?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
Metal oxide nanoparticles are widely used in both consumer products and medical applications, but the knowledge regarding exposure-related health effects is limited. However, it is challenging to investigate nanoparticle interaction processes with biological systems. The overall aim of this project was to improve the possibility to predict exposure-related health effects of metal oxide nanoparticles through interdisciplinary collaboration by combining workflows from the pharmaceutical industry, nanomaterial sciences, and occupational medicine. Specific aims were to investigate nanoparticle-protein interactions and possible adverse immune reactions. Four different metal oxide nanoparticles; CeOx nanocrystals with 5% or 14% Gd, Co3O4, and Fe2O3, were characterized by dynamic light scattering and high-resolution transmission electron microscopy. Nanoparticle-binding proteins were identified and screened for HLA-binding peptides in silico. Monocyte interaction with nanoparticle–protein complexes was assessed in vitro. Herein, for the first time, immunogenic properties of nanoparticle-binding proteins have been characterized. The present study indicates that especially Co3O4-protein complexes can induce both ‘danger signals’, verified by the production of inflammatory cytokines and simultaneously bind autologous proteins, which can be presented as immunogenic epitopes by MHC class II. The clinical relevance of these findings should be further evaluated to investigate the role of metal oxide nanoparticles in the development of autoimmune disease. The general workflow identified experimental difficulties, such as nanoparticle aggregate formation and a lack of protein-free buffers suitable for particle characterization, protein analyses, as well as for cell studies. This confirms the importance of future interdisciplinary collaborations.
Introduction
Metal oxide nanoparticles (MONPs) have a broad spectrum of applications and represent many of today’s commercially produced nanomaterials (Abo-Zeid and Williams Citation2020; Djurišić et al. Citation2015). MONPs, including Fe2O3 and Co3O4, are used as catalysts (Patil et al. Citation2004), as anode materials in lithium batteries in everyday objects, such as telephones and computers, and in sensors detecting pollutants or biomolecules (Li, Xu, and Chen Citation2005). Medical applications include the use of MONPs, especially iron oxide (Nune et al. Citation2009; Wang et al. Citation2014, Citation2015), as a contrast agent in magnetic resonance imaging (MRI) because of their great magnetic properties and that iron is naturally occurring in vivo. Nanoparticles (NPs) of Gadolinium oxide have been investigated for MR contrast enhancement (Ahrén et al. Citation2010, Citation2012; Petoral et al. Citation2009). Cerium oxide NPs with integrated gadolinium are also being pursued as possible contrast agents (Eriksson et al. Citation2018). Thus, exposure to these materials happens either unintentionally, when emerging from the environment, or intentionally when MONPs are used in medical practice.
The unique physical and chemical properties of MONPs compared to their bulk counterparts not only provides new application opportunities but may pose new health risks as well. For instance, studies have shown that various MONPs can induce both acute and chronic inflammation in murine models following inhalation (Cho et al. Citation2010; Sisler et al. Citation2015). Nano-specific toxicity is often ascribed to the increased surface area to volume ratio of the MONPs, which results in higher surface reactivity and consequently triggers the production of reactive oxygen species (ROS) by cells causing oxidative stress (Cappellini et al. Citation2018; Choi and Hu Citation2008; Mirzaei et al. Citation2018). Another important aspect influencing MONP-induced toxicity is the size-related property of them to translocate from lungs or gastrointestinal tract resulting in final residence in various organs and tissues (Buzea, Pacheco, and Robbie Citation2007; Guttenberg et al. Citation2016).
A central aspect of MONP toxicology is to investigate the interaction of MONPs with cells of the immune system. One possibility is that MONPs may be recognized by toll-like receptors (TLRs) shaping both adaptive and innate immune responses. Recently, effects of MONPs on TLR-mRNAs in human monocytes have been suggested (Vasilichin et al. Citation2020). Monocytes are key players and primary initiators of antigen-specific immune responses in acquired immunity (Jakubzick, Randolph, and Henson Citation2017). They act as phagocytizing cells, process antigen, modulate immune responses by production of cytokines and/or chemokines, differentiate into dendritic cells or tissue-associated macrophages, and activate T lymphocytes via major histocompatibility complex (MHC) class II antigen and presentation (Wong et al. Citation2012). Monocyte–MONP interactions are initially determined by the cell’s ability to recognize and take up the MONPs. In a previous paper, we showed that noncoated gadolinium oxide NPs induce ROS production from neutrophils, while biocompatible coating of these MONPs minimizes ROS induction (Abrikossova et al. Citation2012). In biological fluids, such as blood, biomolecules adsorb to the MONP surface leading to formation of a protein corona (Deng et al. Citation2009). This new biological identity impacts the MONPs biological fate and immune responses (Khang et al. Citation2014). For example, recent studies have shown that corona formation and its composition affect cytokine release and apoptosis in monocyte cell lines (Dai et al. Citation2017). Also, different macrophage internalization pathways are initiated depending on which proteins adhere to the NPs (Yan et al. Citation2013).
It appears that most studies of MONP immunotoxicity either look past the possible impact of the protein corona (Chattopadhyay et al. Citation2015; Dankers et al. Citation2018; Giovanni et al. Citation2015; Grosse, Stenvik, and Nilsen Citation2016; Horev-Azaria et al. Citation2013; Hussain et al. Citation2012; Mirzaei et al. Citation2018) or uses fetal bovine serum for corona formation without protein identification (Boyles et al. Citation2015; Dai et al. Citation2017; Gonnissen et al. Citation2016). A noteworthy study by Mirshafiee et al. (Citation2016) showed differential macrophage uptake of silica NPs, which was dependent on whether coronas were formed from human plasma or human serum. Thus, selecting a biologically relevant milieu is fundamental in order to evaluate human relevance. In toxicity evaluations of NPs, it is of main importance to characterize the NPs in use, by means of elemental composition and size distribution. This will enable comparison between studies as well as increase the understanding of the behavior of the NPs during present experimental conditions (Murdock et al. Citation2008; Warheit Citation2008). Moreover, it can be used to assess how NP characteristics affect protein binding and thus the biological identity of the NPs (Lundqvist et al. Citation2008; Lynch et al. Citation2007).
In the present study, the overall aim was to improve the possibility to predict exposure-related health effects of MONPs through interdisciplinary collaboration by combining workflows from the pharmaceutical industry, nanomaterial sciences, and occupational medicine. Specific aims were to investigate NP–protein interactions and possible adverse immune reactions. In this workflow, four different MONPs were characterized and proteins associated to each MONP type after exposure to human plasma were identified. Potential immunogenicity of the NP-binding proteins was analyzed further using an in silico model designed for evaluating immunogenicity of biopharmaceuticals. Only endogenous proteins were candidates for this screening; thus, the relative risk for autoimmune disease was the key question. The effect of MONP–protein complexes on peripheral white blood-cell function, including cytokine release, was investigated using human primary peripheral blood mononuclear cells (PBMCs) from healthy donors.
Material and methods
Metal oxide nanoparticles
Commercial cobalt (II, III) oxide <50 nm (637025-25 G, Co3O4) and iron(III) oxide <50 nm (544884-5 G, Fe2O3) NPs were acquired from Sigma Aldrich (St. Louis, MI, USA). Particle size had been determined by the manufacturer using transmission electron microscopy (TEM). Cerium oxide nanocrystals with 5% or 14% gadolinium (CeOx:Gd5% <10 nm and CeOx:Gd14% <10 nm) were synthesized by wet-chemical synthesis and characterized by DLS in Milli-Q H2O and high-resolution transmission electron microscopy by our team at Molecular Surface Physics & Nanoscience, IFM, Linköping University. Co3O4 and Fe2O3 are common occupational contaminants whereas the CeOx:Gd particles are candidates for medical applications, .
Table 1. The four investigated particle types.
Nanoparticle characterization
For DLS measurements, MONPs were dispersed in Milli-Q H2O, 50% RPMI1640 medium in Milli-Q H2O, or pure RPMI1640 medium, sonicated for 10 minutes and temperature stabilized in a 22°C thermostat bath. The samples were diluted until a signal intensity of about 100 kHz was reached. Measurements were performed at least three times at 22°C, for 60 seconds with an ALV/DLS/SLS-5022F system (ALV-GmbH, Langen Germany) equipped with a 632.8 nm HeNe laser operating at a 90° scattering angle. The samples were polydisperse (polydispersity indexes were above 0.3), therefore, the autocorrelation function was analyzed using contin fit. TEM samples were prepared as follows. Dilute samples of MONPs (MilliQ water for CeOx:Gd5% and CeOx:Gd14% and in ethanol for Co3O4 and Fe2O3) were sonicated for a minimum of 30 minutes and subsequently drop casted onto lacey carbon films supported by a cupper grid. TEM imaging was performed on a FEI TECNAI G2 TF20 UT equipped with a field emission gun and operating at an acceleration voltage of 200 kV. Images were analyzed using DigitalMicrograph 3.21.1374.0 (Gatan Inc.) and ImageJ 1.52i (National Institute of Health, USA).
Blood sampling
Venous blood from healthy adult volunteers was collected into sodium citrate- and sodium heparin tubes for MONP incubation and isolation of PBMCs, respectively. Approximately 15 mL blood was collected from each donor by licensed personnel. All blood donors signed a written consent in which they received information regarding blood donation for research purposes. The use and handling of blood followed Swetox and the Karolinska Institute guidelines.
Nanoparticle/plasma protein incubation prior to identification of adherent proteins
Plasma was recovered from whole blood by centrifugation at 800 g for 10 minutes and stored at −80°C until incubation with MONPs. MONP/plasma protein incubations were essentially performed as previously described by Deng et al. (Citation2009). Modifications included protein-free RPMI1640 medium and the centrifugation step of plasma was omitted to prevent protein structure alterations that may impact the binding properties. In addition, to prevent unspecific protein interactions and facilitate drying of the protein pellet prior to 2DGE, protein precipitation wash buffer was used; dithiothreitol (DTT, Sigma-Aldrich) 20 mM/-acetone.
MONPs were, as described above, dispersed in protein-free RPMI1640 medium (Lonza, Basel, Switzerland) to a final concentration of 2 mg/mL before 1-hour in vitro incubation with 1% autologous plasma (∽600 μg protein/mL) at room temperature. Controls, without MONPs, were prepared to ensure that there was no protein precipitation. Following incubation, samples were centrifuged 40 minutes at 50 000 g at 4°C. The supernatant, containing unbound proteins, was discarded and pellets, consisting of metal oxide NP–protein complexes (MONPPC), were washed using a dithiothreitol (DTT, Sigma-Aldrich) 20 mM/-acetone buffer before further centrifugation for 10 minutes at 50 000 g at 4°C. The supernatant was discarded, and pellets were allowed to air dry prior to protein analyses.
Protein identification by two-dimensional gel electrophoresis (2DGE) and matrix-assisted laser/desorption time-of-flight mass spectrometry (MALDI-TOF MS)
Corona proteins were separated using 2DGE in a setup previously described by Karlsson et al. (Citation2005), essentially according to Görg et al. (Citation2000). Briefly, MONPPC were reconstituted in sample solution consisting of 9 M urea, 65 mM DTT, 2% pharmalyte (GE Healthcare), 4% CHAPS (Sigma Aldrich) and 1% bromophenol blue, incubated 1 hour and centrifuged for 30 minutes at 50 000 g at 4°C. Then, 50 μL of the supernatant comprising denatured proteins was mixed with 300 μL rehydration solution containing 8 M urea, 19 mM DTT, 4% CHAPS, 5.5 mM Orange G (Sigma Aldrich) and 0.5% IPG buffer 3–10 NL (GE Healthcare). The first dimension was performed by in-gel rehydration on pH 3–10 nonlinear IPG strips (GE Healthcare) using an Ettan IPGphor 3 (GE Healthcare) and the following protocol: 30 V for 12 hours, 200 V for 1 hour, 500 V for 1 hour, 1000 V until 1000 Vhr passed, 8000 V until 3975 Vhr passed and 8000 V until 48 000 Vhr passed. Then, proteins were transferred to homogenous home-cast polyacrylamide gels (14% T, 1.5% C) on a Multiphor II (Ge Healthcare, Uppsala, Sweden), which ran at 30 mA, 40–800 V, and 10°C overnight. Proteins were silver stained according to Shevchenko et al. (Citation1996) with minor modifications and visualized using a VersaDoc imaging system 4000 MP from Bio-Rad Laboratories (Hercules, CA, USA). Dominating protein spots were excised from the gel and destained according to Ghafouri et al. (Citation2007). Then, proteins were subjected to in-gel trypsin digestion (Promega corporation, Madison, WA, USA) as described by Karlsson et al. (Citation2005). Tryptic peptides were mixed 1:1 with 2,5-dihydroxybenzoic acid in 70% acetonitrile/0.3% trifluoroacetic acid (20 mg/mL) and identified using a MALDI-TOF MS UltrafleXtreme (Bruker Daltonics, Bremen Germany). Spectra were acquired through 30 000 shots in the mass range 700–3500 kDa and processed with the FlexAnalysis software version 3.4 (Bruker Daltonics). A peptide calibration standard was employed for external calibration and known trypsin autolysis peaks (m/z 842.5100 and 2211.1046) were used during internal calibration. Peptide masses (major peaks) obtained from MALDI-TOF MS analysis were submitted for database search using MS-FIT as search engine. Restrictions were human species, mass tolerance <50 ppm in most searches, trypsin as digestion agent with maximum one missed cleavage, and carbamidomethylation of cysteines was chosen as a fixed modification. Hits were evaluated according to pI, molecular weight, dominating peaks in the spectra, and map matching.
Protein identification by nanoflow liquid chromatography coupled to tandem mass spectrometry (nLC-MS/MS)
Identities of corona proteins were confirmed with a LTQ Orbitrap Velos Pro (Thermo Fisher Scientific, MA, USA). Following 1 hour plasma incubation, the MONPPC and one control without MONPs were reconstituted in 450 μL 6 M urea, incubated for 30 minutes and centrifuged at 50 000 g for 30 minutes at 4°C. The supernatant containing denatured proteins was collected and run through a 3 kDa cutoff spin filter column (Amicon Ultra 3 K device; Merck-Millipore, Germany) to remove urea. Samples were spun at 150 g for 20 minutes, reduced with 0.25 mM DTT in 25 mM ammonium bicarbonate for 15 minutes at 4°C, alkylated with 0.75 mM iodoacetamide in 25 mM ammonium bicarbonate for 15 minutes at 4°C and washed three times with 25 mM ammonium bicarbonate. Spin columns were inverted to collect washed proteins, which were then subjected to tryptic digestion (1:25, trypsin:sample protein by weight). To reduce autocatalytic activity, samples were kept on ice 45 minutes before incubation overnight at 37°C. Resulting peptides were dried using a SpeedVac vacuum system (Savant, Farmingdale, NY) and reconstituted in 0.1% formic acid in water. A total protein concentration of 250 ng was loaded into the nanoflow HPLC system (Easy-nLC; Thermo Fisher Scientific, Massachusetts, USA). A 100 mm*0.75 μm C18 column (Agilent Technologies, Santa Clara, CA, USA) was used for reverse-phase separation of peptides eluted with a linear increase of 2% to 40% acetonitrile for 90 minutes. MaxQuant v1.5.12 (Max Planck Institute of Biochemistry, Germany) was utilized for spectra analysis and searches were made in the human protein database (Swiss-Prot, downloaded 2018-04-25) with a mass tolerance of 4.5 ppm for MS and 20 ppm for MS/MS. Modifications were trypsin as digestion agent and a maximum of 1 missed cleavage.
In vitro evaluation of immunological effects
Endotoxin assessment
Since endotoxins may interfere with the functional immunoassays used, potential presence of endotoxins in the sample preparations were assessed as follows. The endotoxin content of the four different MONP dispersions (without plasma proteins) was assessed using a chromogenic Limulus Amebocyte Lysate (LAL) endpoint assay (Hycult Biotech, Uden, the Netherlands) according to the manufacturer’s instructions with some modifications. The endotoxin content of the four different MONP dispersions was analyzed in relation to the lowest concentration (0.04 EU/mL) of the endotoxin reference for 40 minutes.
Preparation of peripheral blood mononuclear cell culture
Experiments on human monocytes were performed at former Swetox, a subsidiary of the Karolinska Institute between 2014 and 2018. Whole blood samples were collected from three healthy volunteers and PBMCs were isolated by Lymphoprep density gradient centrifugation (Fresenius Kabi Norge AS, Oslo, Norway). Isolated PBMCs were washed twice using phosphate-buffered saline containing 2% human AB + serum (Karolinska University Laboratory, Stockholm, Sweden) for 10 minutes at 300 g. Hereafter, cells were resuspended in complete medium; RPMI1640 (Thermo Fisher scientific, Waltham, MA, USA) supplemented with 100 U/mL penicillin-streptomycin (Thermo Fisher), 2 mM L-glutamine (Thermo Fisher), and 10% human AB + serum (Karolinska University Laboratory), transferred onto 48-well plates (2106 cells/mL, 500 μL/well) and cultured overnight in 37°C, 5% CO2, and 97% humidity before incubation with MONPPC.
Nanoparticle–plasma protein complex/cell culture incubation
MONPs were dispersed in incomplete medium; RPMI1640 (Thermo Fisher), without AB + serum, supplemented with 100 U/mL penicillin–streptomycin (Thermo Fisher), and 2 mM L-glutamine (Thermo Fisher) to a final concentration of 2 mg/mL, followed by 1 hour in vitro incubation with 1% autologous plasma (∼600 µg protein/mL), in separate sets from each individual donor, at room temperature. The stock dispersions of 2 mg/mL MONPPC were then diluted in incomplete medium to concentrations of 0.11 g/mL, 1.1
g/mL and 11
g/mL before 50 μL MONPPC dispersion were added to 500 μL of cell suspension. Thus, final MONP concentrations in the cell cultures were 0.01 μg/mL, 0.1 μg/mL and 1 μg/mL. Control stock dispersion for Co3O4- and Fe2O3 -particles was 1% plasma in complete medium. CeOx:Gd5% and CeOx:Gd14% could not be separated from the water they were synthesized in. Therefore, control stock dispersion for CeOx:Gd5% and CeOx:Gd14% was 1% plasma in 1 mL 65/35 sterile H2O/complete medium. Control stock dispersions were added to the cell cultures in the same proportions (50 μL to 500 μL cell culture) as the MONPPC dispersions. Thus, cells were exposed to <0.5% sterile water. Cells were then incubated with MONPPC and controls for 24 or 72 hours in 37°C, 5% CO2 and 97% humidity. After incubation, the supernatant was collected and stored at −80°C until cytokine analysis. Cells were immediately prepared for flow-cytometry analysis.
Flow cytometry
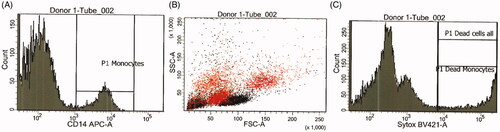
All data were collected on a FACSCanto™ II system (BD Biosciences, Breda, the Netherlands). After the 24-hour exposure to MONPPC, cells were labeled with monocyte-specific mouse anti-human CD14 APC-conjugated antibody (BD Biosciences) and 1 μM SYTOXTM Blue (Thermo Fisher) for the detection of dead cells. CFSE (carboxyfluoresceine, Thermo Fisher) was used to discriminate cells from debris and aggregates. Cells were sampled until approximately 500 CD14+ events were collected. +Monocyte uptake of the MONPPC was measured by gating on live CD14+ cells and calculating side-scatter (SSC) mean values. An increase in SSC from MONPPC-exposed versus non-exposed cultures was used to verify potential particle uptake (Ibuki and Toyooka Citation2012). Data analysis was performed using the BD FACSDiva software version 8.0. CD14+ cell population was identified and gated using an APC-histogram and then further exposed in a SYTOXTM Blue-histogram for determination of monocyte viability, .
Multiplex immunoassay
Cytokine release, following 24- and 72-hour incubation of PBMCs with MONPPC, was assessed using the supernatant from one donor (Donor 1). A 7-Plex Pro Human Cytokine kit (Bio-Rad) was used for detection of the monocyte differentiation marker GM-CSF, the T lymphocyte proliferation marker IL-2, the anti-inflammatory marker IL-10 and the proinflammatory markers IL-1β, IL-6, IL-8, and TNF-α according to the manufacturer’s instruction. The fold change (stimulation index) was determined with respect to control cultures without NP-exposure. A twofold change or above was considered as a positive response. Measurements were performed on a BioPlex® 200 system (Bio-Rad) with the BioPlex Manager software.
Immunogenicity of nanoparticle-binding proteins by in silico MHC class II epitope screening
To investigate the presence of MHC class II epitopes within identified corona-proteins, an in silico screening was performed. Ten of the proteins (Alpha-1-antitrypsin, Alpha-2-HS-glycoprotein, Apolipoprotein A-I, Complement C3, Fibrinogen beta chain, Haptoglobin, Hemopexin, Serotransferrin, Serum amyloid P-component, Transthyretin) identified by MALDI-TOF MS were subjected to MHC class II epitope screening.
Screening for MHC class II binding peptides (T-cell epitopes) was performed using the SciCross Immunogenicity Platform (SCIP). The T-cell epitope prediction algorithms of SCIP are based on statistics and machine learning, including support vector machines (Dönnes and Elofsson Citation2002) and optimization techniques (Schubert et al. Citation2018). The data used to develop these methods originate from both the IEDB database and proprietary data sets. For MHC class II binding peptides, peptides are aligned in order to identify 9-mer binding cores that are used as algorithm training data. The methodology is similar to the Gibbs sampling approach described by Nielsen et al. (Citation2004).
All query protein sequences were divided into 15-mer peptides corresponding to a potential 9-mer core and three flanking residues on each side. Binding properties of every peptide was then predicted for eight alleles, each representing a supertype of the Caucasian Western European population. The predicted binding scores from the eight alleles were combined into an overall immunogenicity score for each peptide. This score ranges from 0 to 100, where 100 means that the peptide is predicted to bind all tested alleles. For each peptide in the protein assessed, it is then possible to assign an immunogenicity score. Stretches along the protein sequence with an immunogenicity score above 50 or 75, denoted as hotspots, were identified. Furthermore, the average immunogenicity score/peptide was determined to get an overall view of the immunogenicity of the query proteins.
Results
Particle characterization
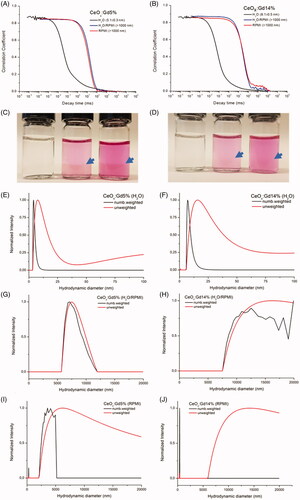
CeOx:Gd5% and CeOx:Gd14% samples were colloidally stable in H2O but aggregated upon RPMI1640 exposure, as indicated by the large shift in correlation function as well as size distributions obtained using DLS, . Particle aggregation in RPMI1640 was visible to the naked eye, . Sample sizes were estimated utilizing CONTIN fit intensity size distributions, . CeOx:Gd5% and CeOx:Gd14% have hydrodynamic diameters less than 10 nm in H2O, whereas particle aggregation in the two RPMI1640-containing media were too pronounced to estimate hydrodynamic diameters besides confirming that the NP aggregates were above 1000 nm.
Figure 2. Correlation functions of (a) CeOx:Gd5% and (b) CeOx:Gd14% in H2O, H2O/RPMI and RPMI. Hydrodynamic diameters from the intensity-weighted distributions are given in brackets. (c) CeOx:Gd5% and (d) CeOx:Gd14% in (from left to right) H2O, H2O/RPMI and RPMI. The blue arrows point at the border of the aggregated nanoparticles. Number weighted and unweighted intensity size distribution of CeOx:Gd5% and CeOx:Gd14% in (e–f) H2O, (g–h) H2O/RPMI and (i–j) RPMI. Note the different x-axis scale bar for graphs e-f) compared to the graphs g-j).
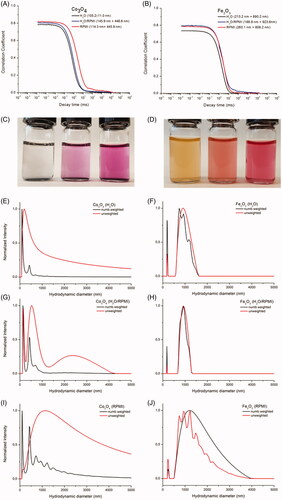
DLS spectra of Co3O4 and Fe2O3 samples are presented in . Sedimentation of Co3O4 NPs was observed. The size distributions of Co3O4 indicated two peaks at about 100 nm and 400 nm in H2O, in . When Co3O4 was dispersed in RPMI1640, the 400 nm distribution became more pronounced suggesting RPMI-induced aggregation, . The increased decay times of Co3O4 in RPMI1640 media further supports this conclusion. Fe2O3 size distribution consisted of one peak with a hydrodynamic diameter of 200 nm and a wider peak exceeding 1000 nm, and thus, aggregation occurs in both H2O and RPMI1640-containing buffers, ). Neither Co3O4 nor Fe2O3 were fully dispersible in any of the medias but were less prone to aggregate in RPMI1640 than CeOx:Gd5% and CeOx:Gd14% as clearly indicated by the decay times of the autocorrelation functions.
Figure 3. Correlation functions of (a) Co3O4 and (b) Fe2O3 in H2O, H2O/RPMI and RPMI. Hydrrodynamic diameters from the intensity weighted distributions are given in brackets. (c) Co3O4 and (d) Fe2O3 in (from left to right) H2O, H2O/RPMI and RPMI. Number weighted and unweighted intensity size distribution of Co3O4 and Fe2O3 in (e–f) H2O, (g–h) H2O/RPMI and (i–j) RPMI.
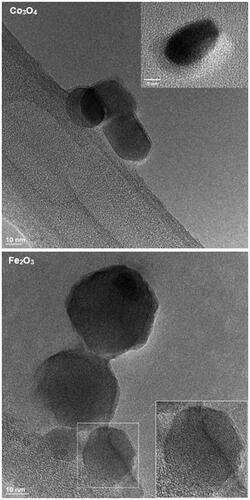
CeOx:Gd5%, CeOx:Gd14%, Co3O4 and Fe2O3 were analyzed using High Resolution Transmission Electron Microscopy (HR-TEM), and . The structure of the individual particles could be clearly resolved, suggesting that they consisted of single crystalline domains. The CeOx:Gd5% and CeOx:Gd14% were all estimated to be 3–5 nm in diameter, in agreement with earlier published data (Eriksson et al. Citation2018). The size of Co3O4 and Fe2O3 were measured to be in the range of 10–40 nm and 10–50 nm, respectively, based on the TEM micrographs below. Both estimates were consistent with the manufacturers information.
Figure 4. TEM image of CeOx:Gd5% and CeOx:Gd14%. Particles are <5 nm, examples of isolated particles where lattice is clearly visible are marked by circles and magnified 3x in insert.
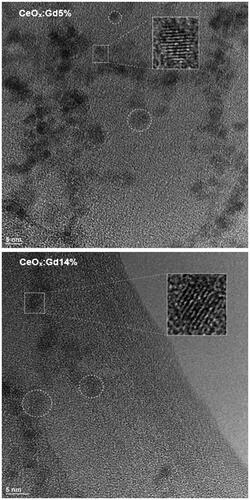
Figure 5. TEM images of Co3O4 and Fe2O3. The Co3O4 particles are of round or oblong shapes in the range of 10-40 nm. Insert shows example of isolated Co3O4 nanoparticle with visible lattice planes. The Fe2O3 particles are in the range of 10-50 nm. Lattice is clearly visible in the lower right particle and magnified 1.5x in insert.
Identification of nanoparticle-binding proteins
Co3O4, Fe2O3, CeOx:Gd5%, CeOx:Gd14% and a control without MONPs were incubated for 1 hour with human plasma. NP-binding proteins were separated by 2DGE, . Dominating spots on gels were excised and a total of 23 proteins were identified through MALDI-TOF MS, , with 19, 13, 14 and 17 proteins identified in Co3O4, Fe2O3, CeOx:Gd5% and CeOx:Gd14% respectively. Identities of 21 proteins were confirmed using nLC-MS/MS.
Figure 6. Identified nanoparticle-binding proteins. A final concentration of 2 mg/mL of Co3O4, Fe2O3, CeOx:Gd5% and CeOx:Gd14% nanoparticles were incubated with 10% plasma for 1 hour prior to analysis of nanoparticle-binding proteins by two-dimensional gel electrophoresis. A plasma incubation without nanoparticles was used as control. Gels were silver stained and dominating protein spots identified using MALDI-TOF MS as seen in .
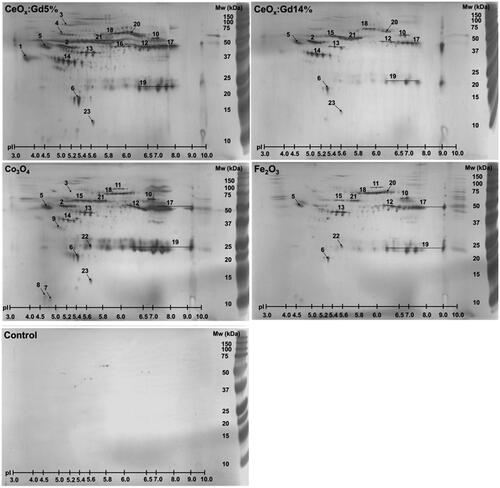
Table 2. Identification of nanoparticle-binding plasma proteins by MALDI-TOF MS following two-dimensional gel electrophoresis (2DGE).
In vitro immunological effects
Monocytes are key players as antigen-presenters and initiate the molecular events leading to generation of acquired immunity. Possible direct cytotoxic effects of MONPs on these cells may contribute to immunosuppression, while stimulatory effects enhance immune activation. Primary human monocytes in PBMC-cultures were assessed for viability following exposure to the MONPPC. Cytokine release was measured to study potentially immunostimulatory effects by the MONPPC on the cells. Prior to cell exposure, all MONP dispersions, without plasma proteins, tested negative for the presence of endotoxins (). The effects of a 24-hour exposure to MONPPC on the viability of monocytes, assessed by flow cytometric analysis of SYTOXTM Blue-labeled cells, are given in . Exposure to the highest (1 µg/mL) concentration of CeOx:Gd5% and CeOx:Gd14% showed a slight increase in cytotoxic effect, at least by a factor of 1.5, while remaining test concentrations and particles showed values comparable to background controls. Potential adherence/uptake of the particles by monocytes was tested by measuring a possible increase in flow-cytometric side-scatter (SSC) since this value is enhanced when cells show a higher degree of granularity (Ibuki and Toyooka Citation2012). Cell-cultures treated with MONPPC generally showed increased SSC-values as compared to background controls in all donors. The most pronounced effect was observed with CeOx:Gd5% and CeOx:Gd14% in all donors, and a dose-response relationship was also indicated, . Analysis of monocyte viability following 72 hours of MONPPC exposure was not possible due to monocyte adherence to the culture plate. Cytokine release following 24- and 72-hour exposure are shown as fold change in , and as absolute values in . The most pronounced cytokine response was detected at 72-hour exposure to 1 μg/mL Co3O4 particles, which resulted in an increase of pro-inflammatory cytokines: IL-1β, IL-6, IL-8 and TNF-α increased approximately 25-fold, 66-fold, 9-fold, and 6-fold, respectively. Cytokines connected to anti-inflammatory processes (IL-10), monocyte differentiation (GM-CSF) and lymphocyte proliferation (IL-2) were all below the detection limit.
Figure 7. Flow-cytometry results showing changes in mean side-scatter (SSC) values for monocytes (CD14+ events) as a result of exposure to the different test materials. An increase in mean SSC, as compared to control, indicate interaction of particles (adhesion/phagocytosis) with the monocytes.
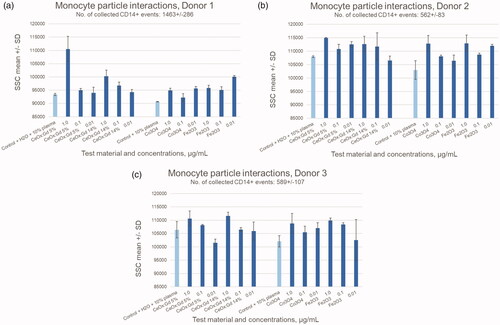
Figure 8. Fold change in proinflammatory cytokines. PBMCs isolated from whole blood were incubated 24 or 72 hours with four different protein-covered nanoparticles (Fe2O3, Co3O4, CeOx:Gd5%, and CeOx:Gd14%) at three different concentrations (1.0, 0.1, and 0.01 μg/mL). The supernatants were collected and cytokines measured using a multiplex bead-based assay. The graphs above illustrate the fold-change of IL-1β, IL-6, IL-8, and TNF-α with respect to controls. Intra assay coefficient of variation (CV) had a range of 0-19 % with a mean ± SD of 4 ± 2.7. A two-fold change (red line) or above was considered significant. Co3O4-exposure resulted in profound fold-changes for all four cytokines following 72-hour incubation.
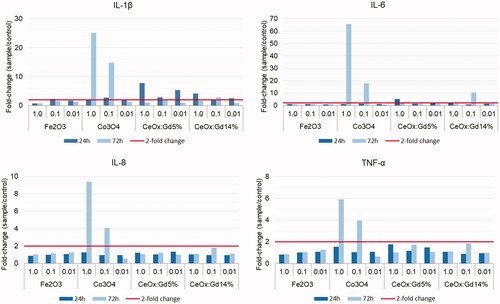
Table 3. Cytotoxicity of test compounds on monocytes in PBMC-cultures. Flow-cytometric results showing percent double-positive (SYTOXTM Blue+/CD14+) cells after 24-hour incubation.
Table 4. Cytokine release in supernatants from PBMCs following 24- and 72-hour exposure to four different protein-covered nanoparticles (Fe2O3, Co3O4, CeOx:Gd5%, and CeOx:Gd14%) at three different concentrations (1.0, 0.1, and 0.01 μg/mL). Values are mean ± SD.
Predicted MHC class II epitopes and potential immunogenicity
Ten proteins that were clearly visible on 2DGE and identified by MALDI-TOF MS were selected for in silico MHC class II epitope screening. Immunogenicity scores were calculated and stretches with high immunogenicity scores, so-called hotspots, were identified. shows an example of an immunogenicity plot for one of the studied proteins. summarizes results obtained with all screened proteins. All proteins except transthyretin, which is also the smallest protein, contained at least one hotspot with immunogenicity score >75. The largest protein, complement C3, showed 14 hotspots >75, whereas alpha-1-antitrypsin showed the highest average score/peptide (28.27). These results indicate potential presence of T-cell epitopes on all the 10 tested proteins.
Figure 9. Immunogenicity score plot. The query protein sequence was divided into 15-mer peptides corresponding to a potential 9-mer core and three flanking residues on each side. Binding properties of every peptide was then predicted for eight alleles, each representing a supertype of the Caucasian Western European population, and the peptide immunogenicity score was calculated. This score ranges from 0 to 100, were 100 means that the peptide binds all tested alleles. Stretches along the protein sequence with an immunogenicity score above 50 or 75 (orange and red line respectively) are defined as hotspots.
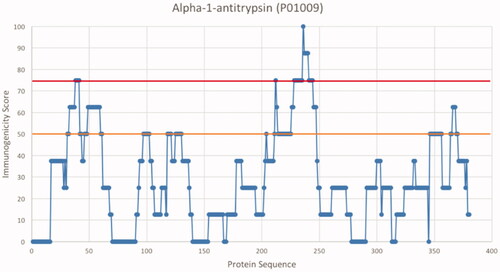
Table 5. Immunogenicity of endogenous nanoparticle-binding proteins.
HLA-DRB1*1101 showed potential presentation of all 27 high-risk peptide clusters found while *0101 only indicated presentation of in total eight of these clusters. Thus, individuals carrying the *0101 profile might show a higher degree of tolerance, while *1101 bearers would theoretically be more at risk for producing autoantibodies against any of the tested proteins. An overview of high-risk cluster binding to specific HLA-types is shown in .
Table 6. Binding of identified peptide high-risk clusters to respectively HLA-DRB1 receptor coded by the given alleles.
Discussion
Intentional and unintentional particle exposure
The range of different types of MONPs present in our environment, produced intentionally or unintentionally, is practically innumerable. Unintentionally produced MONPs formed in occupational settings, such as welding or additive manufacturing, are a major health concern due to the unknown effects of high and/or long-term exposure. Here, exposure routes are mainly inhalation, ingestion or skin contact followed by further distribution via the blood stream. Such metal exposures can be confirmed by metal analyses of urine but still very little is known regarding the relationship between biological levels of metals and adverse health effects. In contrast, intentionally synthesized MONPs with capping and functionalization strategies for biomedical imaging (Ahrén et al. Citation2012; Hu et al. Citation2013) are aimed to be used at low concentrations for a limited time in the blood stream. Moreover, synthesized MONPs are systematically investigated by preclinical and clinical tests before market introduction. However, improved knowledge on the biological responses of such MONPs is still needed. The aim here was to improve the possibility to predict exposure-related health effects of MONPs through interdisciplinary collaboration by combining workflows from the pharmaceutical industry, nanomaterial sciences, and occupational medicine.
The importance of particle characterization under biological relevant conditions
The health concern is that MONPs may have an inherent ability to aggregate in biologically relevant buffers/fluids, which has been highlighted in a recent review by Falahati et al. (Citation2019). Hence, there is a need for improved understanding regarding particle behavior before downstream analyses of particle interacting proteins can be performed, and before physiological effects can be elucidated. In the present pilot study, MONPs were characterized through DLS in both H2O and RPMI1640 as well as through HR-TEM before downstream proteomics and monocyte interaction studies.
In agreement with previously published data by Eriksson et al. (Citation2018), the hydrodynamic diameter of the CeOx:Gd5% and CeOx:Gd14% were below 10 nm in H2O, whereas reduced dispersibility of Co3O4 and Fe2O3 in H2O indicated a process of agglomeration/aggregation. This was shown by the DLS results. Fe2O3 showed two size populations above 100 nm (213 nm and 890 nm), and Co3O4 showed one size population (105 nm). Thus, the hydrodynamic diameters of Fe2O3 and Co3O4 in H2O exceeded those specified by the manufacturer (<50 nm). This occurrence has been reported in earlier characterization studies as well (Lundqvist et al. Citation2008; Adams et al. Citation2006) that concluded that it is necessary to perform your own particle characterization. We did HR-TEM studies to obtain in depth knowledge of the starting materials. HR-TEM allows us to verify the size and crystallinity of the individual particles. The HR-TEM imaging showed that the larger than expected sizes obtained using DLS were due to aggregation.
All particles formed aggregates in RPMI1640. However, Fe2O3 and Co3O4 did not aggregate to the same extent as CeOx:Gd5% and CeOx:Gd14%, whose hydrodynamic diameters increased 100-fold in RPMI1640 as compared to H2O. This behavior is correlated to the fact that the surfaces of CeOx:Gd5% and CeOx:Gd14% were stabilized with acetate and not an organic shielding, such as dextran or poly-ethylene glycol, which are known to prevent aggregation. Toxicity studies of CeOx:Gd with dextran capping is under way (Eriksson et al. Citation2021). However, in this study, the intentions were not to hide the nanomaterial using organic coatings. Rather, the aim was to investigate the cellular response as a function of choice of elemental composition of the NPs. Several studies have revealed that some nanomaterials, but not all, aggregate in biologically relevant media (Cappellini et al. Citation2018; Murdock et al. Citation2008; Ortega et al., Citation2014). Not surprisingly, different media renders different aggregation states and size distributions. Clearly, particle characterization must be carried out in the medium intended for downstream analysis for comparability between measurements as well as correct conclusions regarding health effects. If there are no suitable solutions available for such analyses, then new particle characterization methods need to be developed.
A study by Ortega et al. (Citation2014) suggested that the presence of proteins in the medium (which leads to formation of a protein corona) reduces risk of agglomeration. Indeed, a review by Barbero et al. (Citation2017) pointed out how corona formation increased thermodynamic stability through an electro-steric effect that stabilizes the NPs. However, Zhang et al. (Citation2018) found that proteins could either mitigate or induce agglomeration of different NPs. Hence, particle characterization studies should include proteins as well. In the present study, characterization of corona-covered MONPs was not possible due to the presence of aggregates. In DLS measurements, the scattered light intensity is proportional to the sixth power of the particle radius. Therefore, formation of aggregates drenches out the scattered light of smaller particles and when agglomerates occur, exact size determination becomes difficult because of low peak resolution.
The ratio MONP to plasma used in this toxicity study is chosen in line with previously published work (Deng et al. Citation2009). When trying to optimize these protocols, we found that when particle concentrations were reduced, protein identification was not possible due to low abundance, and when increasing the added plasma volumes protein aggregation occurred in the particle-free control after centrifugation. Therefore, when comparing the mass of MONP (2 mg/mL) to the mass of plasma proteins in use (600 µg/mL), there is a factor of 3.3 in advantage to NPs. One should keep in mind that the density is higher for the particles compared to proteins by a factor of 4–5. The particle to protein ratio used here, may not reflect the authentic case in vivo; however, it is powerful when investigating protein interaction with NP surfaces. Excess of nanomaterial will help to screen and obtain the protein adsorption pattern correlated to the choice of NP. However, the chosen workplan with such dominating fractions of nanomaterial is pushing the limit on dispersibility of MONPs in both H2O and RPMI1640. It has to be noted that what further impacts the possibility to predict whether the current number of proteins will form a complete corona that covers the particles or not is the fact that the particles here are aggregated, which means that the surface area is significantly lower compared to the particles in free form. Due to the above-mentioned uncertainties, we have in this work chosen not to use the term protein corona but instead metal oxide nanoparticle-protein complexes (MONPPC).
Since all MONPs used in the current study aggregated in the RPMI1640 medium, it should be considered whether particle aggregation could be expected in vivo or whether it is merely experimentally conditioned. A review by Buzea, Pacheco, and Robbie (Citation2007) stated that NP agglomerates larger than 100–200 nm in diameter had reduced toxicity compared to their smaller non-agglomerated forms. For studies aiming at mimicking the biological situation, it is therefore necessary to determine whether the particles aggregate in different biological fluids such as pulmonary fluids and blood (and at biologically relevant concentrations) or not, as well as to know how the experimental setup may influence particle behavior, protein interaction and subsequent biological responses.
Studying the nanoparticle-binding proteins
MONPs were incubated 1 hour with proteins to allow formation of a high affinity corona. Proteins identified after isolation and washing, are proposedly those who remain on the MONP surface and share its biological fate (Lundqvist et al. Citation2008).
Isolation of NP-protein complexes by centrifugation is by far the most common approach. It is fast, simple and can be applied to all types of material as compared to approaches such as magnetic separation. Pisani et al. (Citation2017) showed that different separation methods alter the relative abundancy of different corona proteins, but the composition can be determined by either method.
2DGE followed by MALDI-TOF-MS was used to determine protein corona composition and protein identities were further verified by nLC-MS/MS, . Interestingly, all four MONPs had similar protein patterns on the 2DGE gels, . The protein composition differed between the MONPs when identified by MALDI-TOF-MS but not when verified with the more sensitive nLC-MS/MS method. This suggests different levels of protein binding (abundance) but not composition. Exceptions were alpha-2-antiplasmin, not found on Co3O4, and immunoglobulin gamma-1 heavy chain, only found on CeOx:Gd5%. Two proteins identified by MALDI-TOF-MS, immunoglobulin gamma-1 heavy chain and immunoglobulin kappa constant were unverified by nLC-MS/MS. The discrepancies between the two methods may be explained by methodological differences. For example, the selected parent peptide that is sequenced in nLC-MS/MS must be unique for the identified protein and hydrophobicity and abundance may affect which proteins that can be identified by 2DGE and MALDI-TOF-MS. Therefore, these two approaches should be seen as complements to each other.
The human plasma proteome consists of thousands of proteins (Schwenk et al. Citation2017). Nevertheless, the four studied protein coronas consist of the same 23 proteins. The presence of albumin, immunoglobulins, fibrinogen, and apolipoproteins is expected due to their high abundance in plasma and these proteins have often been found in previous protein corona studies as well (Aggarwal et al. Citation2009; Cedervall et al. Citation2007; Deng et al. Citation2009, Lundqvist et al. Citation2017). Moreover, inorganic NPs, such as MONPs, are known to attract adaptor and carrier proteins (Karmali and Simberg Citation2011). Examples of such proteins are transferrin, haptoglobin and alpha-2-HS-glycoprotein, all identified in our study.
Perhaps the similar MONP protein-binding profiles in the present study are because the studied MONPs display similar physical properties. Co2O3 and Fe2O3 are redox-active magnetic metals and the CeOx:Gd5%- and CeOx:Gd14%- particles combine the redox-active properties of CeOx NPs and magnetic properties of Gd. Indeed, Deng et al. (Citation2009) observed similar coronas when comparing TiO2 and SiO2, which have similar physiochemical properties, but a different corona on polymer-coated ZnO NPs. In addition, capping has been shown to drastically change cell response, solubility, and stability. In line, CeOx:Gd NPs have recently been functionalized with dextran by our group, and we have shown that the response is strongly coupled to the capping/coating of the MONPs. The response was highly coupled to Zeta potential and chemical structure at the MONPs surface (Eriksson et al. Citation2021).
However, the formation of aggregates, as discussed above, may also be the reason for the similar protein binding profiles. To understand underlying mechanisms for different protein binding patterns, it is important to be able to characterize MONPs with and without proteins and their behavior in fluids relevant for the physiological environment of the expected exposure.
Monocyte interaction with MONPPC
MONPs may enter the circulatory system either by inhalation (Guttenberg et al. Citation2016), such as during occupational exposure, or by being administered intravenously, such as MONPs engineered for medical purposes. Monocytes are phagocytosing, antigen-presenting cells in the blood and key players in antigen-specific immune responses. Adverse immune reactions following MONP exposure may thus be dictated by monocyte interaction with and uptake of MONPs.
Even though all particles in the current study aggregated in RPMI1640 and had similar corona patterns, different monocyte responses were observed upon exposure to these MONPPC, suggesting that the crude material itself plays a role in toxicity. For example, shows that CeOx:Gd5% and CeOx:Gd14% decreased monocyte viability at the highest tested concentration (1 μg/mL), whereas Fe2O3 and Co3O4 did not. To our knowledge, monocyte interaction with CeOx:Gd-NPs has not previously been tested but cerium oxide NPs have been found to decrease viability at a similar concentration range (Hussain et al. Citation2012; Li et al. Citation2018), which is supporting our findings. However, others have found that concentrations of 200 μg/mL cerium oxide NPs did not affect monocyte proliferation (Lord et al. Citation2012; Ting et al. Citation2013), although the particles used in those studies were coated with APTES and heparin respectively. Additional studies are needed to clarify the relationship between MONP aggregation and monocyte responses.
Interestingly, Co3O4 exposure increased pro-inflammatory cytokines, , which is in line with findings from previous studies using cobalt oxide NPs and primary immune or endothelial cells (Alinovi et al. Citation2015; Chattopadhyay et al. Citation2015). In addition, MONP preparations often contain endotoxins (ETX), pro-inflammatory bacterial cell wall components (Boraschi et al. Citation2017). Since ETX would affect the investigated monocyte response, the ETX content of all particle dispersions used should be determined prior to cell exposures. All MONP dispersions in the present study were tested free of endotoxins and the detected monocyte responses can thereby be ascribed to the investigated MONPPC without endotoxin interference. There was no correlation between cytotoxicity and cytokine responses. The slight increase in toxicity observed with CeOxGd5%- and 14%-particles was not reflected by any cytokine-response and Co3O4-particles showed no cytotoxicity in vitro despite an observed cytokine response. These findings are in line with recent studies showing induction of IL-8 by cobalt via TLR-4 interaction (Anjum et al. Citation2018), while cerium did not augment pro-inflammatory cytokine responses (Hussain et al. Citation2012).
The binding of opsonizing proteins, such as complement C3, complement factor B and immunoglobulins, , suggests that the adhered proteins facilitate monocyte uptake of MONPPC. Indeed, binding of complement C3 to silica NPs has been found to trigger the classical pathway of the complement system and consequently increase cellular uptake by phagocytosis (Tavano et al. Citation2018). Moreover, fibrinogen has been suggested to promote monocyte attachment to foreign substances (Shen and Horbett Citation2001), and Vogt et al. (Citation2015) demonstrated increased monocyte uptake of superparamagnetic iron oxide NPs with fibrinogen-enriched coronas. The results in this study indicate particle-adherence or uptake by monocytes for all MONPPCs tested since flow-cytometric analyses revealed an increase in SSC upon a 24-hour incubation period for all materials.
Predicted MHC class II epitopes on endogenous Nanoparticle-Binding proteins
When innate immune responses fail to clear foreign substances, the adaptive immune system may be activated. This can be achieved through antigen processing and subsequent presentation to CD4+ T lymphocytes through MHC class II complexes on professional phagocytes, such as monocytes. When co-stimulated with inflammatory cytokines, so-called ‘danger signals’, the T lymphocytes, and thus the adaptive immune system, is activated (Gallucci and Matzinger Citation2001).
If the MONPPC are phagocytosed and processed by monocytes, then endogenous proteins bound to the MONP surface could be presented through MHC class II to CD4+ T lymphocytes as antigens resulting in an adaptive immune response toward endogenous proteins. Herein, for the first time, NP-binding proteins have been characterized for their immunogenic properties using current in silico tools normally applied on biopharmaceuticals in the drug-development process used by the pharmaceutical industry.
Under normal conditions, immunologic tolerance toward autologous proteins is established to avoid autoimmunity. However, when an endogenous protein becomes part of a NP-protein complex structural changes may occur (Laera et al. Citation2011). These conformational alterations could potentially contribute to a higher risk for the protein being perceived as nonself. These endogenous proteins may also undergo ‘forced phagocytosis, by being phagocytosed as part of NP–protein complexes.’ If cytokine responses are induced simultaneously, a third factor for creating an antigen-specific response will be present. Taken together, these suggested mechanisms may very well contribute to affected tolerance by inducing immune responses to autologous structures derived from particle-borne proteins, resulting in autoimmunity.
The observed immunogenic properties of the screened proteins in specific HLA-types could induce autoimmune-like conditions in certain individuals. Endogenous proteins are normally not processed by antigen-presenting cells, such as monocytes. If these proteins are, by conjugation to MONPs, carried into the phagocytosis-antigen presenting pathway they will be regarded as “foreign” by the immune system. In fact, Grunewald et al. (Citation2016) has suggested this same mechanism behind vimentin as an autoantigen in patients with lung sarcoidosis. This is based on the finding of vimentin-MHC-class-II-binding and a strong association between MHC class II and T-cell receptors. Our in silico screening showed that alpha-1-antitrypsin (AAT) had higher immunogenicity score than the other tested corona proteins. This is especially interesting, since AAT regulates several proteases and immune processes (Kalfopoulos Citation2017). Moreover, AAT deficiency is related to inflammation and tissue damage in several chronic pulmonary disorders, including asthma and bronchiectasis (Gramegna et al. Citation2018). Even though AAT deficiency is a known hereditary disorder, it raises the question whether these diseases can be induced by particle exposure leading to acquired autoimmunity. For biotherapeutics, it has been shown that aggregates can have a great influence on the uptake and processing by dendritic cells, as well as the subsequent T-cell activation (Rombach-Riegraf et al. Citation2014). In theory, this means that an existing tolerance to a protein could be lost due to changes in antigen processing and presentation caused by NP-protein interaction. This forced antigen-presenting hypothesis requires further evidence by the demonstration of the presence of endogenous protein-specific antibody responses in exposed individuals.
Limitations
There are possible limitations in the current study that must be acknowledged. The NP-plasma protein incubation step by Deng et al. (Citation2009) is highly cited and thereby a precursor to many follow-up studies, but in the particle/protein pellet washing step, as described in the method section, we have chosen to use a wash solution that we have long experience of using prior to 2DGE. It would have been possible to use a different wash solution if we would have identified the proteins using only the less salt-sensitive nLC-MS/MS. However, since we know that both analysis methods have their strengths and drawbacks, we wanted to use both methods as well as identical samples. We do not believe that this washing step may lead to an altered protein corona though we did not find previous reports on interacting human plasma proteins with these particles and are thus unable to completely rule out the possibility. We are also aware that the protein to particle ratio in the study is the opposite of the in vivo situation, but we decided for this approach because we wanted to be as sure as possible that the proteins of relevance for in silico MHC Class-II epitope screening were interacting with the particles. Protein identities may therefore differ from approaches with different protein to particle ratios. The identified problem with particle aggregation in biological buffers is not a novel finding. However, it is a limitation that it was not possible to characterize the particle aggregates in solution with or without proteins using DLS. This is a challenge for future research.
Conclusion
There is an urgent need of biologically relevant in vitro assessments of MONP toxicity. This requires development of both systematic particle characterization, that is, detailed studies of the starting material and thorough investigations of the MONPs in solvents and solutions, and biologically relevant buffers compatible with particle characterization, downstream in vitro analyses, and the MONPs themselves.
Possible immune-stimulating properties of MONPPC are of interest to verify any inducing or potentiating effects of MONPPC on autoimmune disease. For the first time, NP-binding proteins have been characterized for their immunogenic properties. The present study indicates that especially Co3O4 NPs induce ‘danger signals’, verified by production of inflammatory cytokines, and simultaneously bind autologous proteins, which can be presented as immunogenic epitopes by MHC class II. The clinical relevance of these findings should be further evaluated to investigate the role of MONPs in the development autoimmune disease.
| Abbreviations | ||
| MONP | = | metal oxide nanoparticle |
| MONPPC | = | metal oxide nanoparticle-protein complexes |
| MHC | = | major histocompatibility complex |
| PBMCs | = | peripheral-blood mononuclear-cells |
| AAT | = | alpha-1-antitrypsin |
| NP | = | nanoparticle |
| ROS | = | reactive oxygen species |
| ETX | = | endotoxins. |
TNAN-2020-OR-0192-File017.docx
Download MS Word (54.3 KB)Acknowledgments
The authors thanks to Carolina Sandman at former Swetox for performing the experiments on human monocytes.
Disclosure statement
Pierre Dönnes is a majority shareholder of SciCross AB and attests that this report is free of any bias that might be associated with the commercial goals of the company.
Additional information
Funding
References
- Abo-Zeid, Y., and G. R. Williams. 2020. “The Potential anti-Infective Applications of Metal Oxide Nanoparticles: A Systematic Review.” Wiley Interdisciplinary Reviews. Nanomedicine and Nanobiotechnology 12 (2): e1592. doi:https://doi.org/10.1002/wnan.1592.
- Abrikossova, N., C. Skoglund, M. Ahrén, T. Bengtsson, and K. Uvdal. 2012. “Effects of Gadolinium Oxide Nanoparticles on the Oxidative Burst from Human Neutrophil Granulocytes.” Nanotechnology 23 (27): 275101. doi:https://doi.org/10.1088/0957-4484/23/27/275101.
- Adams, L., D. Lyon, A. McIntosh, and P. Alvarez. 2006. “Comparative Toxicity of Nano-Scale TiO2, SiO2 and ZnO Water Suspensions.” Water Science and Technology : a Journal of the International Association on Water Pollution Research 54 (11-12): 327–334. doi:https://doi.org/10.2166/wst.2006.891.
- Aggarwal, P., J. Hall, C. McLeland, M. Dobrovolskaia, and S. McNeil. 2009. “Nanoparticle Interaction with Plasma Proteins as It Relates to Particle Biodistribution, Biocompatibility and Therapeutic Efficacy.” Advanced Drug Delivery Reviews 61 (6): 428–437. doi:https://doi.org/10.1016/j.addr.2009.03.009.
- Ahrén, Maria, Linnéa Selegård, Anna Klasson, Fredrik Söderlind, Natalia Abrikossova, Caroline Skoglund, Torbjörn Bengtsson, Maria Engström, Per-Olov Käll, and Kajsa Uvdal. 2010. “Synthesis and Characterization of PEGylated Gd2O3 Nanoparticles for MRI Contrast Enhancement.” Langmuir: The ACS Journal of Surfaces and Colloids 26 (8): 5753–5762. doi:https://doi.org/10.1021/la903566y.
- Ahrén, M., L. Selegård, F. Söderlind, M. Linares, J. Kauczor, P. Norman, P. Käll, and K. Uvdal. 2012. “A Simple Polyol-Free Synthesis Route to Gd2O3 Nanoparticles for MRI Applications: An Experimental and Theoretical Study.” Journal of Nanoparticle Research 14 (8): 1006. doi:https://doi.org/10.1007/s11051-012-1006-2.
- Alinovi, R., M. Goldoni, S. Pinelli, M. Campanini, I. Aliatis, D. Bersani, P. Lottici, et al. 2015. “Oxidative and Pro-Inflammatory Effects of Cobalt and Titanium Oxide Nanoparticles on Aortic and Venous Endothelial Cells.” Toxicology in Vitro: An International Journal Published in Association with BIBRA 29 (3): 426–437. doi:https://doi.org/10.1016/j.tiv.2014.12.007.
- Anjum, S., A. Mawdesley, H. Lawrence, D. Deehan, J. Kirby, and A. Tyson-Capper. 2018. “The Effect of Cobalt on Inflammatory Cytokine and Adhesion Molecule Expression.” Orthopaedic Proceedings 99-B (SUPP_9): 70–70.
- Barbero, F., L. Russo, M. Vitali, J. Piella, I. Salvo, M. Borrajo, M. Busquets-Fité, et al. 2017. “Formation of the Protein Corona: The Interface between Nanoparticles and the Immune System.” Seminars in Immunology 34: 52–60. doi:https://doi.org/10.1016/j.smim.2017.10.001.
- Boraschi, D., P. Italiani, R. Palomba, P. Decuzzi, A. Duschl, B. Fadeel, and S. Moghimi. 2017. “Nanoparticles and Innate Immunity: new Perspectives on Host Defence.” Seminars in Immunology 34: 33–51. doi:https://doi.org/10.1016/j.smim.2017.08.013.
- Boyles, M., T. Kristl, A. Andosch, M. Zimmermann, N. Tran, E. Casals, M. Himly, et al. 2015. “Chitosan Functionalisation of Gold Nanoparticles Encourages Particle Uptake and Induces Cytotoxicity and Pro-Inflammatory Conditions in Phagocytic Cells, as Well as Enhancing Particle Interactions with Serum Components.” Journal of Nanobiotechnology 13 (1): 84 doi:https://doi.org/10.1186/s12951-015-0146-9.
- Buzea, C., I. Pacheco, and K. Robbie. 2007. “Nanomaterials and Nanoparticles: Sources and Toxicity.” Biointerphases 2(4): MR17–MR71. doi:https://doi.org/10.1116/1.2815690.
- Cappellini, F., Y. Hedberg, S. McCarrick, J. Hedberg, R. Derr, G. Hendriks, I. Odnevall Wallinder, and H. Karlsson. 2018. “Mechanistic Insight into Reactivity and (Geno)Toxicity of Well-Characterized Nanoparticles of Cobalt Metal and Oxides.” Nanotoxicology 12(6): 602–620. doi:https://doi.org/10.1080/17435390.2018.1470694.
- Cedervall, T., I. Lynch, S. Lindman, T. Berggard, E. Thulin, H. Nilsson, K. Dawson, and S. Linse. 2007. “Understanding the Nanoparticle-Protein Corona Using Methods to Quantify Exchange Rates and Affinities of Proteins for Nanoparticles.” Proceedings of the National Academy of Sciences of the United States of America 104 (7): 2050–2055. doi:https://doi.org/10.1073/pnas.0608582104.
- Chattopadhyay, S., S. Dash, S. Tripathy, B. Das, D. Mandal, P. Pramanik, and S. Roy. 2015. “Toxicity of Cobalt Oxide Nanoparticles to Normal Cells; an in Vitro and in Vivo Study.” Chemico-Biological Interactions 226: 58–71. doi:https://doi.org/10.1016/j.cbi.2014.11.016.
- Cho, W., R. Duffin, C. A. Poland, S. E. M. Howie, W. MacNee, M. Bradley, I. L. Megson, and K. Donaldson. 2010. “Metal Oxide Nanoparticles Induce Unique Inflammatory Footprints in the lung: important implications for nanoparticle testing.” Environmental Health Perspectives 118 (12): 1699–1706. pp doi:https://doi.org/10.1289/ehp.1002201.
- Choi, O., and Z. Hu. 2008. “Size Dependent and Reactive Oxygen Species Related Nanosilver Toxicity to Nitrifying Bacteria.” Environmental Science and Technology 42 (12): 4583–4588. doi:https://doi.org/10.1021/es703238h.
- Dai, Q., J. Guo, Y. Yan, C. S. Ang, N. Bertleff-Zieschang, and F. Caruso. 2017. “Cell-Conditioned Protein Coronas on Engineered Particles Influence Immune Responses.” Biomacromolecules 18 (2) : 431–439. doi:https://doi.org/10.1021/acs.biomac.6b01545.
- Dankers, A., C. Kuper, A. Boumeester, B. Fabriek, I. Kooter, M. Gröllers-Mulderij, P. Tromp, I. Nelissen, E. Zondervan-Van Den Beuken, and R. Vandebriel. 2018. “A Practical Approach to Assess Inhalation Toxicity of Metal Oxide Nanoparticles in Vitro.” Journal of Applied Toxicology: JAT 38 (2): 160–171. doi:https://doi.org/10.1002/jat.3518.
- Deng, Z., G. Mortimer, T. Schiller, A. Musumeci, D. Martin, and R. Minchin. 2009. “Differential Plasma Protein Binding to Metal Oxide Nanoparticles.” Nanotechnology 20 (45): 455101. doi:https://doi.org/10.1088/0957-4484/20/45/455101.
- Djurišić, A., Y. Leung, A. Ng, X. Xu, P. Lee, N. Degger, and R. Wu. 2015. “Toxicity of Metal Oxide Nanoparticles: Mechanisms, Characterization, and Avoiding Experimental Artefacts.” Small (Weinheim an Der Bergstrasse, Germany) 11 (1): 26–44. doi:https://doi.org/10.1002/smll.201303947.
- Dönnes, P., and A. Elofsson. 2002. “Prediction of MHC Class I Binding Peptides, Using SVMHC.” BMC Bioinformatics 3 (25): 25. doi:https://doi.org/10.1186/1471-2105-3-25.
- Eriksson, P., M. Assenhöj, C. Brommesson, K. Bunnfors, R. Nosratabadi, S. A. Ljunggren, Z. Hu, H. Karlsson, and K. Uvdal. 2021. “Gd-CeNPs Functionalized with FITC-Conjugated Dextran Show Enhanced T1 Relaxivity, Reduced Protein Adsorption and ROS Scavenging Properties.” Unpublished Manuscript. Linkoping, Sweden: Department of Physics, Chemistry and Biology (IFM), Linkoping University.
- Eriksson, P., A. Tal, A. Skallberg, C. Brommesson, Z. Hu, R. Boyd, W. Olovsson, et al. 2018. “Cerium Oxide Nanoparticles with Antioxidant Capabilities and Gadolinium Integration for MRI Contrast Enhancement.” Scientific Reports 8 (1): 6999. doi:https://doi.org/10.1038/s41598-018-25390-z.
- Falahati, M., F. Attar, M. Sharifi, T. Haertlé, J. Berret, R. Khan, and A. Saboury. 2019. “A Health Concern regarding the Protein Corona, Aggregation and Disaggregation.” Biochimica et Biophysica Acta. General Subjects 1863 (5): 971–991. doi:https://doi.org/10.1016/j.bbagen.2019.02.012.
- Gallucci, S., and P. Matzinger. 2001. “Danger Signals: SOS to the Immune System.” Current Opinion in Immunology 13 (1): 114–119. doi:https://doi.org/10.1016/S0952-7915(00)00191-6.
- Ghafouri, B., H. Karlsson, H. Mörtstedt, A. Lewander, C. Tagesson, and M. Lindahl. 2007. “2,5-Dihydroxybenzoic Acid instead of alpha-cyano-4-hydroxycinnamic acid as matrix in matrix-assisted laser desorption/ionization time-of-flight mass spectrometry for analyses of in-gel digests of silver-stained proteins.” Analytical Biochemistry 371 (1): 121–123. doi:https://doi.org/10.1016/j.ab.2007.07.002.
- Giovanni, M., J. Yue, L. Zhang, J. Xie, C. Ong, and D. Leong. 2015. “Pro-Inflammatory Responses of RAW264.7 Macrophages When Treated with Ultralow Concentrations of Silver, Titanium Dioxide, and Zinc Oxide Nanoparticles.” Journal of Hazardous Materials 297: 146–152. doi:https://doi.org/10.1016/j.jhazmat.2015.04.081.
- Gonnissen, D., Y. Qu, K. Langer, C. Öztürk, Y. Zhao, C. Chen, G. Seebohm, et al. 2016. “Comparison of Cellular Effects of Starch-Coated SPIONs and Poly(Lactic-co-Glycolic Acid) Matrix Nanoparticles on Human Monocytes.” International Journal of Nanomedicine 11: 5221–5236. pp doi:https://doi.org/10.2147/IJN.S106540.
- Görg, A., C. Obermaier, G. Boguth, A. Harder, B. Scheibe, R. Wildgruber, and W. Weiss. 2000. “The Current State of Two-Dimensional Electrophoresis with Immobilized pH Gradients.” Electrophoresis 21 (6): 1037–1053. doi:https://doi.org/10.1002/(SICI)1522-2683(20000401)21:6<1037::AID-ELPS1037>3.0.CO;2-V.
- Gramegna, A., S. Aliberti, M. Confalonieri, A. Corsico, L. Richeldi, C. Vancheri, and F. Blasi. 2018. “Alpha-1 Antitrypsin Deficiency as a Common Treatable Mechanism in Chronic Respiratory Disorders and for Conditions Different from Pulmonary Emphysema? A Commentary on the New European Respiratory Society Statement.” Multidisciplinary Respiratory Medicine 13 (1): 39. doi:https://doi.org/10.1186/s40248-018-0153-4.
- Grosse, S., J. Stenvik, and A. Nilsen. 2016. “Iron Oxide Nanoparticles Modulate Lipopolysaccharide-Induced Inflammatory Responses in Primary Human Monocytes.” International Journal of Nanomedicine 11: 4625–4642. Volume doi:https://doi.org/10.2147/IJN.S113425.
- Grunewald, J., Y. Kaiser, M. Ostadkarampour, N. Rivera, F. Vezzi, B. Lötstedt, R. Olsen, et al. 2016. “T-Cell Receptor–HLA-DRB1 Associations Suggest Specific Antigens in Pulmonary Sarcoidosis.” European Respiratory Journal 47 (3): 898–909. doi:https://doi.org/10.1183/13993003.01209-2015.
- Guttenberg, M., L. Bezerra, N. Neu-Baker, M. del Pilar Sosa Idelchik, A. Elder, G. Oberdörster, and S. Brenner. 2016. “Biodistribution of Inhaled Metal Oxide Nanoparticles Mimicking Occupational Exposure: A Preliminary Investigation Using Enhanced Darkfield Microscopy.” Journal of Biophotonics 9 (10): 987–993. doi:https://doi.org/10.1002/jbio.201600125.
- Horev-Azaria, L., G. Baldi, D. Beno, D. Bonacchi, U. Golla-Schindler, J. Kirkpatrick, S. Kolle, et al. 2013. “Predictive Toxicology of Cobalt Ferrite Nanoparticles: comparative in-Vitro Study of Different Cellular Models Using Methods of Knowledge Discovery from Data.” Particle and Fibre Toxicology 10 (1): 32. doi:https://doi.org/10.1186/1743-8977-10-32.
- Hu, Z., M. Ahrén, L. Selegård, C. Skoglund, F. Söderlind, M. Engström, X. Zhang, and K. Uvdal. 2013. “Highly Water-Dispersible Surface-Modified Gd(2)O(3) Nanoparticles for Potential Dual-modal Bioimaging.” Chemistry (Weinheim an Der Bergstrasse, Germany) 19 (38): 12658–12667. doi:https://doi.org/10.1002/chem.201301687.
- Hussain, S., F. Al-Nsour, A. B. Rice, J. Marshburn, Z. Ji, J. I. Zink, B. Yingling, N. J. Walker, and S. Garantziotis. 2012. “Cerium Dioxide Nanoparticles Do Not Modulate the Lipopolysaccharide-Induced Inflammatory Response in Human Monocytes.” International Journal of Nanomedicine 7: 1387–1397.
- Hussain, S., F. Al-Nsour, A. Rice, J. Marshburn, B. Yingling, Z. Ji, J. Zink, N. Walker, and S. Garantziotis. 2012. “Cerium Dioxide Nanoparticles Induce Apoptosis and Autophagy in Human Peripheral Blood Monocytes.” ACS Nano 6 (7): 5820–5829. doi:https://doi.org/10.1021/nn302235u.
- Ibuki, Y., and T. Toyooka. 2012. “Nanoparticle Uptake Measured by Flow Cytometry.” In Nanotoxicity. Methods in Molecular Biology (Methods and Protocols), edited by Reineke J. vol 926. Totowa, NJ: Humana Press. https://doi.org/https://doi.org/10.1007/978-1-62703-002-1_11.
- Jakubzick, C., G. Randolph, and P. Henson. 2017. “Monocyte Differentiation and Antigen-Presenting Functions.” Nature Reviews. Immunology 17 (6): 349–362. doi:https://doi.org/10.1038/nri.2017.28.
- Kalfopoulos , 2017. “Pathophysiology of Alpha-1 Antitrypsin Lung Disease.” In Alpha-1 Antitrypsin Deficiency. Methods in Molecular Biology, edited by Borel F., Mueller C. vol 1639. New York, NY: Humana Press.
- Karlsson, Helen, Per Leanderson, Christer Tagesson, and Mats Lindahl. 2005. “Lipoproteomics I: Mapping of Proteins in Low-Density Lipoprotein Using Two-Dimensional Gel Electrophoresis and Mass Spectrometry.” Proteomics 5 (2): 551–565. doi:https://doi.org/10.1002/pmic.200300938.
- Karmali, P., and D. Simberg. 2011. “Interactions of Nanoparticles with Plasma Proteins: implication on Clearance and Toxicity of Drug Delivery Systems.” Expert Opinion on Drug Delivery 8 (3): 343–357. doi:https://doi.org/10.1517/17425247.2011.554818.
- Khang, D., Y. Lee, E. Choi, T. Webster, and S. Kim. 2014. “Effect of the Protein Corona on Nanoparticles for Modulating Cytotoxicity and Immunotoxicity.” International Journal of Nanomedicine 10 (1): 97–113.
- Laera, S., G. Ceccone, F. Rossi, D. Gilliland, R. Hussain, G. Siligardi, and L. Calzolai. 2011. “Measuring Protein Structure and Stability of Protein-nanoparticle Systems with Synchrotron Radiation Circular Dichroism.” Nano Letters 11 (10): 4480–4484. doi:https://doi.org/10.1021/nl202909s.
- Li, C., X. Shi, L. Bao, J. Yang, A. Damirin, and J. Zhang. 2018. “The Correlation between Multiple Variable Factors and the Autocatalytic Properties of Cerium Oxide Nanoparticles Based on Cell Viability.” New Journal of Chemistry 42 (12): 9975–9986. doi:https://doi.org/10.1039/C8NJ00293B.
- Li, W., L. Xu, and J. Chen. 2005. “Co3O4 Nanomaterials in Lithium-Ion Batteries and Gas Sensors.” Advanced Functional Materials 15 (5): 851–857. doi:https://doi.org/10.1002/adfm.200400429.
- Lord, M., M. Jung, W. Teoh, C. Gunawan, J. Vassie, R. Amal, and J. Whitelock. 2012. “Cellular Uptake and Reactive Oxygen Species Modulation of Cerium Oxide Nanoparticles in Human Monocyte Cell Line U937.” Biomaterials 33 (31): 7915–7924. doi:https://doi.org/10.1016/j.biomaterials.2012.07.024.
- Lundqvist, M., C. Augustsson, M. Lilja, K. Lundkvist, B. Dahlbäck, S. Linse, and T. Cedervall. 2017. “The Nanoparticle Protein Corona Formed in Human Blood or Human Blood Fractions.” PLoS One 12 (4): e0175871 doi:https://doi.org/10.1371/journal.pone.0175871.
- Lundqvist, M., J. Stigler, G. Elia, I. Lynch, T. Cedervall, and K. Dawson. 2008. “Nanoparticle Size and Surface Properties Determine the Protein Corona with Possible Implications for Biological impacts.” Proceedings of the National Academy of Sciences of the United States of America 105 (38): 14265–14270. doi:https://doi.org/10.1073/pnas.0805135105.
- Lynch, I., T. Cedervall, M. Lundqvist, C. Cabaleiro-Lago, S. Linse, and K. Dawson. 2007. “The nanoparticle-protein complex as a biological entity; a complex fluids and surface science challenge for the 21st century.” Advances in Colloid and Interface Science 134-135: 167–174. doi:https://doi.org/10.1016/j.cis.2007.04.021.
- Mirshafiee, V., R. Kim, M. Mahmoudi, and M. Kraft. 2016. “The Importance of Selecting a Proper Biological Milieu for Protein Corona Analysis in Vitro: Human Plasma versus Human Serum.” The International Journal of Biochemistry & Cell Biology 75: 188–195. doi:https://doi.org/10.1016/j.biocel.2015.11.019.
- Mirzaei, S., Z. Hadadi, F. Attar, S. Mousavi, S. Zargar, A. Tajik, A. Saboury, S. Rezayat, and M. Falahati. 2018. “ROS-Mediated Heme Degradation and Cytotoxicity Induced by Iron Nanoparticles: hemoglobin and Lymphocyte Cells as Targets.” Journal of Biomolecular Structure and Dynamics 36 (16): 4235–4245. doi:https://doi.org/10.1080/07391102.2017.1411832.
- Murdock, R., L. Braydich-Stolle, A. Schrand, J. Schlager, and S. Hussain. 2008. “Characterization of Nanomaterial Dispersion in Solution Prior to in Vitro Exposure Using Dynamic Light Scattering Technique.” Toxicological Sciences: An Official Journal of the Society of Toxicology 101 (2): 239–253. doi:https://doi.org/10.1093/toxsci/kfm240.
- Nielsen, M., C. Lundegaard, P. Worning, C. Hvid, K. Lamberth, S. Buus, S. Brunak, and O. Lund. 2004. “Improved Prediction of MHC Class I and Class II Epitopes Using a Novel Gibbs Sampling Approach.” Bioinformatics (Oxford, England) 20 (9): 1388–1397. doi:https://doi.org/10.1093/bioinformatics/bth100.
- Nune, S., P. Gunda, P. Thallapally, Y. Lin, M. Forrest, and C. Berkland. 2009. “Nanoparticles for Biomedical Imaging.” Expert Opinion on Drug Delivery 6 (11): 1175–1194. doi:https://doi.org/10.1517/17425240903229031.
- Ortega, R., C. Bresson, C. Darolles, C. Gautier, S. Roudeau, L. Perrin, M. Janin, et al. 2014. “Low-Solubility Particles and a Trojan-Horse Type Mechanism of Toxicity: The Case of Cobalt Oxide on Human Lung Cells.” Particle and Fibre Toxicology 11 (1): 14 doi:https://doi.org/10.1186/1743-8977-11-14.
- Patil, N., B. Uphade, D. McCulloh, S. Bhargava, and V. Choudhary. 2004. “Styrene Epoxidation over Gold Supported on Different Transition Metal Oxides Prepared by Homogeneous Deposition–Precipitation.” Catalysis Communications 5 (11): 681–685. doi:https://doi.org/10.1016/j.catcom.2004.08.006.
- Petoral, R., F. Söderlind, A. Klasson, A. Suska, M. Fortin, N. Abrikossova, L. Selegård, P. Käll, M. Engström, and K. Uvdal. 2009. “Synthesis and Characterization of Tb3+-Doped Gd2O3 Nanocrystals: A Bifunctional Material with Combined Fluorescent Labeling and MRI Contrast Agent Properties.” The Journal of Physical Chemistry C 113 (17): 6913–6920. doi:https://doi.org/10.1021/jp808708m.
- Pisani, C., J. Gaillard, C. Dorandeu, C. Charnay, Y. Guari, J. Chopineau, J. Devoisselle, J. Armengaud, and O. Prat. 2017. “Experimental Separation Steps Influence the Protein Content of Corona around Mesoporous Silica nanoparticles.” Nanoscale 9 (18): 5769–5772. doi:https://doi.org/10.1039/c7nr01654a.
- Rombach-Riegraf, V., A. Karle, B. Wolf, L. Sordé, S. Koepke, S. Gottlieb, J. Krieg, et al. 2014. “Aggregation of Human Recombinant Monoclonal Antibodies Influences the Capacity of Dendritic Cells to Stimulate Adaptive T-Cell Responses in Vitro.” PLoS One 9 (1): e86322 doi:https://doi.org/10.1371/journal.pone.0086322.
- Schubert, B., C. Schärfe, P. Dönnes, T. Hopf, D. Marks, and O. Kohlbacher. 2018. “Population-specific design of de-immunized protein biotherapeutics.” PLoS Computational Biology 14 (3): e1005983 doi:https://doi.org/10.1371/journal.pcbi.1005983.
- Schwenk, J., G. Omenn, Z. Sun, D. Campbell, M. Baker, C. Overall, R. Aebersold, R. Moritz, and E. Deutsch. 2017. “The Human Plasma Proteome Draft of 2017: Building on the Human Plasma PeptideAtlas from Mass Spectrometry and Complementary Assays.” Journal of Proteome Research 16 (12): 4299–4310. doi:https://doi.org/10.1021/acs.jproteome.7b00467.
- Shen, M., and T. Horbett. 2001. “The Effects of Surface Chemistry and Adsorbed Proteins on Monocyte/Macrophage Adhesion to Chemically Modified Polystyrene Surfaces.” Journal of Biomedical Materials Research 57 (3): 336–345. doi:https://doi.org/10.1002/1097-4636(20011205)57:3<336::AID-JBM1176>3.0.CO;2-E.
- Shevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. “Mass Spectrometric Sequencing of Proteins Silver-stained Polyacrylamide Gels.” Analytical Chemistry 68 (5): 850–858. doi:https://doi.org/10.1021/ac950914h.
- Sisler, J., R. Li, W. McKinney, R. Mercer, Z. Ji, T. Xia, X. Wang, et al. 2015. “Differential Pulmonary Effects of CoO and La2O3 Metal Oxide Nanoparticle Responses during Aerosolized Inhalation in Mice.” Particle and Fibre Toxicology 13(1). doi:https://doi.org/10.1186/s12989-016-0155-3.
- Tavano, R., L. Gabrielli, E. Lubian, C. Fedeli, S. Visentin, P. Polverino De Laureto, G. Arrigoni, et al. 2018. “C1q-Mediated Complement Activation and C3 Opsonization Trigger Recognition of Stealth Poly(2-Methyl-2-Oxazoline)-Coated Silica Nanoparticles by Human Phagocytes.” ACS Nano 12 (6): 5834–5847. doi:https://doi.org/10.1021/acsnano.8b01806.
- Ting, S., J. Whitelock, R. Tomic, C. Gunawan, W. Teoh, R. Amal, and M. Lord. 2013. “Cellular Uptake and Activity of Heparin Functionalised Cerium Oxide Nanoparticles in Monocytes.” Biomaterials 34 (17): 4377–4386. doi:https://doi.org/10.1016/j.biomaterials.2013.02.042.
- Vasilichin, V. A., S. A. Tsymbal, A. F. Fakhardo, E. Anastasova, A. S. Marchenko, A. A. Shtil, V. Vinogradov, and E. I. Koshel. 2020. “Effects of Metal Oxide Nanoparticles on Toll-Like Receptor mRNAs in Human Monocytes.” Nanomaterials (Basel) 10 (1): 127. doi:https://doi.org/10.3390/nano10010127.
- Vogt, C., M. Pernemalm, P. Kohonen, S. Laurent, K. Hultenby, M. Vahter, J. Lehtiö, M. Toprak, and B. Fadeel. 2015. “Proteomics Analysis Reveals Distinct Corona Composition on Magnetic Nanoparticles with Different Surface Coatings: Implications for Interactions with Primary Human Macrophages.” PLoS One 10 (10): e0129008 doi:https://doi.org/10.1371/journal.pone.0129008.
- Wang, G., X. Zhang, Y. Liu, Z. Hu, X. Mei, and K. Uvdal. 2015. “Magneto-fluorescent Nanoparticles with High-intensity NIR emission, T1- and T2-weighted MR for Multimodal Specific Tumor Imaging.” Journals of Material Chemistry B 3 (15): 3072–3080. doi:https://doi.org/10.1039/c5tb00155b.
- Wang, G., X. Zhang, A. Skallberg, Y. Liu, Z. Hu, X. Mei, and K. Uvdal. 2014. “One-Step Synthesis of Water-Dispersible Ultra-Small Fe3O4 Nanoparticles as Contrast Agents for T1 and T2 Magnetic Resonance Imaging.” Nanoscale 6 (5): 2953–2963. doi:https://doi.org/10.1039/c3nr05550g.
- Warheit, D. 2008. “How Meaningful Are the Results of Nanotoxicity Studies in the Absence of Adequate Material Characterization?” Toxicological Sciences: An Official Journal of the Society of Toxicology 101 (2): 183–185. doi:https://doi.org/10.1093/toxsci/kfm279.
- Wong, K., W. Yeap, J. Tai, J. Ong, T. Dang, and S. Wong. 2012. “The Three Human Monocyte Subsets: implications for Health and Disease.” Immunologic Research 53 (1-3): 41–57. doi:https://doi.org/10.1007/s12026-012-8297-3.
- Yan, Yan,Katelyn T. Gause, Marloes M. J. Kamphuis, Ching-Seng Ang, Neil M. O'Brien-Simpson, Jason C. Lenzo, Eric C. Reynolds, Edouard C. Nice, and Frank Caruso. 2013. “Differential Roles of the Protein Corona in the Cellular Uptake of Nanoporous Polymer Particles by Monocyte and Macrophage Cell Lines.” ACS Nano 7 (12): 10960–10970. doi:https://doi.org/10.1021/nn404481f.
- Zhang, P., X. Xu, Y. Chen, M. Xiao, B. Feng, K. Tian, Y. Chen, and Y. Dai. 2018. “Protein Corona between Nanoparticles and Bacterial Proteins in Activated Sludge: Characterization and Effect on Nanoparticle Aggregation.” Bioresource Technology 250: 10–16. doi:https://doi.org/10.1016/j.biortech.2017.11.008.