 ratio and then injected subcutaneously into the dorsal flank of the mice. The mice were maintained with care for 3 weeks in animal house and then sacrificed. The tumour xenografts were then collected, and their weight and volume were examined. The excised tumours were further processed for protein expression analysis through immuno-histochemical fluorescence study.
2.8. Statistical analysis
The SPSS 22.0 software was used for carrying out the statistical analyses of the experimental data. The experiments were performed using three replicates and final values represent the mean ± standard deviation. One-way ANOVA and student’s t-test were performed for the analysis of statistical difference. The p < 0.05 was taken to reveal the statistically significant difference.
3. Results
3.1. The miRNA-128 has significant down-regulation in colon cancer
Real-time PCR-based expression analysis was used to look into the expression profile of miRNA-128 in colon cancer. The results showed that the colon cancer tissues possess significantly (P < 0.05) lower transcript levels of miRNA-128 in comparison to the normal corresponding colon tissues (Figure (A)). To spot whether similar is true for the miRNA-128 expression in colon cancer cell lines, the expression analysis was extended to the cancerous and normal epithelial colon cell lines. All the cancer cell lines (HT-29, RKO, SW480 and SW948) were seen to exhibit significantly (P < 0.05) lower miRNA-128 expression when compared with the normal epithelial colon cell line (CCD-18Co) (Figure (B)). Therefore, it can be stated that human colon cancer is associated with transcriptional down-regulation of miRNA-128 suggesting possible regulatory role of miRNA-128 in colon cancer mechanics. Since, lowest expression of miR-128 was observed in HT-29 cell lines, this cell line was used for further experimentation.
Figure 1. Over-expression of colon cancer repressed miRNA-128 reduced the proliferation of colon cancer cells. (A) RT-PCR expression analysis of miRNA-128 in colon cancer and corresponding normal human tissues; (B) RT-PCR expression analysis of miRNA-128 in colon cancer cell lines (HT-29, RKO, SW480 and SW948) and normal colon epithelial cell line, CCD-18Co; (C) RT-PCR expression analysis of miRNA-128 in colon cancer cell line, HT-29 transfected with miR-128 mimics or miR-NC; (D) MTT assay for the estimation of viability of HT-29 cancer cells transfected with miR-128 mimics or miR-NC. The experiments were performed in triplicate and expressed as mean ± SD (*P < 0.05).
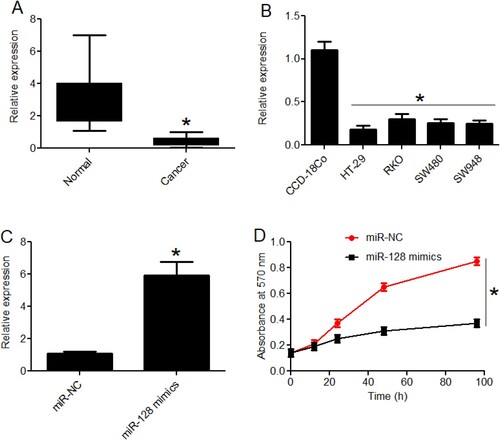
3.2. The over-expression of miRNA-128 inhibited cancer cell growth through apoptosis
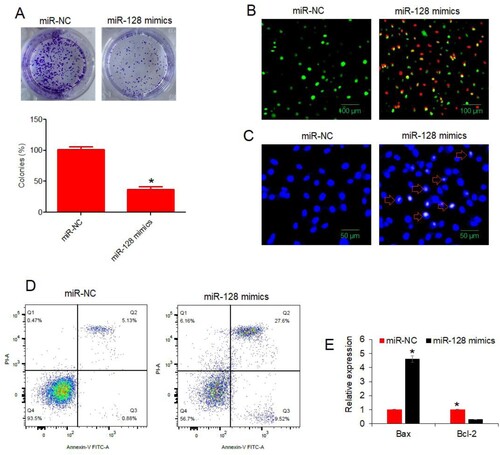
To characterize the molecular role of miRNA-128 in colon cancer, miRNA-128 was re-expressed in HT-29 cancer cells. The over-expression of miRNA-128 in HT-29 cancer cells was confirmed through RT-PCR. The miR-128 mimics transfected cancer cells were shown to possess almost 6 times higher miRNA-128 expression than the normal control transfected cells (Figure (C)). When the viability of cancer cells over-expressing miRNA-128 was analysed, the cancer cells showed significant (P < 0.05) decline in the viabilities and effects were particularly evident when higher incubation periods were used (Figure (D)). Similarly, the colony formation was reduced by more than 50% under miRNA-128 over-expression (Figure (A)). The reason hypothesized for decrease in HT-29 cell viability was the cell apoptosis. To validate the assumption, the apoptosis of HT-29 cancer cells over-expressing miRNA-128 was analysed and compared with the normal control cells through DAPI and AO/EB staining methods and flow cytometry. From both AO/EB and DAPI staining procedures, it was confirmed that colon cancer cells exhibited nuclear deformation under miRNA-128 up-regulation (Figure (B,C)). The picture was clearer when flow cytometric apoptosis analysis was performed. The HT-29 cancer cellular cultures over-expressing miRNA-128 were found to have significantly (P < 0.05) higher percentage of apoptotic cells (Figure (D)). The percentage of apoptosis was found to be 6.01% and 37.02% for miR-NC and miR-128 mimics transfected HT-29 cancer cells respectively. Moreover, the expression analysis of Bax and Bcl-2 apoptotic marker genes showed that Bax expression increased while Bcl-2 expression decreased under miRNA-128 over-expression (Figure (E)). To sum up, the results indicate that over-expression of miRNA-128 in colon cancer cells induced apoptosis and resulted into the decline of cancer cell viability, in vitro.
Figure 2. miR-128 over-expression declined colony formation and induced apoptosis in colon cancer cells. (A) Analysis of colony formation from HT-29 cancer cells transfected with miR-128 mimics or miR-NC; (B) AO/EB staining analysis of HT-29 cancer cells transfected with miR-128 mimics or miR-NC. Green, yellow and red colors indicate normal, early apoptotic and late apoptotic cells, respectively; (C) DAPI staining analysis of HT-29 cancer cells transfected with miR-128 mimics or miR-NC. Arrows indicate apoptotic cells; (D) flow cytometric examination of apoptosis of HT-29 cancer cells transfected with miR-128 mimics or miR-NC. The experiments were performed in triplicate and expressed as mean ± SD (*P < 0.05).
3.3. The miRNA-128 regulates colon cancer tumourigenesis by targeting Rho family GTPase-3 (RND3)
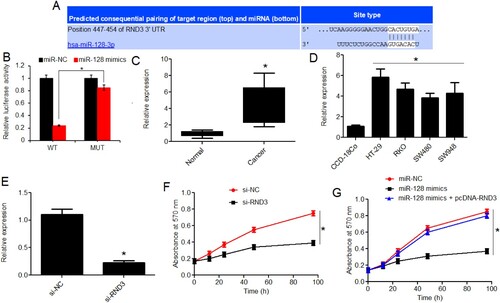
The bioinformatics target identification through TragetScan database showed that miRNA-128 interacts with a specific nucleotide segment in the 3′-UTR of RND3 transcript to repress their expression at translational level (Figure (A)). To confirm this interactional repression of RND3 by miRNA-128 in colon cancer, the dual luciferase reporter assay was performed. The decline in the luciferase activity confirmed the interaction of miRNA-128 with 3′-UTR of RND3 when the predicted interaction site was used in the native state (Figure (B)). As expected, the RND3 transcript level was shown to be significantly (P < 0.05) higher in colon cancer tissues in comparison to normal colon tissues (Figure (C)). Further support was gained from the expression analysis of RND3 in colon cancer cell lines in comparison with normal colon cell line. RND3 was found to have significantly (P < 0.05) higher expression in all the cancer cell lines owing to lesser transcript abundance of miRNA-128 in them (Figure (D)). Now, to evaluate if post-transcriptional targeting of RND3 by miRNA-128 in colon cancer had any bearing on the proliferation of colon cancer cells, the experimental knockdown of miRNA-128 was performed in HT-29 colon cancer cells (Figure (E)). RND3 gene silencing resulted in reduction of cancer cell proliferation in the same way as under miRNA-128 over-expression (Figure (F)). In contrary, RND3 over-expression could avoid the growth inhibitory effects of miR-128 over-expression on the colon cancer cells (Figure (G)). The results thus indicated that the colon cancer growth regulatory role of miRNA-128 is exerted through translational repression of Rho family GTPase-3 (RND3).
Figure 3. Rho family GTPase-3 (RND3) is targeted by miRNA-128 and modulates the growth regulatory role of miRNA-128 in colon cancer. (A) Bioinformatics target prediction of miRNA-128 in colon cancer; (B) dual luciferase reporter assay for the validation of miRNA-128 interaction with 3′-UTR of RND3 in colon cancer; (C) RT-PCR expression analysis of RND3 in colon cancer and corresponding normal human tissues; (D) RT-PCR expression analysis of RND3 in colon cancer cell lines (HT-29, RKO, SW480 and SW948) and normal colon epithelial cell line, CCD-18Co; (E) RT-PCR expression analysis of RND3 in colon cancer cell line, HT-29 transfected with si-RND3 or si-NC; (F) MTT assay for the estimation of viability of HT-29 cancer cells transfected with si-RND3 or si-NC; (G) MTT assay for the estimation of viability of HT-29 cancer cells transfected with miR-NC, miR-128 mimics or miR-128 mimics plus pcDNA-RND3. The experiments were performed in triplicate and expressed as mean ± SD (*P < 0.05).
3.4. The miRNA-128 intra-tumour over-expression hampered tumour growth in mice xenografts
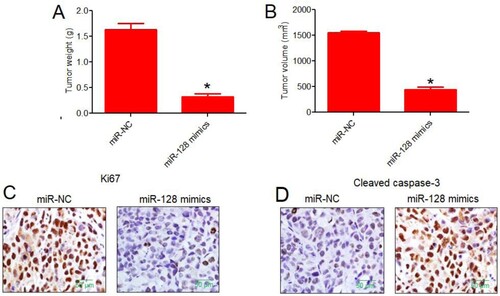
To investigate whether the in vitro results correlate with the in vivo role of miRNA-128 in colon cancer, the xenograft tumour were developed in mice. After sacrificing the mice, the mice inducted for tumourigenesis with colon cancer cells over-expressing miRNA-128 exhibited significantly lower tumour size. The average tumour weight was also found to be fairly low under miRNA-128 up-regulation (Figure (A)). Similar inhibitory effect was observed on the tumour volume under miRNA-128 over-expression (Figure (B)). The immuno-fluorescence study on the mice tumours revealed that mice tumours exhibited lower proliferation rate under miRNA-128 evident from lower Ki67 proliferation marker expression (Figure (C)). The level of apoptosis was confirmed to be higher in mice tumours over-expressing miRNA-128 (Figure (D)). Collectively, the results of the in vivo mice study specify that miRNA-128 is crucial for the colon cancer tumourigenesis and as such it may serve as vital therapeutic target against this serious malignancy in near future.
Figure 4. In vivo mice tumourigenesis study signified the tumour-suppressive role of miRNA-128 in colon cancer. (A) Average weight (g) of xenograft mice tumours collected from mice injected with HT-29 cancer cells over-expressing miRNA-128 or normal control HT-29 cancer cells; (B) average volume (mm3) of xenograft mice tumours collected from mice injected with HT-29 cancer cells over-expressing miRNA-128 or normal control HT-29 cancer cells; (C) immuno-histochemical fluorescence analysis of Ki67, proliferation marker from xenograft mice tumours collected from mice injected with HT-29 cancer cells over-expressing miRNA-128 or normal control HT-29 cancer cells; (D) immuno-histochemical fluorescence analysis of cleaved caspase-3, apoptosis marker from xenograft mice tumours collected from mice injected with HT-29 cancer cells over-expressing miRNA-128 or normal control HT-29 cancer cells. The experiments were performed in triplicate and expressed as mean ± SD (*P < 0.05).
4. Discussion
In depth understanding of progression of human colon cancer is lacking and the same is very crucial for formulating better preventive and curative strategies against this deadly disorder. Although the survival rates have enhanced to some extent during the recent era, colon cancer is still a serious health disorder [Citation3]. As is true for other human cancers, the prognosis of colon cancer at earlier stages is very difficult and in most of the cases, the patients suffering from this disease are found to exhibit lymphatic node metastasis at the time of diagnosis [Citation17]. Taken these hindrances in mind, continuous efforts are being made to explore the alternative prognostic and treatment possibilities so as to curb the menace of colon cancer more effectively. The present research investigation represents an endeavor towards this scientific objective. The micro-RNAs (miRNAs) have emerged as important regulators of various eukaryotic physiological processes [Citation6]. Besides, the miRNAs also regulate the key pathological aspects of several human diseases [Citation18]. The role of miRNAs in human cancer is known to everyone [Citation19]. The dysregulation of miRNAs has been elucidated to be associated with number of human cancers [Citation9,Citation20]. Previously, the colon cancer was reported to exhibit significant level of miRNA-128 repression [Citation11]. The results of the present study also deduced the similar transcriptional down-regulation of miRNA-128 in colon cancer. The study further insights into the regulation of colon cancer cell proliferation by miRNA-128. The re-expression of miRNA-128 intensively diminished the colon cancer cell proliferation in vitro, and these anti-proliferative effects of miRNA-128 suggest the tumour-suppressive role of miRNA-128 in colon cancer which in addition reflects the already established role of miRNA-128 in human cancers [Citation21]. In a recent study, miR-128 has been shown to suppress the proliferation of anaplastic thyroid cancer [Citation22]. Similarly, miR-128 has been found to suppress the growth of the esophageal squamous-cell cancer [Citation23]. The reason behind this proliferation turn down was proved to be the induction of apoptosis in colon cancer cells over-expressing miRNA-128. Similar mechanism of proliferation decline was reported by the researchers for miRNA-128 in the previous studies also [Citation24,Citation25]. The miRNAs are seen to act at translational level to repress the expression of a great number of protein coding genes [Citation7]. For, instance miR-128 has been shown to target LIMK1, ZEB1, VEGF-C, RhoE and BMI-1 [Citation12–15,Citation20]. However, the present study for the first time reported RND3 as the molecular target of miRNA-128 in colon cancer. The over-expression of RND3 in colon cancer, resulting from one of the possible reasons of alleviation of miRNA-128, might be responsible for the enhanced cell proliferation and metastatic behavior of colon cancer cells as has been reported earlier [Citation26]. The miRNA-128/RND3 axis was shown to possess considerable growth regulatory role in colon cancer. The tumour-suppressive function of miRNA-128 in colon cancer was indicated in a clearer way from the mice in vivo tumourigenesis study and it was shown that miRNA-128 up-regulation allowed limited cell proliferation and higher apoptosis in tumour cells. Summing up, the results of present study are suggestive of tumour-suppressive role of miRNA-128 in colon cancer and highlighted the therapeutic potential of this vital regulatory molecule. However, more studies on higher number of human tissues and cell lines are required to establish miR-128 as a molecular therapeutic target for the management of colon cancer. Additionally, the evaluation of expression of miR-128 at different stages of colon cancer will also help to unveil its prognostic value.
5. Conclusion
To conclude, the present study showed miRNA-128 repression is associated with the growth and proliferation of colon cancer. In addition, the results indicated the tumour-suppressive role of miRNA-128 and discovered the RND3 mediated down-stream signaling of miRNA-128 in colon cancer. The study further proposed that miRNA-128 might be utilized as vital therapeutic target for the management of human colon cancer.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Zhao Y, Hu X, Zuo X, et al. Chemopreventive effects of some popular phytochemicals on human colon cancer: a review. Food Funct. 2018;9(9):4548–4568.
- Zhang B, Fang C, Deng D, et al. Research progress on common adverse events caused by targeted therapy for colorectal cancer. Oncol Lett. 2018;16(1):27–33.
- Benson AB, Venook AP, Cederquist L, et al. Colon cancer, version 1.2017, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Net. 2017;15(3):370–398.
- Vannini I, Fanini F, Fabbri M. Emerging roles of microRNAs in cancer. Curr Opin Genet Dev. 2018;48:128–133.
- Lan J, Huang Z, Han J, et al. Redox regulation of microRNAs in cancer. Cancer Lett. 2018;418:250–259.
- Herranz H, Cohen SM. MicroRNAs and gene regulatory networks: managing the impact of noise in biological systems. Genes Dev. 2010;24(13):1339–1344.
- Fabian MR, Sundermeier TR, Sonenberg N. Understanding how miRNAs post-transcriptionally regulate gene expression. InmiRNA regulation of the translational machinery. Berlin: Springer; 2010. p. 1–20.
- Miska EA. How microRNAs control cell division, differentiation and death. Curr Opin Genet Dev. 2005;15(5):563–568.
- Melo SA, Esteller M. Dysregulation of microRNAs in cancer: playing with fire. FEBS Lett. 2011;585(13):2087–2099.
- Shah MY, Calin GA. MicroRNAs as therapeutic targets in human cancers. Wiley Interdiscip Rev: RNA. 2014;5(4):537–548.
- Wu L, Shi B, Huang K, et al. MicroRNA-128 suppresses cell growth and metastasis in colorectal carcinoma by targeting IRS1. Oncol Rep. 2015;34(5):2797–2805.
- Zhao J, Li D, Fang L. MiR-128-3p suppresses breast cancer cellular progression via targeting LIMK1. Biomed Pharmacother. 2019;115:108947.
- Guo Y, Yue P, Wang Y, et al. PCAT-1 contributes to cisplatin resistance in gastric cancer through miR-128/ZEB1 axis. Biomed Pharmacother. 2019;118:109255.
- Li B, Chen H, Wu N, et al. Deregulation of miR-128 in ovarian cancer promotes cisplatin resistance. Int J Gynecol Cancer. 2014;24:8.
- Zhou XU, Qi L, Tong S, et al. miR-128 downregulation promotes growth and metastasis of bladder cancer cells and involves VEGF-C upregulation. Oncol Lett. 2015;10(5):3183–3190.
- Shang C, Hong Y, Guo Y, et al. miR-128 regulates the apoptosis and proliferation of glioma cells by targeting RhoE. Oncol Lett. 2016;11(1):904–908.
- Wolpin BM, Mayer RJ. Systemic treatment of colorectal cancer. Gastroenterology. 2008;134(5):1296–1310.
- Osman A. MicroRNAs in health and disease–basic science and clinical applications. Clin Lab. 2012;58(5–6):393.
- Ors-Kumoglu G, Gulce-Iz S, Biray-Avci C. Therapeutic microRNAs in human cancer. Cytotechnology. 2019;71(1):411–425.
- Chen PS, Su JL, Hung MC. Dysregulation of microRNAs in cancer. J Biomed Sci. 2012;19(1):1–8.
- Jin M, Zhang T, Liu C, et al. miRNA-128 suppresses prostate cancer by inhibiting BMI-1 to inhibit tumor-initiating cells. Cancer Res. 2014;74(15):4183–4195.
- Chen J, Zhao D, Meng Q. Knockdown of HCP5 exerts tumor-suppressive functions by up-regulating tumor suppressor miR-128-3p in anaplastic thyroid cancer. Biomed Pharmacother. 2019;116:108966.
- Zhao L, Li R, Xu S, et al. Tumor suppressor miR-128-3p inhibits metastasis and epithelial–mesenchymal transition by targeting ZEB1 in esophageal squamous-cell cancer. Acta Biochim Biophys Sin. 2018;50(2):171–180.
- Zhao D, Han W, Liu X, et al. Micro RNA-128 promotes apoptosis in lung cancer by directly targeting NIMA-related kinase 2. Thorac Cancer. 2017;8(4):304–311.
- Cui J, Zhao Y, Sethi P, et al. Micro-RNA-128 (miRNA-128) down-regulation in glioblastoma targets ARP5 (ANGPTL6), Bmi-1 and E2F-3a, key regulators of brain cell proliferation. J Neuro-Oncol. 2010;98(3):297–304.
- Paysan L, Piquet L, Saltel F, et al. Rnd3 in cancer: a review of the evidence for tumor promoter or suppressor. Mol Cancer Res. 2016;14(11):1033–1044.
