ABSTRACT
Introduction
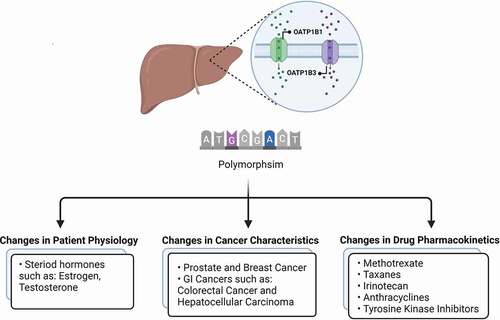
Members of the solute carrier family of organic anion transporting polypeptides are responsible for the cellular uptake of a broad range of endogenous compounds and xenobiotics in multiple tissues. In particular, the polymorphic transporters OATP1B1 and OATP1B3 are highly expressed in the liver and have been identified as critical regulators of hepatic elimination. As these transporters are also expressed in cancer cells, the function alteration of these proteins have important consequences for an individual’s susceptibility to certain drug-induced side effects, drug–drug interactions, and treatment efficacy.
Areas covered
In this mini-review, we provide an update of this rapidly emerging field, with specific emphasis on the direct contribution of genetic variants in OATP1B1 and OATP1B3 to the transport of anticancer drugs, the role of these carriers in regulation of their disposition and toxicity profiles, and recent advances in attempts to integrate information on transport function in patients to derive individualized treatment strategies.
Expert opinion
Based on currently available data, it appears imperative that different aspects of disease, physiology, and drugs of relevance should be evaluated along with an individual’s genetic signature, and that tools such as biomarker levels can be implemented to achieve the most reliable prediction of clinically relevant pharmacodynamic endpoints.
1. Introduction
Genetic differences are a leading cause of substantial inter-individual variability in drug response and toxicity reported in most patient populations, and cause drug plasma concentrations to vary up to 600 fold in patients of the same bodyweight receiving the same standard dose [Citation1–3]. Since pharmacokinetic characteristics of a drug are mainly determined by the expression and activity of drug-metabolizing enzymes and/or drug transporters, different expression/activity levels in these proteins are often the culprit in variable drug response and toxicity in individuals.
The solute carrier (SLC) superfamily of transporters includes more than 400 members, of which many are localized in the cell membrane of organs involving in absorption (intestine), distribution (brain) and elimination (kidney, liver) [Citation4]. While the main function of these transporters is to facilitate the entry and removal of essential nutrients and endogenous toxins, many SLC superfamily members also contribute greatly to the cellular and systemic distribution of many therapeutic drugs [Citation5–7]. The Organic Anion Transporting Polypeptides (OATPs) subfamily appears to be particularly relevant in the cellular uptake of a broad spectrum of pharmaceutical agents [Citation8–10], including drugs and xenobiotics that exhibit structural similarity with bile salts, bilirubin, anionic peptides, steroid conjugates, and thyroid hormones [Citation11]. Several OATP variants have been identified and associated with significant alteration on the pharmacokinetic profile of their drug substrates, and as such, identifying these genetic polymorphisms should be considered in designing individualized drug therapy regimens [Citation12].
Many reviews and research articles have been published on this subject in recent years [Citation13,Citation14], which covered different single nucleotide polymorphisms (SNPs) and their effects on the distribution of various drugs. In the present mini-review, we highlight recent advances in the influence of polymorphisms in the OATP1B1 and OATP1B3 (gene names, SLCO1B1 and SLCO1B3), which are among the most highly expressed membrane transporters in the liver [Citation15,Citation16], on cancer pharmacotherapy. We focused on articles reported in the past 5 years in order to avoid redundancy with existing literature reviews, and categorized these based on the way they alter cancer therapy and seek to explore the idea that changing our approach to identifying these SNPs can be useful in determining clinical relevance and their applicability in predicting pharmacokinetic characteristics of anticancer drugs. A scheme of discussed topics can be found in .
2. Pharmacogenetics variations related to changes in patient physiology
Many endogenous substances such as Estradiol-17β-glucuronide, Estrone-3-sulfate, Testosterone, Thyroxine (T4), Triiodothyronine (T3), DHEA-S, Prostaglandin E2 and bile acids such as Cholate, Taurocholate, Tauroursodeoxycholate are substrates for OATPs [Citation17], and therefore when the function of transporters is altered due to genetic polymorphisms, changes in the circulating levels of these substances could occur, which may in turn affect the body’s response to certain substrate drugs.
For example, since the distribution and elimination of estrone and other testosterone compounds are dependent on transport by OATPs, it could indicate that function of drugs with similar structure that are used to treat cancers with sex hormone dysregulation also rely on OATP transport and their polymorphisms.
Breast cancer, one of the most prevalent cancers, makes up a large portion of sex hormone-related cancers – as estrogen-dependent breast cancer is its most common type [Citation18]. Currently, one of the standards of care for patients with estrogen-dependent breast cancer is a therapeutic remedy with the aromatase inhibitor (AI) exemestane. Aromatase inhibitors are used to control breast cancer, specifically in postmenopausal women, by decreasing systemic, endogenous estrogens through the inhibition of estrogen production. This starves the tumor of its necessary endogenous growth signals, preventing the continuing growth of ER+ breast cancers [Citation19,Citation20]. It has been clinically noted that there has been significant interindividual variability in patients’ response to exemestane. Steroid compounds, which include hormonal molecules, are known to be substrates for OATPs, leading to the hypothesis that genetic polymorphism may be a significant factor in this interpatient variability. While a multitude of SNPs in OATP1B1 and OATP1B3 are known, only OATP1B1 rs4149056, OATP1B1 rs2306283, OATP1B3 rs4149117, and OATP1B3 rs7311358 have been associated with significant clinical effects on variable drug transport [Citation21–24]. Furthermore, SLCO1B1*5 (rs4149056) is correlated with an increased systemic estrogen concentration before treatment with an AI and increased exemestane concentration during treatment with AI [Citation22,Citation25]. A retrospective, open-label, pharmacogenetic study of exemestane and its major metabolite, 17-hydroexemestane, carried out by Gregory et al. [Citation22], reported that exemestane metabolism in vivo is correlated with OATP1B1 rs4149056C > T [Citation20]. This conclusion was derived from a study involving 14 healthy postmenopausal female volunteers receiving a single dose of oral exemestane in whom the subjects carrying the OATP1B1 rs4149056C > T genotype had a statistically significant difference in the plasma levels of exemestane and its metabolite although this was not observed for the other genotype groups (OATP1B1 rs2306283 G > A, OATP1B3 rs4149117G > T, and OATP1B3 rs7311358A > G) [Citation22]. Dempsey et al. [Citation26] also evaluated the influence of OATP1B1 variants on relevant phenotypes in 503 postmenopausal women receiving exemestane and letrozole, another AI aromatase inhibitor that is a suspected substrate for OATPs. Within their study, the authors observed that there was a 51% increase in estrone sulfate pretreatment concentrations in the SLCO1B1*5 (rs4149056) genotype group. In another study, the (C) allele of rs10841753, in contrast, was associated with a 20% plasma decrease in pretreatment levels of estrone sulfate albeit it was not clinically significant [Citation26]. This data support the notion that OATPs and their polymorphisms are involved in the endogenous distribution of estrones and other steroidal molecules within the body. Dempsey et al. noted further that, after 3 months of AI treatment, SLCO1B1*5 (rs4149056) carriers are more likely to maintain detectable estrone concentrations. This implies that these patients have worse outcomes than those not carrying this allele [Citation26]. These recent studies show a strong association between polymorphic variants in OATP1B1 and the disposition of AIs [Citation22].
Continuing with estrogen-dependent breast cancer, a common standard of care as recommended by the National Comprehensive Cancer Network is tamoxifen. This common post-operative endocrine therapy is recommended regardless of menstruation, age, tumor size, or lymph status [Citation27]. For tamoxifen, and its active form endoxifen, both isoforms of tamoxifen, Z and E, are estrogen receptor agonists, while the Z form is notably more effective [Citation28]. As agonists, these molecules compete with endogenous estrogen for the estrogen receptor-binding sites, and this competition significantly inhibits the growth of breast cancer cells. However, it is important to note, similar to exemestane, that significant differences in response to tamoxifen have been noted that cannot be explained exclusively by altered organ function or differences in age and lifestyle, and that genetic polymorphism in OATP transporters potential contributes to changes in response. In contrast to the above study of exemestane, the research group Gao et al. [Citation29] used an in vitro model to study the effects of genetic polymorphisms on tamoxifen uptake via standard MTT assay. The authors found that following 24 or 48 hours of exposure to tamoxifen, cells expressing SLCO1b1*1a, SLCO1b1*1b rs2306283) or SLCO1B1*5 (rs4149056) are associated with increased cellular uptake of tamoxifen as compared to negative controls, and that levels were lower in the OATP1B1* 5 (521 CC) group than in the OATP1B1* 1a or OATP1B1* 1b (388 GG) groups [Citation29]. This observation is consistent with the notion that OATP1B1*5 represents a common variant associated with decreased function and differential ability cause drug-induced toxicity. Although the allelic distribution is known to vary widely in different populations, the degree to which OATP variants contribute to interpopulation differences in response to tamoxifen in clinical practice remains to be established [Citation15,Citation30].
These recent studies with AIs and tamoxifen serve to illustrate the correlation of the distribution and elimination of hormones via OATPs – which can be further extrapolated to the drugs that affect these estrones and androgens. Within different racial populations, OATPs maintain a high genetic polymorphic variability, meaning that specific SNP may affect the function of the transporter and that this information should be taken into account when administering therapeutics such as AIs and tamoxifen [Citation29].
3. Pharmacogenetics variations related to changes in cancer characteristics
The concentration of anticancer drugs in cancer cells is an important component of their pharmacological response as it accounts for their net interaction with intracellular targets. Therefore, reduced intracellular levels of an active agent due to mechanisms such as reduced activity in expression/function of the transporters involved in uptake, can lead to chemoresistance [Citation31]. As mentioned in the previous section, OATP1B1 and OATP1B3 are also involved in transport of many endogenous substances and nutrients. This recognition has led to the thesis that, depending on individual characteristics of the cancer cell, genetic variants affecting SLC genes may significantly alter cancer prognosis as well as therapeutic responsiveness to anticancer drugs. Changes in disease intensity that are dependent on OATP1B1 or OATP1B3 polymorphism have also been reported fin the context of other therapeutic areas, including tuberculosis and cardiovascular disease [Citation32,Citation33]. While the effects of different polymorphisms on uptake and sensitivity to different drugs are mainly discussed in the next section, we will here review some recent insights into general correlations between treatment of cancers and OATP polymorphism.
Under normal physiological conditons, OATP1B1 is almost exclusively expressed at the sinusoidal membrane of hepatocytes while OATP1B3 is also found, albeit to a much lower degree, in other tissues, such as colon, prostate, and testis. In cancer tissue, both transporters are expressed in certain hepatocellular carcinomas and cholangiocarcinomas, whereas expression of OATP1B1 has been detected in certain colon and ovarian cancers. Furthermore, OATP1B3 has been detected in some cancers along the carcinomas of the head and neck, and cancers in GI tract, such as esophageal cancers and adenocarcinomas of stomach, colorectal, and pancreas, but also in certain lung, testicular, prostate, ovarian, and breast cancers [Citation34,Citation35].
Low expression of OATP1B3 has been correlated with a poor prognosis in hepatocellular carcinomas (HCC), where patients expressing high tumoral levels of OATP1B3 experienced significantly longer overall survival (OS) and disease-free survival (DFS) compared to patients with low expression (33.0% vs 12.9%, P = 0.001; 18.8% vs 5.3%, P < 0.0001) [Citation36]. Although the mechanistic basis of this variable expression in unclear, it is conceivable that particular decreased function haplotypes of OATP1B3 associated with the SNPs p.Met233Ile (c.699 G > A), p.Gly522Ser (c.1564 G > A) and p.Val560Ala (c.1679 T > C) might similarly correlate with survival outcomes. Although most prior studies have suggested that these SNPs are not strong predictors of the pharmacokinetics and response to OATP1B3 substrates in vivo [Citation31,Citation37], prospective validation studies are required to provide definitive answers to the role of OATP1B3 variants in HCC. In several other cancer indications, including multiple myeloma, data from recent clinical studies have suggested that the OATP1B1 (388 G) variant does not significantly contribute to treatment outcomes [Citation38]. Interestingly, in patients with colorectal cancer, the rs60571683 SNPs in SLCO1B3 was reported as an N6-methyladenosine (m6A)-associated SNP promote development of colorectal cancer [Citation39].
According to Huang et al. when three SNPs in SLCO/OATP gene were studied in 137 metastatic CRC patients, in SLCO1B1 gene was associated with a higher rapid response rate and also served as an independent prognostic factor for a longer progression-free survival (PFS) [Citation40,Citation41].
For prostate cancer, in a meta analysis by Shen et al. on 840 patients from Japan, USA, and China, the risk of developing prostate cancer in SLCO1B3 (rs4149117, GG) was the highest when compared with SLCO1B3 rs4149117, rs4149117, and rs4149117 with RR = 0.44, 0.37, and 0.07, respectively. Two or five years risk free symptoms (RFS) of patients was also higher with SLCO1B3 (rs4149117, GG) compared to rs4149117. Nonetheless, SLCO1B3 (rs4149117) did not show any significant increase in 10 years RFS of prostate cancer compared to other genotypes [Citation42].
In another study, de novo OATP1B3 expression in prostate cancer was shown to be linked to greater androgen uptake and therefore greater OATP1B3 activity results in the development of androgen deprivation therapy resistance and shorter overall survival in castration-resistant prostate cancer patients [Citation43].
For breast cancer progression, current data on the clinical significance of SLC transporters are mixed. While in one study expression of SLCO1B3 was shown to be associated with tumors of higher histological grade, adverse survival and increased risk of early recurrence [Citation44,Citation45]. In an immunohistological analysis of 102 breast carcinomas, SLCO1B3 expression was inversely correlated with tumor size and was shown to be associated with a decreased risk of recurrence [Citation46]. For ER+ tumors, SLCO1B3 expression was linked to a good prognosis [Citation46]. More data are needed to improve our understanding of the role of polymorphism in these transporters in breast cancer treatment [Citation45]. Interestingly, a cancer-specific type OATP1B3 (Ct-OATP1B3) has also been reported, which lacks 28 amino acids at the N-terminus and has been detected in certain colon and lung tumors [Citation47]. Although Ct-OATP1B3 could potentially serve as an early cancer biomarker, the full-length, liver type OATP1B3 appears to be the predominant form in HCC as well as ovarian, prostate, bladder and breast cancers [Citation31].
4. Pharmacogenetics variations related to changes in drug pharmacokinetics
A wealth of information has been generated over the past decades supporting the thesis that variants in the SLCO1B1 and SLCO1B3 genes are strongly associated with the pharmacokinetic profiles of several drugs and drug classes in oncology, including methotrexate, taxanes, irinotecan, and tyrosine kinase inhibitors. In this section, an update is presented on this rapidly evolving field.
4.1. Methotrexate
Methotrexate (MTX) is an antimetabolite chemotherapeutic drug [Citation48], that is primarily excreted through the kidneys, but also undergoes hepatic uptake and enterohepatic recirculation, both of which contribute to its overall disposition [Citation49]. MTX was among the first anticancer drug substrates of OATPs and is known to be transported into the liver by OATP1B1 and OATP1B3, transporters that have a profound impact on MTX disposition [Citation50–52].
In the past five years, a great deal of attention has been given to the importance of SLCO1B1 variants in predicting MTX clearance and toxicity, and several studies have specifically investigated the influence of OATP1B1 polymorphism on outcome of treatment with MTX. Among the 12 polymorphisms analyzed in 322 Chinese children treated with high dose MTX for acute lymphoblastic leukemia (ALL), the SLCO1B1 rs10841753 variant was correlated significantly with plasma MTX levels at 48 hours, and children with SLCO1B1 rs4149056 CC genotype had a worse outcome compared to children with TT or TC genotypes [Citation53]. In another Chinese study examining the effect of OATP1B1 polymorphism on MTX pharmacokinetics, it was revealed that the OATP1B1-388 G > A SNP reduces MTX clearance, suggesting that patients with SLCO1B1 AG or GG should receive a lower dose of MTX. Although the elimination of MTX is correlated with by creatinine clearance, in this trial, creatinine clearance was not associated with MTX clearance, presumably because values of creatinine clearance were maintained within a narrow range [Citation54]. It is further noteworthy that the observations made in patients with ALL are not necessarily universally applicable to other MTX-sensitive diseases, as SNPs in SLCO1B1 were not substantially linked with MTX clearance in patients with osteosarcoma [Citation55].
In addition to providing a mechanistic explanation for variability in the pharmacokinetics of MTX, genetic variation in the OATP1B1 transporter has also been connection with changes in the MTX toxicity. For example, the SLCO1B1 rs2306283 variant tripled the probability of MTX-induced neurotoxicity in Brazilian ALL pediatric patients who received high dose MTX during consolidation [Citation56]. At the same time, among the fifteen SNPs analyzed by Eldem et al. [Citation57], the rs4149056 CC and rs11045879 variant alleles were found to be linked to worse MTX tolerance in Turkish ALL children who received MTX as maintenance therapy. However, drug tolerance in this trial was determined based on the reported tolerated doses, clinical findings, and adverse events, although a better association between the polymorphism and drug response might be established if drug and metabolite concentrations were examined. Furthermore, the genetic factors involved in delayed MTX elimination in children with ALL were investigated and the proportion of subjects with the SLCO1B1 rs2306283 SNP in the delayed and normal elimination groups was found to be different. In comparison to GG carriers, SLCO1B1 rs2306283 AA carriers had a considerably delayed MTX elimination, which was linked to an elevated risk of oral mucositis, hepatotoxicity, and myelosuppression in the delayed clearance group [Citation58]. A retrospective analysis of data obtained from 63 patients with B cell lymphoma has shown that the SLCO1B1 rs11045879 CT genotype resulted in higher MTX concentrations and the CC genotype increased the risk of developing neutropenia [Citation59].
To explain the association between SLCO1B1 SNPs and methotrexate toxicity, Martinez et al. [Citation60] investigated the mechanism involved in the effect of OATP1B1 polymorphism on MTX elimination. This research has shown that c.388A>G and c.521 T > C variant alleles resulted in impaired OATP1B1 function and caused a buildup of endogenous metabolites, which are known substrates of renal organic anion transporters and may interfere with the OAT1 and OAT3-mediated renal elimination of MTX. These results are consistent with the findings of Schulte et al. [Citation61], where the presence of the SLCO1B1 521 T > C genotype was associated with lower MTX clearance in children and young adults with leukemia or lymphoma. However, in 18 Thai patients who received MTX as part of low-risk gestational trophoblastic neoplasia management SLCO1B1 521 T > C genotype was not shown to be linked to MTX response or toxicity [Citation62].
4.2. Taxanes
Paclitaxel and docetaxel are widely used chemotherapy agents and cabazitaxel has been approved by the FDA to treat hormone-refractory prostate cancer [Citation63]. Paclitaxel and docetaxel, but not cabazitaxel, were found to be substrates of OATP1B1, OATP1B3 [Citation64,Citation65]. Therefore, attempts have been made to assess the impact of SLCO1B1 and SLCO1B1 variants on taxenes therapeutic use.
In a Malaysian study assessing the association between SLCO1B3 polymorphism and docetaxel nonhematological side effects, the SLCO1B3 334 T > G genotype was not found to be linked to docetaxel induced fatigue, nausea, oral mucositis, or vomiting [Citation64]. Similarly, neither docetaxel sensitivity nor the frequency of neutropenia was shown to be correlated with SLCO1B3 (rs11045585) variant [Citation66]. However, a cabazitaxel genetic association study in advanced prostate cancer patients showed that SLCO1B1*15 carriers had a decreased incidence of leukopenia and neutropenia, but no variations in cabazitaxel pharmacokinetics or overall survival were reported. While this study is the largest genetic association research in cabazitaxel-treated patients to date, the limited amount of data in the pharmacokinetic studies and the low minor allele frequencies may have affected the reliability of the findings [Citation67].
4.3. Irinotecan
Irinotecan is a prodrug that is converted to active metabolite 7-ethyl-10-hydroxycamptothecin (SN-38) via hepatic carboxylesterase [Citation68]. OATP1B1 and OATP1B3 have been shown to modulate the disposition of irinotecan and its active metabolite SN-38 in vitro and in vivo [Citation69–71].
Diarrhea is known to be the dose-limiting side effect of irinotecan, and while Han et al. [Citation72] showed that patients with the OATP1B1*15 haplotype had a substantial increase in SN-38 area under the curve, Lu et al. [Citation73] found no significant correlation between the SLCO1B1*15 genotype and the occurrence of delayed diarrhea. Small sample size with a total of 27 patients might contribute to this discrepancy.
4.4. Anthracyclines
Previous in vitro and animal research has found that OATP1B1 and OATP1B3 facilitate anthracycline transport, particularly for doxorubicin [Citation74] and daunorubicin [Citation75], although the role of OATPs in idarubicin transport is still undefined. Recent studies have evaluated the association of SNPs in anthracycline transporter genes with the efficacy and toxicity of idarubicin-based chemotherapy in AML. Although no single or combinations of variations were predictive of complete remission, combinations of rs4149056 CC and ABCB1 (triple variant haplotype, rs1128503) were associated with higher incidence of death. In addition, several combinations of SLCO1B1 and SLC22A16 with ABCB1 SNPs were linked to greater toxicities, including nephrotoxicity and hepatotoxicity, neutropenia, and mucositis [Citation76]. These preliminary observations warrant further validation in larger patient populations in order to confirm their potential clinical significance.
4.5. Tyrosine kinase inhibitors
Tyrosine kinase inhibitors (TKIs) are targeted agents that impair cell singling pathways and induce cellular death by inhibiting tyrosine kinase enzymes [Citation77]. Several studies have demonstrated a contribution of OATP1B1 and OATP1B3 to the cellular uptake of TKIs [Citation78–82] and these studies support the thesis that polymorphic variants in the associated genes may influence the outcome of TKI-based therapy. It has been reported that the absence of SLCO1B1 521 T > C and SLCO1B1*1b resulted in increased regorafenib concentrations compared to other genotypes, and was linked to a higher incidence of regorafenib-related side effects, such as anemia and hepatic injury [Citation83,Citation84]. On the other hand, the SLCO1B1 388A>G genotype was not correlated with the plasma concentration of regorafenib or any of its metabolites; in view of the fact that regorafenib is extensively bound to human plasma proteins, analysis of unbound regorafenib concentrations might be indicated and such studies could be employed in the future to reexamine associations of OATP1B1 variants with regorafenib pharmacokinetics [Citation85]. In the case of imatinib, studies have indicated that imatinib plasma levels were not statistically significantly correlated with the SLCO1B3 (334 T > G and 699 G > A) genotype [Citation86], although associations of this variant may exist in relation to the efficacy of imatinib in the treatment of chronic myeloid leukemia [Citation87]. Conversely, these genotypes have also been linked to a higher likelihood of developing imatinib resistance [Citation88]. In patients with gastrointestinal stromal tumors treated with sunitinib, the presence of the T-allele in SLCO1B3 rs4149117 was shown to be predictive for OS. These results can highlight the beneficial role of administering sunitinib in selected patients on the basis of an individual’s genotype [Citation89].
5. Conclusion
OATP1B1 and OATP1B3 are important members of the SLC superfamily of transporters, and polymorphic variants of these proteins have profound implications for the pharmacokinetic profile of an increasingly large number of oncology drugs (). Although many studies have examined the effects of different SNPs in the SLCO1B1 and SLCO1B3 genes on phenotypes associated with anticancer drugs, several of these reports suffer from limitations that preclude immediate implementation of this knowledge in the clinic. These limitations include small sample sizes, lack of validation cohorts, absence of mechanistic studies that confirm reliability and plausibility of the findings, a relatively small effect size that is only observable in large cohorts of patients, and a general a failure to include patients from various ethnic backgrounds. In addition, differences in the methods employed, the selection of limited variants, and the use of varying treatment protocols employing single-agent or polypharmacy approaches used to evaluate the significance of these genetic associations is so extensive that, at present, the FDA only considers the importance of these transporters in connection with the pharmacotherapy of statins, elagolix, and viloxazine [Citation90] .For these agents, the available evidence from non-clinical in vivo studies and clinical ex vivo studies strongly support the thesis that OATP1B1 and/or OATP1B3 function presents a rate-limiting step in drug elimination, and it is likely that this feature is a prerequisite for drugs to be sensitive to the presence of damaging variants in these transporters in a manner that warrants clinical consideration.
Table 1. Characteristics of each SLCO1B1 and SLCO1B3 single nucleotide polymorphisms (SNPs) discussed in this review.
In order to address in a more reliable and effective manner the degree to which oncology drugs are liable to OATP1B1 and OATP1B3 variants, the implementation of endogenous biomarker tests for these transporters has been proposed. For example, levels of coproporphyrin (CP)-I and CP-III have been shown to be a potentially useful marker for detection of altered function in subjects carrying the OATP1B1 genotype c.521 T > C (OATP1B1-Val174Ala) [Citation91]. It has been suggested that measurement of CP-I levels along with genotyping in a PBPK model can be employed to evaluate the impact of SLCO1B1 genotype, ethnicity, and sex on an individual’s OATP1B1 function to allow for more accurate predictions of drug–drug interactions [Citation92]. Other studies in different ethnic groups have confirmed the sensitivity and specificity of CP-1 as an endogenous probe for phenotyping OATP1B1 [Citation93–96] and similar cases have been built for use of the endogenous biomarkers glycochenodeoxycholate and glycodeoxycholate 3-O-glucuronides (GCDCA-3G) [Citation95,Citation96]. Although endogenous biomarkers are useful tools in prediction of function of OATP1B1 and OATP1B3, the substrate specificity of different variants of these transporters can pose limitations in their use. For instance, the OATP1B1*1b variant is conserved for its transport function toward the uptake of estradiol, estradiol-17β-D-glucuronide, and several statins [Citation97–102]; however, its activity in transporting bromosulfophthalein (BSP) and cholyltaurine is altered. In contrast, OATP1B1*5 is associated with reduced uptake of these estrogens, erythromycin, and several statins, and similar observations have been made for OATP1B3 [Citation101]. It should also be noted that in vitro findings about the uptake activities of OATP1B3 variants are not always in line with findings made in vivo [Citation11,Citation102].
6. Expert opinion
It is expected that, within the next decade, the importance of OATP1B1 and OATP1B3 transporters in the pharmacokinetic and toxicity profile of the classic cytotoxic anticancer drugs will be more clearly defined. However, recent studies suggest that this field of research is equally important for small-molecule targeted drugs and may be an important consideration during the various development stages of new agents. Based on currently available knowledge accumulated over the past decade in the field of OATP1B1 and OATP1B3 genetics, it appears imperative that different aspects of disease, physiology, and drugs of relevance should be evaluated along with an individual’s genetic signature, and that tools such as biomarker levels can be implemented to achieve the most reliable prediction of clinically relevant pharmacodynamic endpoints. Indeed, it is expected that successful identification and development of predictive biomarkers of OATP1B1 and OATP1B3 in the near future will have a profound impact on attempts to further optimize the rational use of cancer chemotherapeutic agents. Such biomarker strategies will allow optimization of rationally designed combinatorial treatments, and open new avenues to enhance the therapeutic window of chemotherapy-based regimens in a routine clinical setting. This in turn would have a significant impact on the morbidity and mortality associated with the usage of drugs in the treatment of diverse malignant diseases.
Article highlights
The polymorphic transporters OATP1B1 and OATP1B3 have important consequences on pharmacokinetic of drugs and treatment efficacy.
The direct contribution of polymorphism in these transporters in drug disposition with emphasis on patient’s physiology, cancer type and drug characteristics are explored in this mini review.
Currently, available knowledge accumulated over the past decade suggests that different aspects of disease, physiology, and drugs of relevance should be evaluated along with an individual’s genetic signature.
Tools such as biomarker levels can be implemented to achieve the most reliable prediction of clinically relevant pharmacodynamic endpoints.
Such biomarker strategies will allow optimization of rationally designed combinatorial treatments, and potentially enhance the therapeutic window of chemotherapy-based regimens in a routine clinical setting.
This box summarizes key points contained in the article.
Declaration of interest
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
Reviewer disclosures
Peer reviewers on this manuscript have no relevant financial or other relationships to disclose.
Additional information
Funding
References
- Evans WE, Relling MV. Moving towards individualized medicine with pharmacogenomics. Nature. 2004;429(6990):464–468.
- Evans WE, McLeod HL. Pharmacogenomics—drug disposition, drug targets, and side effects. N Engl J Med. 2003;348(6):538–549.
- Weinshilboum R. Inheritance and drug response. N Engl J Med. 2003;348(6):529–537.
- Roth M, Obaidat A, Hagenbuch B. OATPs, OATs and OCTs: the organic anion and cation transporters of the SLCO and SLC22A gene superfamilies. BrJ Pharmacol. 2012;165(5):1260–1287.
- Koepsell H, Lips K, Volk C. Polyspecific organic cation transporters: structure, function, physiological roles, and biopharmaceutical implications. Pharm Res. 2007 Jul;24(7):1227–1251.
- Zhang L, Brett CM, Giacomini KM. Role of organic cation transporters in drug absorption and elimination. Annu Rev Pharmacol Toxicol. 1998;38(1):431–460.
- Gründemann D, Gorboulev V, Gambaryan S, et al. Drug excretion mediated by a new prototype of polyspecific transporter. Nature. 1994;372(6506):549–552.
- Schaller L, Lauschke VM. The genetic landscape of the human solute carrier (SLC) transporter superfamily. Hum Genet. 2019;138(11):1359–1377.
- He L, Vasiliou K, Nebert DW. Analysis and update of the human solute carrier (SLC) gene superfamily. Hum Genomics. 2009;3(2):1–12.
- Koepsell H. The SLC22 family with transporters of organic cations, anions and zwitterions. Mol Aspects Med. 2013;34(2–3):413–435.
- Zhou F, Zhu L, Wang K, et al. Recent advance in the pharmacogenomics of human Solute Carrier Transporters (SLCs) in drug disposition. Adv Drug Deliv Rev. 2017 Jul;116:21–36.
- Clarke JD, Cherrington NJ. Genetics or environment in drug transport: the case of organic anion transporting polypeptides and adverse drug reactions. Expert Opin Drug Metab Toxicol. 2012;8(3):349–360.
- Brecht K, Schäfer AM, H. E MZS. Uptake transporters of the SLC21, SLC22A, and SLC15A families in anticancer therapy—modulators of cellular entry or pharmacokinetics? Cancers (Basel). 2020;12(8):2263.
- Nie Y, Yang J, Liu S, et al. Genetic polymorphisms of human hepatic OATPs: functional consequences and effect on drug pharmacokinetics. Xenobiotica. 2019;50(3): 297–317.
- König J, Seithel A, Gradhand U, et al. Pharmacogenomics of human OATP transporters. Naunyn-Schmiedeberg’s Arch Pharmacol. 2006;372(6):432–443.
- Kalliokoski A, Niemi M. Impact of OATP transporters on pharmacokinetics. Br J Pharmacol. 2009 Oct;158(3):693–705.
- Obaidat A, Roth M, Hagenbuch B. The expression and function of organic anion transporting polypeptides in normal tissues and in cancer. Annu Rev Pharmacol Toxicol. 2012;52(1):135–151.
- Clemons M, Goss P. Estrogen and the risk of breast cancer. N Engl J Med. 2001 Jan;344(4):276–285.
- Geisler J, Haynes B, Anker G, et al. Influence of letrozole and anastrozole on total body aromatization and plasma estrogen levels in postmenopausal breast cancer patients evaluated in a randomized, cross-over study. J clin oncol. 2002 Feb;20(3):751–757.
- Geisler J, King N, Anker G, et al. In vivo inhibition of aromatization by exemestane, a novel irreversible aromatase inhibitor, in postmenopausal breast cancer patients. Clin Cancer Res. 1998 Sep;4(9):2089–2093.
- Ramsey LB, Johnson SG, Caudle KE, et al. The clinical pharmacogenetics implementation consortium guideline for SLCO1B1 and simvastatin-induced myopathy: 2014 update. Clin Pharmacol Ther. 2014 Oct;96(4):423–428.
- Gregory BJ, Chen SM, Murphy MA, et al. Impact of the OATP1B1 c.521T>C single nucleotide polymorphism on the pharmacokinetics of exemestane in healthy post-menopausal female volunteers. J Clin Pharm Ther. 2017;42(5):547–553.
- Boivin -A-A, Cardinal H, Barama A, et al. Organic anion transporting polypeptide 1B1 (OATP1B1) and OATP1B3: genetic variability and haplotype analysis in white Canadians. Drug Metab Pharmacokinet. 2010;25(5):508–515.
- Giacomini KM, et al. Membrane transporters in drug development. Nat Rev Drug Discov. 2010;9(3):215–236.
- Dudenkov TM, Ingle JN, Buzdar AU, et al. SLCO1B1 polymorphisms and plasma estrone conjugates in postmenopausal women with ER+ breast cancer: genome-wide association studies of the estrone pathway. Breast Cancer Res Treat. 2017 Jul;164(1):189–199.
- Dempsey JM, Kidwell KM, Gersch CL, et al. Effects of SLCO1B1 polymorphisms on plasma estrogen concentrations in women with breast cancer receiving aromatase inhibitors exemestane and letrozole. Pharmacogenomics. 2019 Jun;20(8):571–580.
- Tryfonidis K, Basaran G, Bogaerts J, et al. A European organisation for research and treatment of cancer randomized, double-blind, placebo-controlled, multicentre phase II trial of anastrozole in combination with gefitinib or placebo in hormone receptor-positive advanced breast cancer (NCT00066378. Eur J Cancer. 2016 Jan;53:144–154.
- Pu Z, Zhang X, Chen Q, et al. Establishment of an expression platform of OATP1B1 388GG and 521CC genetic polymorphism and the therapeutic effect of tamoxifen in MCF-7 cells. Oncol Rep. 2015;33(5):2420–2428.
- Gao CM, Pu Z, He C, et al. Effect of OATP1B1 genetic polymorphism on the uptake of tamoxifen and its metabolite, endoxifen. Oncol Rep. 2017;38(2):1124–1132.
- Wang T, Zhou Y, Cao G. Pharmacogenetics of tamoxifen therapy in Asian populations: from genetic polymorphism to clinical outcomes. Eur J Clin Pharmacol. 2021 Aug;77(8):1095–1111.
- Alonso-Peña M, Espinosa-Escudero RA, Soto-Muñiz M, et al. Role of transportome in the pharmacogenomics of hepatocellular carcinoma and hepatobiliary cancer. Pharmacogenomics. 2019 Aug;20(13):957–970.
- Ang SA, Nugroho AK, Sadewa AH, et al. The SLCO1B1*15 haplotype associated with lower clinical outcome in Indonesian tuberculosis patients. J Med Sci. 2018;50:50–59.
- Zhong Z, Wu H, Li B, et al. Analysis of SLCO1B1 and APOE genetic polymorphisms in a large ethnic Hakka population in southern China. J Clin Lab Anal. 2018 Jul;32(6):e22408–e22408.
- Thakkar N, Lockhart AC, Lee W. Role of organic anion-transporting polypeptides (OATPs) in cancer therapy. AAPS J. 2015 May;17(3):535–545.
- Al-Abdulla R, Perez-Silva L, Abete L, et al. Unraveling ‘the cancer genome atlas’ information on the role of SLC transporters in anticancer drug uptake. Expert Rev Clin Pharmacol. 2019 Apr;12(4):329–341.
- Chen S, Li K, Jiang J, et al. Low expression of organic anion-transporting polypeptide 1B3 predicts a poor prognosis in hepatocellular carcinoma. World J Surg Oncol. 2020;18(1):127.
- Gong IY, Kim RB. Impact of genetic variation in OATP transporters to drug disposition and response. Drug Metab Pharmacokinet. 2013;28(1):4–18.
- Niebudek K, Balcerczak E, Mirowski M, et al. Association of ABCB1 T-129C polymorphism and multiple myeloma risk in Polish population. Pol J Pathol. 2018;69(4):405–409.
- Zhao H, Jiang J, Wang M, et al. Genome-wide identification of m6a-associated single-nucleotide polymorphisms in colorectal cancer. Pharmgenomics Pers Med. 2021 Jul;14:887–892.
- Huang L, Zhang T, Xie C, et al. SLCO1B1 and SLC19A1 gene variants and irinotecan-induced rapid response and survival: a prospective multicenter pharmacogenetics study of metastatic colorectal cancer. PLoS One. 2013;8(10):e77223.
- Moradi-Marjaneh R, Khazaei M, Seifi S, et al. Pharmacogenetics of anticancer drug sensitivity and toxicity in colorectal cancer. Curr Pharm Des. 2018;24(23):2710–2718.
- Shen Q, Liang Y, Li H, et al. Association between SLCO1B3 gene polymorphism and prostate cancer risk: a meta-analytic study. Bangladesh J Pharmacol. 2018 Mar 25;13(1 SE–Meta–Analysis):98–105.
- Sissung TM, Ley AM, Strope JD, et al. Differential expression of OATP1B3 mediates unconjugated testosterone influx. Mol Cancer Res. 2017 Aug;15(8):1096–1105.
- Zhang H, Rakha EA, Ball GR, et al. The proteins FABP7 and OATP2 are associated with the basal phenotype and patient outcome in human breast cancer. Breast Cancer Res Treat. 2010;121(1):41–51.
- Sutherland R, Meeson A, Lowes S. Solute transporters and malignancy: establishing the role of uptake transporters in breast cancer and breast cancer metastasis. Cancer Metastasis Rev. 2020 Sep;39(3):919–932.
- Muto M, Onogawa T, Suzuki T, et al. Human liver‐specific organic anion transporter‐2 is a potent prognostic factor for human breast carcinoma. Cancer Sci. 2007;98(10):1570–1576.
- Sun Y, Furihata T, Ishii S, et al. Unique expression features of cancer-type organic anion transporting polypeptide 1B3 mRNA expression in human colon and lung cancers. Clin Transl Med. 2014;3(1):1–12.
- Cronstein BN. THE MECHANISM OF ACTION OF METHOTREXATE. Rheum Dis Clin North Am. 1997 Nov;23(4):739–755.
- Shen DD, Azarnoff DL. Clinical pharmacokinetics of methotrexate. Clin Pharmacokinet. 1978;3(1):1–13.
- Durmus S, Lozano-Mena G, Van Esch A, et al. Preclinical mouse models to study human OATP1B1- and OATP1B3-mediated drug–drug interactions in vivo. Mol Pharm. 2015;12(12):4259–4269.
- Ramsey LB, et al. Rare versus common variants in pharmacogenetics: SLCO1B1 variation and methotrexate disposition. Genome Res. 2012 Jan;22(1):1–8.
- Ramsey LB, Panetta JC, Smith C, et al. Genome-wide study of methotrexate clearance replicates SLCO1B1. Blood. 2013;121(6):898–904.
- Liu SG, Gao C, Zhang R-D, et al. Polymorphisms in methotrexate transporters and their relationship to plasma methotrexate levels, toxicity of high-dose methotrexate, and outcome of pediatric acute lymphoblastic leukemia. Oncotarget. 2017;8(23):37761–37772.
- Wang Z, Zhang N, Chen C, et al. Influence of the OATP polymorphism on the population pharmacokinetics of methotrexate in Chinese patients. Curr Drug Metab. 2019;20(7):592–600.
- Lui G, Treluyer J-M, Fresneau B, et al. A pharmacokinetic and pharmacogenetic analysis of osteosarcoma patients treated with high-dose methotrexate: data from the OS2006/sarcoma-09 trial. Pharmacogenomics J Clin Pharmacol. 2018;58(12):1541–1549.
- de Carvalho DC, Wanderley AV, Dos Santos AMR, et al. Pharmacogenomics and variations in the risk of toxicity during the consolidation/maintenance phases of the treatment of pediatric B-cell leukemia patients from an admixed population in the Brazilian Amazon. Leuk Res. 2018;74(August):10–13.
- Eldem I, Yavuz D, Cumaoğullari Ö, et al. SLCO1B1 polymorphisms are associated with drug intolerance in childhood leukemia maintenance therapy. J Pediatr Hematol Oncol. 2018;40(5):e289–e294.
- Cheng Y, Chen M-H, Zhuang Q, et al. Genetic factors involved in delayed methotrexate elimination in children with acute lymphoblastic leukemia. Pediatr Blood Cancer. 2021;68(5):1–12.
- Huang S, Jin L, Yang J, et al. Study on relationships of tumor status and gene polymorphism with blood concentration of MTX and toxicities in 63 pediatric mature B cell lymphoma in Chinese population. Technol Cancer Res Treat. 2021;20:1–8.
- Martinez D, Muhrez K, Woillard J-B, et al. Endogenous metabolites-mediated communication between OAT1/OAT3 and OATP1B1 may explain the association between SLCO1B1 SNPs and methotrexate toxicity. Clin Pharmacol Ther. 2018;104(4):687–698.
- Schulte RR, Choi L, Utreja N, et al. Effect of SLCO1B1 polymorphisms on high-dose methotrexate clearance in children and young adults with leukemia and lymphoblastic lymphoma. iClin Transl Sci. 2021;14(1):343–353.
- Srisuttayasathien M, Areepium N, Lertkhachonsuk R. ABCB1 and SLCO1B1 gene polymorphisms predict methotrexate-resistant for low-risk gestational trophoblastic neoplasia. Per Med. 2021;18(3):107–114.
- De Bono JS, Oudard S, Ozguroglu M, et al. Prednisone plus cabazitaxel or mitoxantrone for metastatic castration-resistant prostate cancer progressing after docetaxel treatment: a randomised open-label trial. Lancet. 2010;376(9747):1147–1154.
- Walton GD, Hu S, Gui C, et al. Influence of drug formulation on OATP1B-mediated transport of paclitaxel. Cancer Res. 2014;74(11):3137–3145.
- De Graan AM, Lancaster CS, Obaidat A, et al. Influence of polymorphic OATP1B-Type carriers on the disposition of docetaxel. Clin Cancer Res. 2012;18(16):4433–4440.
- Sone K, Oguri T, Uemura T, et al. Genetic variation in the ATP binding cassette transporter ABCC10 is associated with neutropenia for docetaxel in Japanese lung cancer patients cohort. BMC Cancer. 2019;19(1):1–9.
- Belderbos BPS, de With M, Singh RK, et al. The influence of single-nucleotide polymorphisms on overall survival and toxicity in cabazitaxel-treated patients with metastatic castration-resistant prostate cancer. Cancer Chemother Pharmacol. 2020;85(3):547–553.
- De Man FM, Goey AKL, Van Schaik RHN, et al. Individualization of irinotecan treatment: a review of pharmacokinetics, pharmacodynamics, and pharmacogenetics. Clin Pharmacokinet. 2018;57:1229–1254.
- Nozawa T, Minami H, Sugiura S, et al. Role of organic anion transporter OATP1B1 (OATP-C) in hepatic uptake of irinotecan and its active metabolite, 7-ethyl-10-hydroxycamptothecin: in vitro evidence and effect of single nucleotide polymorphisms. Drug Metab Dispos. 2005;33(3):434–439.
- Teft WA, Welch S, Lenehan J, et al. OATP1B1 and tumour OATP1B3 modulate exposure, toxicity, and survival after irinotecan-based chemotherapy. Br J Cancer. 2015;112(5):857–865.
- Iusuf D, Ludwig M, Elbatsh A, et al. OATP1A/1B transporters affect irinotecan and SN-38 pharmacokinetics and carboxylesterase expression in knockout and humanized transgenic Mice. Mol Cancer Ther. 2014;13(2):492–503.
- Han JY, Lim H-S, Shin ES, et al. Influence of the organic anion-transporting polypeptide 1B1 (OATP1B1) polymorphisms on irinotecan-pharmacokinetics and clinical outcome of patients with advanced non-small cell lung cancer. Lung Cancer. 2008;59(1):69–75.
- Lu H, Qin J, Han N, et al. Banxia xiexin decoction is effective to prevent and control irinotecan-induced delayed diarrhea in recurrent small cell lung cancer. Integr Cancer Ther. 2018;17(4):1109–1114.
- Lee HH, Leake BF, Kim RB, et al. Contribution of organic anion-transporting polypeptides 1A/1B to doxorubicin uptake and clearance. Mol Pharmacol Mol Pharmacol. 2017;91:14–24.
- Drenberg CD, Paugh SW, Pounds SB, et al. Inherited variation in OATP1B1 is associated with treatment outcome in acute myeloid leukemia. Clin Pharmacol Ther. 2016;99(6):651–660.
- Megías-Vericat JE, Martínez-Cuadrón D, Herrero MJ, et al. Impact of combinations of single-nucleotide polymorphisms of anthracycline transporter genes upon the efficacy and toxicity of induction chemotherapy in acute myeloid leukemia. Leuk Lymphoma. 2021;62(3):659–668.
- Jiao Q, Bi L, Ren Y, et al. Advances in studies of tyrosine kinase inhibitors and their acquired resistance. Mol Cancer. 2018;17(36)
- Hu S, Franke RM, Filipski KK, et al. Interaction of imatinib with human organic ion carriers. Clin Cancer Res. 2008;14(10):3141–3148.
- Zimmerman EI, Hu S, Roberts JL, et al. Contribution of OATP1B1 and OATP1B3 to the disposition of sorafenib and sorafenib-glucuronide. Clin Cancer Res. 2013;19(6):1458–1466.
- Vasilyeva A, Durmus S, Li L, et al. Hepatocellular shuttling and recirculation of sorafenib-glucuronide is dependent on Abcc2, Abcc3, and Oatp1a/1b. Cancer Res. 2015;75(13):2729–2736.
- Bauer M, Matsuda A, Wulkersdorfer B, et al. Influence of OATPs on hepatic disposition of erlotinib measured with positron emission tomography. Clin Pharmacol Ther. 2018;104(1):139–147.
- Ohya H, Shibayama Y, Ogura J, et al. Regorafenib is transported by the organic anion transporter 1B1 and the multidrug resistance protein 2. Biol Pharm Bull. 2015;38(4):582–586.
- Maeda A, Ando H, Ura T, et al. Association between ABCG2 and SLCO1B1 polymorphisms and adverse drug reactions to regorafenib: a preliminary study. Int J Clin Pharmacol Ther. 2017;55(5):409–415.
- Maeda A, Irie K, Ando H, et al. Associations among regorafenib concentrations, severe adverse reactions, and ABCG2 and OATP1B1 polymorphisms. Cancer Chemother Pharmacol. 2019;83(1):107–113.
- Kobayashi K, Sugiyama E, Shinozaki E, et al. Associations among plasma concentrations of regorafenib and its metabolites, adverse events, and ABCG2 polymorphisms in patients with metastatic colorectal cancers. Cancer Chemother Pharmacol. 2021;87(6):767–777.
- Wang Q, Jiang Z-P, Zeng J, et al. Effects of trough concentration and solute carrier polymorphisms on imatinib efficacy in Chinese patients with chronic myeloid leukemia. J Pharm Pharm Sci. 2020;23(1):1–9.
- Omran MM, Abdelfattah R, Moussa HS, et al. Association of the trough, peak/trough ratio of imatinib, pyridine–N-oxide imatinib and ABCG2 SNPs 34 G>A and SLCO1B3 334 T>G with imatinib response in Egyptian chronic myeloid leukemia patients. Front Oncol. 2020;10(August):1–12.
- Mohammadi F, Rostami G, Assad D, et al. Association of SLC22A1,SLCO1B3 drug transporter polymorphisms and smoking with disease risk and cytogenetic response to imatinib in patients with chronic myeloid leukemia. Lab Med. 2021;52(6):584–596.
- Kloth JSL, Verboom MC, Swen JJ, et al. Genetic polymorphisms as predictive biomarker of survival in patients with gastrointestinal stromal tumors treated with sunitinib. Pharmacogenomics J. 2018 Jan;18(1):49–55.
- “Table of pharmacogenomic biomarkers in drug labeling.”
- Giacomini KM, Yee SW, Ratain MJ, et al. Pharmacogenomics and patient care: one size does not fit all. Sci Transl Med. 2012 Sep;4(153): 153-ps18.
- Takita H, Barnett S, Zhang Y, et al. PBPK model of coproporphyrin i: evaluation of the impact of SLCO1B1 genotype, ethnicity, and sex on its inter-individual variability. CPT Pharmacometrics Syst Pharmacol. 2021 Feb;10(2):137–147.
- Suzuki Y, Sasamoto Y, Koyama T, et al. Relationship of hemoglobin level and plasma coproporphyrin-I concentrations as an endogenous probe for phenotyping OATP1B. iClin Transl Sci. 2021 Jul;14(4):1403–1411.
- Suzuki Y, Sasamoto Y, Koyama T, et al. Substantially increased plasma coproporphyrin-i concentrations associated with OATP1B1*15 Allele in Japanese general population. iClin Transl Sci. 2021 Jan;14(1):382–388.
- Neuvonen M, et al. Identification of glycochenodeoxycholate 3-O-glucuronide and glycodeoxycholate 3-O-glucuronide as highly sensitive and specific OATP1B1 biomarkers. Clin Pharmacol Ther. 2021 Mar;109(3):646–657.
- Takehara I, Terashima H, Nakayama T, et al. Investigation of glycochenodeoxycholate sulfate and chenodeoxycholate glucuronide as surrogate endogenous probes for drug interaction studies of OATP1B1 and OATP1B3 in healthy Japanese volunteers. Pharm Res. 2017 Aug;34(8):1601–1614.
- Kameyama Y, Yamashita K, Kobayashi K, et al. Functional characterization of SLCO1B1 (OATP-C) variants, SLCO1B1*5, SLCO1B1*15 and SLCO1B1*15+C1007G, by using transient expression systems of HeLa and HEK293 cells. Pharmacogenet Genomics. 2005 Jul;15(7):513–522.
- Michalski C, Cui Y, Nies AT, et al. A naturally occurring mutation in the SLC21A6Gene causing impaired membrane localization of the hepatocyte uptake transporter. J Biol Chem. 2002;277(45):43058–43063.
- Nozawa T, Nakajima M, Tamai I, et al. Genetic polymorphisms of human organic anion transporters OATP-C (SLC21A6) and OATP-B (SLC21A9): allele frequencies in the Japanese population and functional analysis. J Pharmacol Exp Ther. 2002;302(2):804–813.
- Tirona RG, Leake BF, Merino G, et al. Polymorphisms in OATP-C: identification of multiple allelic variants associated with altered transport activity among European-and African-Americans. J Biol Chem. 2001;276(38):35669–35675.
- Letschert K, Keppler D, König J. Mutations in the SLCO1B3 gene affecting the substrate specificity of the hepatocellular uptake transporter OATP1B3 (OATP8). Pharmacogenet Genomics. 2004;14(7):441–452.
- Schwarz UI, Meyer Zu Schwabedissen HE, Tirona RG, et al., Identification of novel functional organic anion-transporting polypeptide 1B3 (OATP1B3) polymorphisms and assessment of substrate specificity. Pharmacogenet Genomics. 2011;21(3): 103.