Abstract
Plastic nanoparticles are widely spread in the biosphere, but health risk associated with their effect on the human organism has not yet been assessed. The purpose of this study was to determine the genotoxic potential of non-functionalized polystyrene nanoparticles (PS-NPs) of different diameters of 29, 44, and 72 nm in human peripheral blood mononuclear cells (PBMCs) (in vitro). To select non-cytotoxic concentrations of tested PS-NPs, we analyzed metabolic activity of PBMCs incubated with these particles in concentrations ranging from 0.001 to 1000 µg/mL. Then, PS-NPs were used in concentrations from 0.0001 to 100 μg/mL and incubated with tested cells for 24 h. Physico-chemical properties of PS-NPs in media and suspension were analyzed using dynamic light scattering (DLS), atomic force microscopy (AFM), scanning electron microscopy (SEM) and zeta potential. For the first time, we investigated the mechanism of genotoxic action of PS-NPs based on detection of single/double DNA strand-breaks and 8-oxo-2′-deoxyguanosine (8-oxodG) formation, as well as determination of oxidative modification of purines and pyrimidines and repair efficiency of DNA damage. Obtained results have shown that PS-NPs caused a decrease in PBMCs metabolic activity, increased single/double-strand break formation, oxidized purines and pyrimidines and increased 8oxodG levels. The resulting damage was completely repaired in the case of the largest PS-NPs. It was also found that extent of genotoxic changes in PBMCs depended on the size of tested particles and their ζ-potential value.
1. Introduction
Long-term exposure to certain chemicals can cause DNA damage. The most common types of DNA lesions are single strand breaks (SSBs), double strand breaks (DSBs), oxidative modification of DNA bases, and adducts formation (Christmann and Kaina Citation2013). They can be induced both by exogenous factors: UV radiation, ionization, and various types of chemicals, and endogenous factors, such as metabolic and replication stresses resulting in the formation of reactive oxygen species (ROS) (Felgentreff et al. Citation2021).
Nanoplastics (NPs) and microplastics (MPs) resulting from decomposition of large pieces of plastic are pollutants of global concern. Until recently, it has been considered that the diameter of plastic NPs should not exceed 100 nm, although in the latest reports the diameter of 1000 nm has been assumed as the upper limit (Gigault et al. Citation2018; Rakowski and Grzelak Citation2020; Mitrano, Wick, and Nowack Citation2021). MPs are plastic particles up to 5 mm in diameter (Hartmann et al. Citation2019). In this work, we have adopted a traditional size of particles referring to NPs, which should not exceed a diameter of above 100 nm. Among the above-mentioned particles, polypropylene (PP) and polyethylene (PE) NPs are commonly found in the environment, while polystyrene nanoparticles (PS-NPs) are determined in smaller amounts. Polystyrene is produced by polymerization of styrene monomers and it is the fifth leading thermoplastic material on the global market. This polymer is widely used in the production of food packaging, automotive industry, electronics, household appliances, and more (ChemicalSafetyFacts.org Citation2020). The key to understanding the potential toxicity of NPs is their physico-chemical properties, that is, size, shape, and surface charge. We used PS-NPs because they are commercially available in different size and well-characterized by their size, shape, and lack of functionalization to understand the principles of polymer chemistry. Additionally, using scanning electron microscopy (SEM), atomic force microscopy (AFM), and dynamic light scattering (DLS), we confirmed the size and shape of the tested particles. Additionally, we performed zeta potential measurements.
More and more studies on PS-NPs have focused on the assessment of their toxic effects to humans. This is a consequence of previous reports stating that they can penetrate into human organisms, translocate through biological barriers, enter cells environment (Xu et al. Citation2019; Z. Liu et al. Citation2021; Hwang et al. Citation2022) and interact with the cells of the immune system (including lymphocytes) (Rubio et al. Citation2020).
For this reason, PS-NPs as a model of non-functionalized particles with the smallest diameters (29, 44, and 72 nm) penetrating into the cells were selected for more detailed analyzes and evaluation of molecular mechanism of their genotoxic action.
As shown by numerous studies on animals, plastic particles (especially NPs) after penetrating organisms, may be accumulated in trace amounts in the spleen, liver, thymus, lungs, heart, reproductive organs, and the brain (European Food Safety Authority (EFSA) Citation2016; Prüst, Meijer, and Westerink Citation2020; Hwang et al. Citation2022). The main source of human exposure to plastic particles is consumption of contaminated food, primarily seafood (Hantoro et al. Citation2019; Toussaint et al. Citation2019). In addition, microplastics have also been detected in milk, honey, table salt, and mineral water (Auta, Emenike, and Fauziah Citation2017; Horton et al. Citation2017; Cox et al. Citation2019, Kutralam-Muniasamy et al. Citation2020). Campanale et al. (Citation2020) proved that plastic particles with a diameter less than 2.5 µm can reach the gastrointestinal (GI) tract by ingestion and be internalized by the GI cells by endocytosis. Plastic particles with a diameter of 50–500 µm were found in feces, and their number was approximately 20 per 10 µg of feces. Thanks to this analysis, it was estimated that 90,000 of these particles are released in the feces annually (van Raamsdonk et al. Citation2020). Other studies have indicated the presence of 12 microplastics fragments ranging from 5 to 10 µm in diameter in women placentas (Ragusa et al. Citation2021).
Plastic particles also enter the human body through the respiratory system and, to a lesser extent, through the skin (Enyoh et al. Citation2020). Particles have also been shown to be adsorbed by the lung epithelium (Asgharian, Hofmann, and Miller Citation2001; Smith et al. Citation2002). Enaud et al. (Citation2020) proved that particles, after getting into the circulatory system, showed immunological effect on the so-called gut–lung axis. It has been estimated that the daily human exposure to plastic particles ranges from 26 to 130 (Facciolà et al. Citation2021). Recently, Leslie et al. (Citation2022) showed the presence of plastic particles in human blood. Blood from 22 volunteers was tested, and the presence of particles was found in 17 of tested participants. The concentration of these xenobiotics reached as much as 12 µg/mL of blood, while the concentration of the analyzed PS alone was 4.8 µg/mL. In this study on a small group of donors, the mean blood concentration of plastic particles was 1.6 µg/mL (Leslie et al. Citation2022). However, a question arises here whether the presence of such high particle concentrations in the blood of people is an indisputable fact or an effect of contamination of the sample during collection and preparation, which has yet to be clarified. Under conditions of high exposure or high individual sensitivity, MPs can cause inflammatory changes resulting from their possible interaction with tissues. There are a small number of publications on the interaction of PS-NPs with leukocytes. Rubio et al. (Citation2020) studied the effect of PS-NPs with a diameter of about 50 nm on the population of immune cells. They used three different human leukocyte lines for analysis, including Raji-B (B lymphocytes), TK6 (lymphoblasts), and THP-1 (monocytes). The study showed low toxicity, ROS production, and genotoxicity of NPs in Raji-B and TK6 cells with less uptake of NPs. No side effects were observed in monocytes, although uptake of the tested particles was higher in this cell type. In another study, mice were exposed to MPs with a diameter of 10–150 µm at concentrations of 20 and 200 µg/g for 5 weeks. In tested animals, a decreased percentage of regulatory T cells and an increased percentage of Th17 cells in splenocytes were noted. In addition, there was a change in the level of inflammatory interleukin 1α (IL1α) and the granulocyte colony stimulating factor G-CSF (Li et al. Citation2020).
The results we have obtained so far, have shown that tested PS-NPs disturbed the redox balance, increased the total level of ROS and highly ROS (a hydroxyl radical), as well as induced lipid and protein oxidation in human PBMCs (Kik et al. Citation2021). Our recent study on the genotoxic potential of PS-NPs is justified considering the results obtained so far, which demonstrated an increase in ROS, in particular hydroxyl radical level, which may contribute to DNA damage.
The aim of this research was to determine the mechanism of interaction of plastic PS-NPs with human peripheral blood mononuclear cells (PBMCs). PBMCs due to large nucleus, and a key role playing in the immune system are very often used in the studies that evaluate genotoxic potential of various xenobiotics. Therefore, we assessed the effect of PS-NPs of different diameters (29, 44, and 72 nm) on DNA damage and repair in human PBMCs. Genotoxic damage can contribute to disorders of the immune system, which can lead to the development of cancer or autoimmune diseases (e.g. asthma, allergy; Farhat et al. Citation2011). In addition, PBMCs participate in the transport of xenobiotics that is why the analysis of the mechanism of interaction of PS-NPs with this cell type is fully justified (Santovito, Cervella, and Delpero Citation2014).
For this research, we used non-functionalized PS-NPs of different (small) diameters, which, as shown in previous studies, can enter the cells (Z. Liu et al. Citation2021) and are well-characterized in terms of their size and shape, in order to check whether these NPs can cause genotoxic effects. Our research can therefore be the starting point for further analyzes of environmentally relevant particles. In order to assess genotoxic potential of tested PS-NPs, we used mainly comet assay (single-cell gel electrophoresis) and chromatography (8-oxodG level). The cells were incubated with PS-NPs in concentrations ranging from 0.0001 to 100 µg/mL for 24 h. Preliminary studies also assessed the effect of PS-NPs (0.0001–1000 µg/mL) on the metabolic activity of PBMCs after 24 h exposure in order to select their concentrations that do not alter cell viability, and consequently may be used in genotoxicity tests. A range of PS-NPs concentrations was used, which corresponded (with the exception of 100 µg/mL) to plastic levels that were found in human blood (Leslie et al. Citation2022).
2. Materials and methods
2.1. Chemical standards
Standards of non-functionalized PS-NPs were purchased from Polysciences Europe GmbH. Restriction enzymes, that is, Endonuclease III (EndoIII) and DNA-formamidopirymidyne glycosidase (Fpg) were used for detection of oxidatively modified purines and pyrimidines, respectively (New England Biolabs). Nuclease P1 and Shrimp Alkaline Phosphatase (rSAP) were used to detect 8-OHdG and were purchased from New England Biolabs. 3-(4,5-dimethythiazol-2-yl)-2,5 diphenyl-tetrazolium bromide (MTT), DMF, and sodium dodecyl sulfate (SDS) were used to assess the metabolic activity of PBMCs and were purchased in Sigma-Aldrich (St Louis, MA, USA) and (ROTH). Other reagents: Lymphocyte Separation Medium (1.077 g/mL) and RPMI medium 1640 were bought in Biowest and Biotech. Type I agarose and type XI agarose, EDTA, Triton, fetal bovine serum needed for the assessment of DNA damage were supplied from Sigma-Aldrich, USA. The remaining reagents, that is, NaCl, NaOH, TRIS, sodium acetate, and HEPES were purchased from POCh (Poland) and Roth (Germany).
2.2. Physico-chemical characterization of polystyrene nanoparticles
Taking into account recent reports indicating that in order to obtain comparable results, in-depth characterization of material (microplastics) is needed (Ramsperger et al. Citation2022), we decided to perform physico-chemical tests of PS-NPs. We took photos using AFM and SEM and assessed PS-NPs hydrodynamic size using the DLS technique. PS-NPs were diluted in water and in RPMI medium to study their size by DLS and zeta potential. Non-functionalized PS-NPs suspensions of various diameters were diluted in RPMI medium to study their biological effects, while tested NPs diluted in water were used to assess their physical properties by AFM and SEM.
Diluted water suspensions of PS-NPs were deposited on silicon wafers prior to AFM and SEM imaging. A piece of silicon wafer was cleaned in ethanol and dried in a flow of pure air. Freshly cleaned wafers were placed vertically in a cuvette containing diluted water suspension of PS-NPs of given size and allowed to slowly evaporate for 2–3 days. Next, wafers with deposited NPs were withdrawn and dried in ambient conditions.
The AFM measurements were performed on Bruker Dimension Icon (Billerica, Massachusetts, USA) operating in tapping mode in ambient air. A non-contact Tespa V2 cantilever from Bruker operating at resonance frequency ν = 324 kHz was used. Typical image size was 2 μm × 2 μm.
Field Emission Scanning Electron Microscopy (FE-SEM) imaging was carried out using a FEI NovaNano SEM 450 microscope (Hillsboro, OR, USA) equipped with a Schottky gun operating in immersion mode using a through lens detector (TLD) at 5 kV voltage. Typical magnification was 150,000×.
Dynamic Light Scattering (DLS) measurements - stability and hydrodynamic size of PS-NPs labeled as A (29 nm), B (44 nm), and C (72 nm) were performed using a DLS (Litesizer 500, Particle Analyzer, Anton Paar, Graz, Austria), equipped with a laser of wavelength of 658 nm as the light source and scattering angle θ = 173°. Samples in water were measured without filtering in a quartz cuvette at 25 °C, while in RPMI at 37 °C.
The zeta potentials of the PS-NPs (c = 500 μg/mL) were measured using a Malvern Instruments Zetasizer Nano-ZS (Malvern Instruments Ltd., Malvern, UK). Samples in electric field were prepared in capillary cells (DTS1061). Measurement was carried out at 37 °C in water and RPMI at pH 7.4 with 5 repetitions. The zeta potential value was calculated directly from the Helmholtz–Smoluchowski equation using Malvern software (Sze et al. Citation2003).
2.3. Biological material
Human peripheral blood mononuclear cells (PBMCs) were used for this study. PBMCs were isolated from the buffy coat purchased at the Regional Center for Blood Donation and Treatment in Lodz, Poland. The purchase of blood for research is possible thanks to the contract concluded between the Institute of Biophysics of Environmental Pollution, University of Lodz, and the aforementioned Blood Donation Center. Blood Bank employees receive a coat of leukocytes and platelets through the preparation of whole blood from healthy, nonsmoking donors aged 18–30 years. The blood bank in Lodz is authorized to collect blood and separate its components based on the accreditation of the Minister of Health (No. BA/2/2004). The experiments described in this study were approved by the Bioethics Committee of University of Lodz (Resolution No. 8/KBBN-UŁ/II/2019 (08/04/2019)).
2.3.1. PBMCS isolation
The isolation of PBMCs took place in several stages. In the first stage, the buffy coat was centrifuged (600×g, 10 min, 20 °C), which allowed for the removal of plasma and collection of the resulting PBMCs layer. Further isolation was performed by centrifugation (600×g, 30 min, 20 °C) of the blood on a Lymphocyte Separation Medium with a density of 1.077 g/mL. After centrifugation, the resulting lymphocyte ring was collected, to which 3 mL of erythrocyte lysis buffer (150 mM NH4Cl, 10 mM NaHCO3, 1 mM EDTA, pH 7.4) was added. The samples were incubated for 5 min at 20 °C and supplemented with 6.5 mL of PBS. The cells were centrifuged again (200×g, 15 min, 20 °C). In the final step, the supernatant was collected, and the cells attached to the bottom of the tube were washed twice with RPMI medium containing L-glutamine and 10% fetal bovine serum. Then, centrifugation was performed (200×g, 15 min, 20 °C). The final PBMCs density used for the experiments was 5 × 104 cells/mL.
2.3.2. Isolation of DNA and determination of the 8-oxo-2′-deoxyguanosine (8-oxodG) in DNA isolates
The analyzes were performed using a method described earlier by Gackowski et al. (Citation2016) and Starczak et al. (Citation2021) with some modifications. DNA extraction and hydrolysis to deoxynucleosides: briefly, a pellet of frozen cells was dispersed in ice-cold buffer B, pH 8.0 containing Tris-HCl (10 mmol/L), Na2EDTA (5 mmol/L), and deferoxamine mesylate (0.15 mmol/L). SDS solution was added (to a final concentration of 0.5%), and the mixture was gently mixed using a polypropylene Pasteur pipette. Samples were incubated at 37 °C for 30 min. Proteinase K was added to a final concentration of 1 mg/mL and incubated at 37 °C for 1 h. The mixture was cooled to 4 °C, transferred to Phase Lock Gel – Light tubes. The mixture of phenol, chloroform, and isoamyl alcohol (25:24:1) was added (1:1 v/v) and vortexed vigorously. After extraction, the aqueous phase was treated with a chloroform:isoamyl alcohol mixture (24:1). The supernatant was treated with two volumes of cold ethanol to precipitate high molecular weight nucleic acids. The precipitate was removed with a plastic spatula, washed with 70% (v/v) ethanol and dissolved in 50 µL Milli-Q grade deionized water. The samples were mixed with 200 mM ammonium acetate containing 0.2 mM ZnCl2, pH 4.6 (1:1 v/v). Nuclease P1 (200 U, New England Biolabs, Ipswich, MA, USA) and tetrahydrouridine (10 µg/sample) were added to the mixture and incubated at 37 °C for 1 h. Subsequently, 10% (v/v) NH4OH and 6 U of shrimp alkaline phosphatase (rSAP, New England Biolabs, Ipswich, MA, USA) were added, and the samples were incubated for 1 h at 37 °C. Finally, all the hydrolysates were ultra-filtered prior to injection.
2.4. Biological properties of PS-NPs
2.4.1. Analysis of cell viability
The cytotoxic effect of the investigated PS-NPs was determined by a standard MTT method, based on the reduction of yellow 3-(4,5-dimethythiazol-2-yl)-2,5 diphenyl-tetrazolium bromide (MTT) by cellular reductase, mainly mitochondria succinate dehydrogenase. The reduction of MTT to formazan is possible due to the NAD(P)H-dependent oxidoreductase enzymes that are present in living cells. Insoluble formazan crystals are dissolved in the solubilization buffer. The study was performed in a 96-well plate after 24 h of incubation of PS-NPs with PBMCs at a density of 1 × 105 cells/well. After this time, 20 µL of MTT was added and the cells were incubated for 3–4 h. Then 100 µL of 20% SDS and 50% DMF were added and the incubation was also carried out for 24 h. In the next step, the contents of the wells were poured into the sink and 100 µL of DMSO was added per well. The absorbance was measured at 570 nm with a spectrophotometer.
2.4.2. Single cell gel electrophoresis (comet assay), as a technique for detecting single/double DNA strand breaks and their repair
Using this technique, the cells were immersed in low melting point agarose and placed on microscopic slides coated with normal melting point agarose and then lysed. Slides were then dipped in the expanding buffer. Released DNA was submitted to 20 min electrophoresis in alkaline conditions in which 17 V and 32–115 A current were applied. In neutral conditions electrophoresis was carried out for 60 min at the 9 V and 98 A current. After electrophoresis, all slides were dried and stained with 40 µL DAPI at 2 µg/mL, centrifuged prior to use for 15 min at 219×g at 4 °C. A Zeiss Axio Scope. A1 fluorescence microscope (Carl Zeiss Microscopy GmbH, Germany) with an AxioCam MR microscope camera was used to observe the comets. DNA damage was viewed using the ZEN blue program, at 20× magnification. LUCIA Comet Assay v.7.60 (Laboratory Imaging, Prague, Czech Republic) software was used to count the comets. For each concentration, at least 50 comets were counted, so, for one donor the total number of comets was approx. 1000 (7 concentrations × 50 comets × 3 types of NPs). The mean value of DNA in the comet’s tail was taken as the DNA damage index (Tail DNA%; Tice et al. Citation2000; Woźniak and Błasiak Citation2003).
2.4.3. Single and double DNA strand breaks measured by the alkaline and neutral versions of the comet test
The comet assay enables the identification of single/double DNA strand-breaks according to the procedure of Singh et al. (Citation1988) and Singh and Stephens (Citation1997) with some modifications (Klaude et al. Citation1996). After 24 h of exposure of PBMCs to PS-NPs, the samples were centrifuged at 112×g for 5 min at 4 °C. The cells were immersed in 0.75% low melting point (LMP) agarose and placed on microscopic slides coated with 0.5% normal melting point (NMP) agarose. The slides prepared in this way were placed in the lysis buffer (2.5 M NaCl, 0.1 M EDTA, 10 mM Tris, 1% Triton X-100, pH 10) for 1.5 h at 4 °C. After the lysis had been completed, the slides were rinsed 2–3 times with the expanding buffer (300 mM NaOH and 1 mM EDTA, pH > 13) and left in this buffer for 20 min. Then, fresh electrophoretic buffer (300 mM NaOH and 1 mM EDTA) was prepared and electrophoresis in alkaline conditions was performed for 20 min. For the detection of double-stranded lesions, the neutral version of the comet assay was applied, with a buffer consisting of 100 mM of TRIS and 300 mM of sodium acetate, pH 9. The duration of electrophoresis in the neutral version was 60 min.
2.4.4. Oxidative modification of purines and pyrimidines
Modification of the comet assay using restriction enzymes, i.e. endonuclease III (EndoIII) and formamidopirymidyne DNA glycosylase (Fpg) allows for detection of oxidatively modified purines and pyrimidines, respectively. The course of the experiment after the 24-h incubation of the tested cells with NPs was the same as in section 2.4.1, up to cell lysis. After lysis, slides were washed several times with HEPES buffer (40 mM HEPES-KOH, 0.5 mM Na2EDTA, 0.1 KCl, 0.2 mg/mL BSA, pH 9). Then, 50 µL of a buffer containing 1 U EndoIII or Fpg was applied to each slide. Slides were covered with coverslips and incubated at 37 °C for 30 min in a moist chamber. Finally, the coverslips used in the previous step were removed. The obtained results of DNA strand breaks formation and DNA base oxidation were expressed as percent of DNA in the comet tail versus the concentration of the individual tested substance. For this analysis, the alkaline version of the comet assay was used. Based on literature data, we decided to use 1 U of each enzyme per gel, which guaranteed their utilization in excess (therefore, the calibration curve was not prepared; Czarny et al. Citation2015).
2.4.5. Detection of 8-oxodG: 2D-UPLC-MS/MS analysis
DNA hydrolysates were spiked with an internal standard [15N5]-8-oxo-2′-deoxyguanosine ([15N5]-8-oxodG) at a volumetric ratio of 4:1 to a final concentration of 50 fmol/µL. Chromatographic separation was performed with a Waters ACQUITY 2D-UPLC system with a photodiode array detector for the first dimension of the 2D-chromatography (used for quantification of the unmodified deoxonucleosides) and a Xevo TQ-S tandem quadrupole mass spectrometer (used for the second dimension of the 2D-chromatography to analyze 8-oxodG). The at-column dilution technique was used between the first and second dimensions to improve the retention on the trap/transfer column. Separation was performed with a gradient elution for 10 min using a mobile phase of 0.05% acetate (A) and acetonitrile (B) (0.7–5% B for 5 min, column washing with 30% acetonitrile and re-equilibration with 99% A for 3.6 min). The flow rate for the second dimension was 0.3 mL/min. The separation was performed with a gradient elution for 10 min using a mobile phase of 0.01% acetate (A) and methanol (B) (1–50% B for 4 min, isocratic flow of 50% B for 1.5 min, and re-equilibration with 99% A until the next injection). All samples were analyzed with three to five technical replicates, of which the technical mean was used for further calculation. Mass spectrometric detection was performed using a Waters Xevo TQ-S or TQ-XS tandem quadrupole mass spectrometer equipped with an electrospray ionization source. Collision-induced dissociation was obtained using argon 6.0 at 3 × 10−6 bar pressure as the collision gas. Transition patterns for all the analyzed compounds and the specific detector settings were determined using the MassLynx 4.1 IntelliStart feature set in a quantitative mode to ensure the best signal-to-noise ratio and a resolution of 1 at MS1 and 0.75 at MS2 (Gackowski et al. Citation2016).
2.4.6. DNA repair
RPMI 1640 medium with l-glutamine heated to 37 °C was added to cells exposed to PS-NPs for 24 h. DNA repair was assessed by the extent of residual DNA damage detection at each time point (0, 30, 60, 90, and 120 min) using the alkaline version of the comet assay. The samples were prepared as described above.
2.5. Statistical analysis
The Shapiro-Wilk test was used to verify the results for normality. In the next step, one-way analysis of variance (ANOVA) and Tuckey’s post-hoc test were performed to assess the significance of differences between the means. Reproducibility of the results was obtained by carrying out tests in five replications (blood from five donors). From the results, average of at least three repetitions is drawn. Only for chromatographic analysis, the unpaired Student’s t test (two-tailed) was used. The results are presented as mean ± SD. All tests were performed at the level of significance of the data α = 0.05. Statistical analyzes were performed with the Statistica 13 software (StatSoft Inc., Tulsa, OK, USA).
3. Results
3.1. Physico-chemical characterization of PS
In order to confirm the shape, size, and behavior of PS-NPs in a suspension and as-deposited on the substrate surface, two types of microscopy techniques, dynamic scattering physical method and analysis of zeta potential were used.
3.1.1. Ps-NPs characterization by DLS
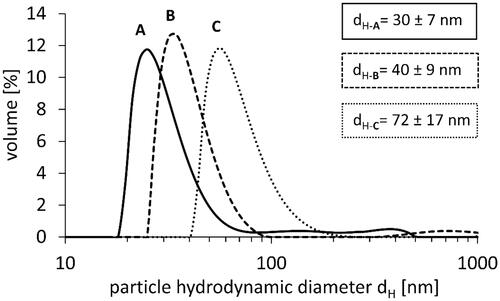
DLS measurements were performed to characterize the hydrodynamic diameter and size distribution of PS-NPs in water (). The measurements in water revealed that PS-NPs formed stable suspensions, which exhibited homogenous size distribution. The hydrodynamic particle size in water (dH) equaled to dH-A = 30 ± 7 nm, dH-B = 40 ± 9 nm, and dH-C = 72 ± 17 nm, and corresponded well to size of NPs provided by the manufacturer: 29, 44, and 72 nm, respectively.
We obtained different results when analyzing PS-NPs in the cellular medium containing albumin. The hydrodynamic particle size (dH) in RPMI medium equaled to dH-A = 95 ± 4 nm, dH-B = 60 ± 5 nm, and dH-C = 63 ± 5 nm.
The agglomeration of PS-NPs would be visible in DLS measurements as additional peaks at larger dimensions (sizes). In our measurements in water and in buffer (Kik et al. Citation2021) such peaks were not visible and only signals from particles having defined size were visible indicating that PS-NPs did not form agglomerates in water.
Differently, agglomeration of PS-NPs in RPMI medium was observed, especially in the case of the smallest particles. For 26 nm PS-NPs, we observed 3 peaks and a diameter 3 times greater than declared by the manufacturer ().
Table 1. The size of PS-NPs obtained by measurement with DLS, AFM, and SEM techniques.
3.1.2. Ps-NPs characterization by AFM
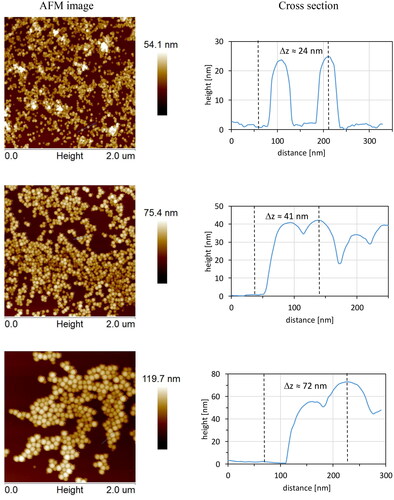
AFM images of PS-NPs deposited on Si surface showing their morphology and approximated size are presented in . It was found that PS-NPs were unevenly distributed over the Si surface. Most of them formed local close-packed agglomerates, due to the strong inter-particle and surface-particle attracting interactions. However, part of the particles were also visible as single objects. Estimated heights of selected PS-NPs (marked by blue lines in AFM images and presented as cross-sectional graphs) were: 24, 44, and 72 nm. AFM imaging revealed that the size of PS-NPs did not change much independently whether the measurement was performed on samples prepared as a deposit on the surface in the air or in a suspension by DLS. Moreover, obtained size was practically the same as a size of PS-NPs obtained in DLS technique in water.
3.1.3. Ps-NPs characterization by SEM
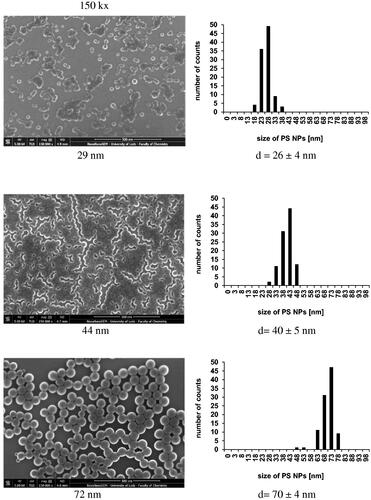
Further analysis of PS-NPs deposited on Si wafer was performed with the use of FE-SEM. presents images of PS-NPs at magnifications of 150 k× (left column) and PS-NPs size distribution (right column). It was found that PS-NPs form various flat nanostructures depending of their size. Small-size PS-NPs (29 nm) were visible mostly as separated single objects and as agglomerates exhibiting irregular shapes at low number. PS-NPs having medium size formed closely packed meander-like structures. Finally, the largest PS-NPs formed two-dimensional islands having the size of several microns built of closely inter-connected particles. The organization of these particles on the surface indicated for strong interactions of PS-NPs with a substrate but also between individual particles due to adhesive interactions occurring during and after deposition. The average size distribution of PS-NPs from SEM images is presented on histograms and corresponds to the results obtained in DLS and AFM measurements. The size of NPs obtained by application of various techniques is gathered in .
3.1.4. Differences in particles’ ζ-potentials
We measured the ζ-potentials at pH 7.4 of each particle type after their incubation in water and in RPMI medium (). The incubation of all tested PS-NPs in RPMI medium showed a significant change in ζ-potential dependly on the particle size from −41 ± 3 mV (for the smallest particles of 29 nm) to −56 ± 2 mV (for the largest particles of 71 nm). In contrast, the absolute value of ζ-potential in the water slightly lowered with increasing diameter from −40 ± 1 mV (for the smallest particles of 29 nm) to −36 ± 1 mV (for the largest particles of 72 nm; ).
Table 2. The level of ζ-potential of PS-NPs in water and in cell medium (RPMI), pH 7.4.
3.2. MTT assay
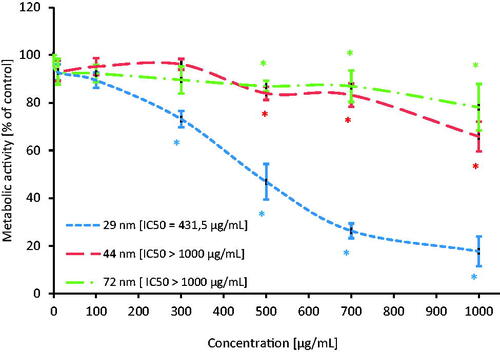
Metabolic activity of PBMCs after 24-h exposure to PS-NPs was assessed by means of the tetrazole salt reduction test (MTT test). The research showed a statistically significant decrease in metabolic activity for all tested particles. NPs with a diameter of 29 nm caused a decrease in the tested parameter from the concentration of 300 µg/mL, while NPs with a diameter of 44 and 72 nm from the concentration of 500 µg/mL depleted the examined parameter (). The IC50 concentration was determined (). The IC50 concentration for the smallest NPs was 431.5 µM, while for the 44 and 72 nm NPs it was over 1000 µM. Based on determined viability, PS-NPs at the concentrations that did not significantly change cell viability (≤ 100 µg/mL) were taken for further analysis.
3.3. Single and double DNA strand break: alkaline and neutral version of the comet assay
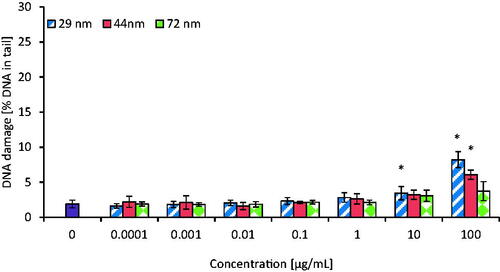
Formation of single (SSBs) and double DNA strand breaks (DSBs) was assessed in PBMCs incubated for 24 h with PS-NPs in concentrations ranging of 0.0001–100 µg/mL. All tested NPs caused DNA single/double-strand breaks formation. NPs with a diameter of 29 nm caused statistically significant changes in DNA integrity from the concentration of 0.01 µg/mL, while NPs with a diameter of 44 and 72 nm from the concentrations of 0.1 and 10 µg/mL caused DNA damage, respectively ().
Figure 5. The level of DNA strand-breaks in human PBMCs incubated for 24 h with PS-NPs of 29, 44, and 72 nm in diameter in the range of concentrations of 0.0001-100 µg/mL. Statistically significant changes for p < 0.05* (n = 5).
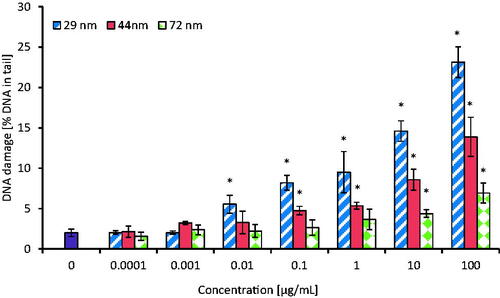
The level of DNA damage in tested cells (expressed as % of DNA in comet tail) after their exposure to the highest concentration (100 µg/mL) of PS-NPs was 23.15 ± 1.89 for 29 nm NPs, as well as 13.88 ± 2.43 and 6.93 ± 1.23 for 44 and 72 nm PS-NPs, respectively, as shown in .
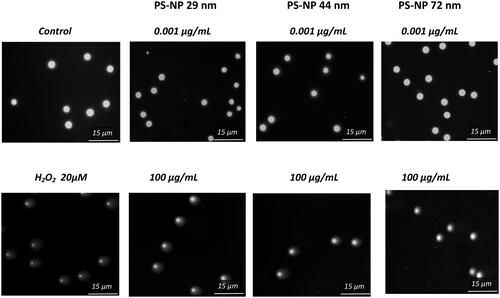
Figure 6. Selected photos showing the level of single and double strand-breaks (DNA in comet tail) in human PBMCs incubated for 24 h with PS-NPs of 29, 44, and 72 nm in diameter at two selected concentrations of 0.001 and 100 µg/mL versus the control. Photos were taken with a Zeiss Axio Scope. A1 fluorescence microscope.
DSBs, which are one of the most destructive forms of DNA helix damage, were detected after exposure of PBMCs to the smallest NPs (29 nm) at 10 and 100 µg/mL (% of DNA in comet tail − 3.44 ± 0.93 and 8.22 ± 1.13, respectively) and PS-NPs of intermediate size (44 nm) at 100 µg/mL (% of DNA in comet tail − 6.08 ± 0.66). No changes in double-stranded lesions were found after PBMCs treatment with NPs with a diameter of 72 nm (% of DNA in the tail − 3.74 ± 1.36 versus control cells − 1.91 ± 0.54%; ). The comparison of the tested substances showed that PS-NPs with a diameter of 29 nm caused the highest level of DNA strand breaks formation in human PBMCs.
3.4. Oxidative modifications of purines and pyrimidines
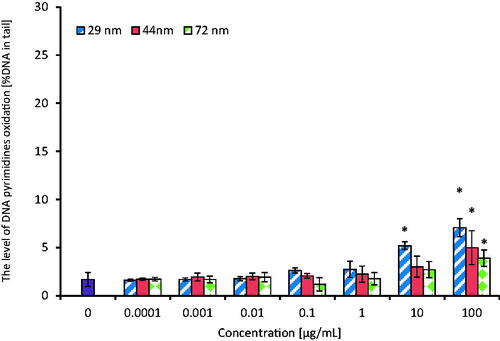
Detection of oxidative DNA damage involving purines and pyrimidines oxidation was done after exposure of PBMCs to PS-NPs in concentrations range from 0.0001 to 100 µg/mL for 24 h. The cells were then treated with repair enzymes: enodunuclease III (EndoIII) or formamidopyrimidine N-glycosylase (Fpg). The analysis of this parameter showed the formation of oxidized purine and pyrimidine bases in PBMCs after their treatment with tested PS-NPs with a higher level of oxidative damage detected by the inclusion of Fpg ( and ). A statistically significant increase in the level of purine oxidation occurred in PBMCs treated with the smallest NPs (29 nm) from the concentration of 0.01 µg/mL, while bigger particles (44 and 72 nm) increased this parameter from 10 and 100 µg/mL, respectively. After treatment of the cells with EndoIII, the increase in the level of pyrimidine oxidation occurred at higher PS-NPs concentrations in comparison to samples treated with Fpg. It was found that NPs of 29 nm increased pyrimidine oxidation from 10 µg/mL, while NPs of bigger sizes (44 and 72 nm) raised this parameter from 100 µg/mL. The presence of oxidized bases was expressed as % of DNA in the comet tail.
Figure 8. The level of DNA purines oxidation in human PBMCs (analysis by means of alkaline version of the comet assay with formamidopyrimidine-DNA glycosylase). The cells were incubated for 24 h with PS-NPs of 29, 44, and 72 nm in diameter in the range of concentrations of 0.0001–100 µg/mL. The value of comet tail (damaged DNA) in the presence of either enzyme for different concentrations of PS-NPs was reduced by the value obtained in comet assay without the enzyme (value for enzymatic buffer for the appropriate concentration of PS-NPs). Statistically significant changes for p < 0.05* (n = 5).
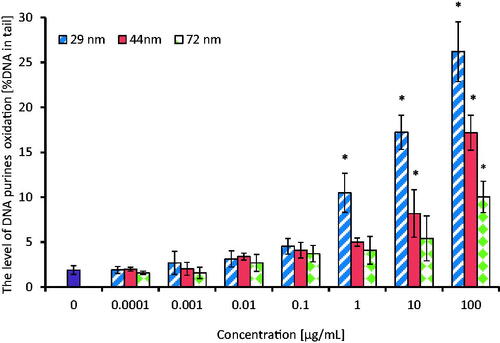
Figure 9. The level of DNA pyrimidines oxidation in human PBMCs (analysis by means of alkaline version of the comet assay with endonuclease III). PBMCs were incubated for 24 h with PS-NPs of 29, 44, and 72 nm in diameter in the range of concentrations of 0.0001–100 µg/mL. The value of comet tail (damaged DNA) in the presence of either enzyme for different concentrations of PS-NPs was reduced by the value obtained in comet assay without the enzyme (value for enzymatic buffer for the appropriate concentration of PS-NPs). Statistically significant changes for p < 0.05* (n = 5).
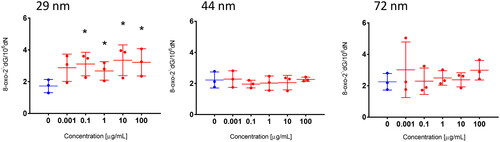
3.5. Detection of 8-oxodG
The most important DNA oxidation biomarkers are oxidized products of guanine and deoxyguanosine, which include 8-oxo-2′-deoxyguanosine (8-oxodG). To detect 8-oxodG, PBMCs were exposed to PS-NPs in concentration range from 0.0001 to 100 µg/mL for 24 h. The cells were subsequently frozen. In the next stage, DNA isolation was performed, as described in the Section 2.1.2, and the tested parameter was determined in the obtained DNA isolates. Two-dimensional (2D) liquid chromatography was used for the 8-oxodG analysis. Changes in the tested parameter were observed only after PBMCs exposure to the smallest size NPs. A statistically significant increase in 8-oxodG occurred from their concentration of 0.1 µg/mL ().
3.6. DNA repair
The time course of DNA damage repair kinetics of SSBs and DSBs was carried out after 24 h of the exposure of PBMCs to PS-NPs, which, after particles removal, were resuspended in heated RPMI 1640 medium with l-glutamine. The cells were post-incubated at 37 °C at selected time intervals (30, 60, 90, and 120 min). The resulting DNA damage caused by the largest NPs was completely repaired, and significant extent of DNA damage repair was observed in PBMCs treated with NPs with a diameter of 29 and 44 nm. A statistically significant decrease in DNA damage occurred in PBMCs treated with 29 and 44 nm NPs after 30 min of post-incubation, while in the case of 72 nm particles after 60 min of post-incubation ().
Figure 11. Time course of the repair kinetics of DNA damage (SSBs and DSBs), measured as DNA in comet tail of PBMCs treated for 24 h with PS-NPs of 29, 44, and 72 nm in diameter in the concentration of 100 μg/mL, and then post-incubated for 2 h in medium deprived of tested particles. (*) Statistically significant different from control (p < 0.05). (#) Statistically significant different from time "0" for individual PS-NPs size (p < 0.05). Each value represents the mean ± SD (n = 5).
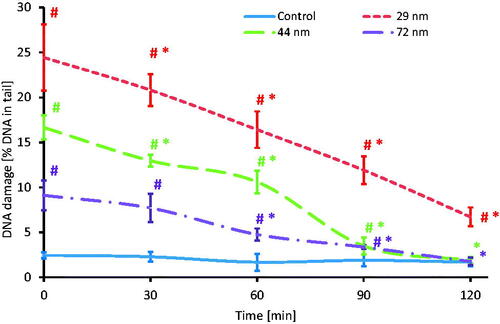
DNA damage in PBMCs, amounting to 24.43% (t = 0 min) caused by the smallest NPs, decreased to 6.72% (t = 120 min), which showed that there was no complete DNA repair.
For particles with a diameter of 44 nm, DNA damage decreased from 16.65 (t = 0 min) to 1.88% (t = 120 min), which was comparable to damage level (2.45%) in control samples. Similarly, slight damage induced by 72 nm NPs was completely repaired: from 9.11 (t = 0 min) to 1.75% (t = 120 min).
4. Discussion
Environmental xenobiotics can cause genome instability as a result of significant chromosome rearrangements, such as polyploidy, aneuploidy, gene amplification, DNA strand breaks, and disruptions in repair of these breaks. In addition, they induce changes in genes that regulate cell growth and proliferation, and thus may lead to neoplastic transformations (Tubbs and Nussenzweig Citation2017; Alimba et al. Citation2021), therefore the mechanism of genotoxic action of PS-NPs on PBMCs appears to be key in assessing their toxicity.
Damage to human DNA leads to disturbances in numerous cellular processes, which may result in various disorders, including cancer development. The assessment of genotoxic potential of xenobiotics influencing human organisms is crucial for the evaluation of human safety. Some reports have shown carcinogenic effect of styrene, but there are no available data regarding its polymer derivative - polystyrene (Huff and Infante Citation2011). Animal studies on carcinogenic potential of styrene have provided conflicting results and limited evidence. There are several epidemiological studies suggesting a possible association between the exposure to styrene and an increased risk of leukemia and lymphoma in humans (Thompson et al. Citation2016). However, those pieces of evidence are not unequivocal, because of the simultaneous exposure of humans to numerous other chemicals, and insufficient data concerning their levels and exposure times.
There is no study that has assessed genotoxic mechanism of action of PS-NPs or other plastic NPs in any cell type. Therefore, we decided for the first time to determine genotoxic effect of non-functionalized PS-NPs of different diameters on human PBMCs, focusing on single and double strand-breaks formation, oxidative damage to purines and pyrimidines, and changes in 8-oxo-2′-deoxyguanosine level and repair capacity of the resulting DNA damage.
PS-NPs may accumulate in the human body and exert toxicity by inducing or enhancing an immune response. Chronic exposure is anticipated to be of greater concern due to the accumulative effects of PS-NPs action that could occur in the human body. This is expected to be dose-dependent, and a robust evidence base of exposure levels is currently lacking (Wright and Kelly Citation2017). The exposure of humans to plastic and PS (determined even in the placenta of women, Ragusa et al. Citation2021) and far insufficient data on the adverse effects of these substances, persuade us to conduct studies that assess toxic effects of PS-NPs on human blood cells and describe the underlying mechanisms of their action. PS particles are usually used as model substances in the studies of the effect of characteristic particle surfaces on various biological parameters, because they can be easily synthesized over a broad range of sizes. NPs are characterized by a higher surface in relation to their volume, which has an important effect on their reactivity (Xia et al. Citation2008).
Questions about the genotoxicity of plastic particles are constantly arising. Avio et al. (Citation2015) suggest that MPs can induce genomic instability through DNA damage, such as DNA strand breaks and micronucleus formation or chromosomal aberrations. According to some scientific reports, MPs can cause DNA damage both through direct interactions with genetic material and indirect interactions, such as ROS or toxic ions formation. It was shown that exposure of NIH 3T3 cells to cationic functionalized PS-NPs caused changes during mitosis in their cell cycle, leading to an extension of the G0/G1 phase, which in turn led to DNA damage (Hu and Palić Citation2020). In other studies, increased chromosomal aberrations have been observed in plant root meristematic cells (Maity et al. Citation2020) and mammalian cell lines (Poma et al. Citation2019; Rubio et al. Citation2020). Alimba et al. (Citation2021) proved that plastic particles could reduce gene transcription and increase DNA fragmentation. Recent studies by Sarma et al. (Citation2022) showed a reduction in the mitotic index in PBMCs after exposure to PS-NPs at high concentrations (500, 1000, and 2000 µg/mL). They also observed a significant increase in micronucleus formation, percent of cytostasis, and a decrease in the nuclear division rate. This analysis showed that PS-NPs cytotoxicity was related to oxidative stress, genotoxic activity, and genomic instability. Other studies have shown DNA damage in the blood cells of marine organisms, including Scrobicularia plana and Mytilus galloprovincialis after exposure to PS and polyethylene microparticles at 0.5, 5, and 50 µg/mL (Avio et al. Citation2015; Ribeiro et al. Citation2017).
The above reports provide evidence that MPs and NPs can directly or indirectly interact with DNA and cause damage to the genetic material in various organisms. So far, a lot of studies have been done on indirect mechanisms of NPs–DNA interaction, including PS-NPs, which have shown oxidative stress induction by generating intracellular ROS (mainly in animals) (Sun et al. Citation2018; Liu et al. Citation2020; L. Liu et al. Citation2021; Sarasamma et al. Citation2020; CitationHorie and Tabei 2021). Similarly, our previous studies revealed that the exposure of PBMCs to PS-NPs increased ROS, including hydroxyl radical level and caused lipids and protein oxidation that could lead to loss of homeostasis, and consequently DNA damage (Kik et al. Citation2021). Verster and Bouwman (Citation2018) proved that most of the studied lung tissues with neoplastic changes contained microplastics. Taking into account the above mentioned studies, which have shown the induction of ROS by PS-NPs, which are responsible for numerous DNA lesions, we decided to evaluate genotoxic properties of these substances.
An increase in ROS level, especially the hydroxyl radical, may result in the formation of an 8-oxo-2′-deoxyguanosine (8-oxodG) derivative. Therefore, using the 2D-UPLC-MS/MS analysis, we determined 8-oxodG formation in PBMCs exposed to PS-NPs. We observed 8-oxodG formation, but only under the influence of the smallest NPs with a diameter of 29 nm from the concentration of 0.1 µg/mL (). 8-oxodG is considered to be the most sensitive and useful marker of DNA oxidative damage, which can be found in many tissues and fluids because it readily penetrates into the blood (Nemtsova et al. Citation2022). It is well-established that 8-oxodG is highly mutagenic; it mispairs with adenine during DNA replication and causes GC to AT conversion, which is the most frequent type of spontaneous mutation (Dizdaroglu et al. Citation2002; Ma et al. Citation2016). 8-oxodG is by far the most extensively studied change occurs as a consequence of oxidative DNA damage (Mangal et al. Citation2009). Sokmen et al. (Citation2019) found that PS-NPs with a diameter of 20 nm were dispersed throughout the entire brain area of Danio rerio. Moreover, in these fish species, 8-oxodG occurred in neuronal cells of the brain tissue.
It is known that there is a link between DNA strand breakage and DNA base modifications (Toyokuni and Sagripanti Citation1996). Therefore, we analyzed DNA single and double-strand breaks formation using the alkaline version of the comet assay, and exclusively double-strand breaks using the neutral version of this assay. For this purpose, we used PS-NPs in the concentrations range from 0.0001 to 100 µg/mL. We showed that 24 h of incubation of PBMCs with PS-NPs (29 nm) from the concentration of 0.01 µg/mL led to significant static increase in DNA damage (SSBs and DSBs formation) in the alkaline conditions. For PS-NPs with a diameter of 44 and 72 nm, the increase in this parameter was observed from their concentration of 0.1 and 10 µg/mL, respectively (). On the other hand, using the neutral version of the comet assay, we observed an increase in the level of very undesirable DSBs in PBMCs exposed to the smallest NPs from their concentration of 10 µg/mL. These results correlated with the formation of 8-oxodG only in tests with PS-NPs with a diameter of 29 nm ().
In the next stage of the study, we examined the level of oxidative damage to DNA purines and pyrimidines with modified comet assay using the enzymes Fpg and EndoIII. We showed much greater damage to purines than to pyrimidines caused by all tested NPs. The most profound changes were found in purines of PBMCs exposed to the smallest NPs, which also correlated with DSBs and 8-oxodG formation ().
DNA damage in human white blood cells, such as lymphocytes, monocytes, and polymorphonuclear cells after ex vivo exposure to various PS-NPs concentrations (0–100 µg/mL) was also observed by Ballesteros et al. (Citation2020). They showed notable differences in the basal levels of DNA damage between various cells subpopulations. The highest level of DNA strand breaks measured with the alkaline version of the comet assay was observed in polymorphonuclear cells (% of tail DNA − 36.86 ± 9.94), while damage in monocytes and lymphocytes was 29.31 ± 9.39 % and 20.24 ± 3.61 %, respectively, after exposure to PS-NPs at their concentrations of <50 µg/mL. At the concentrations of 50 and 100 µg/mL, DNA breakage increased only in monocytes and polymorphonuclear cells.
Similarly, the analysis by Zheng, Yuan, and Chunguang (Citation2019) showed DNA damage in rat liver cells (C57BL6-J) exposed to PS-NPs at 5 and 10 µM. Other researchers detected DNA damage in the Hs27 (human fibroblasts) cell line exposed to PS-NPs at 75 µg/mL using the cytokine block micronucleus (CBMN) assay (Poma et al. Citation2019). In turn, Vecchiotti et al. (Citation2021) showed the formation of micronuclei in colorectal adenocarcinoma cells (HCT116) exposed to high concentrations (800 and 1200 µg/mL) of PS-NPs (100 nm), which indicated their low genotoxic potential. In addition, they observed formation of nuclear buds and protrusions of nucleoplasm. Other studies used white carp (Ctenopharyngodon idella), which was exposed to PS-NPs at 0.04, 34, and 34 µg/L for 20 days. The results showed DNA damage in red blood cells of studied fish (Guimarães et al. Citation2021). Subsequent studies conducted by Alaraby et al. (Citation2022) revealed a significant increase in the level of DNA damage in Drosophila larvae exposed to PS particles at doses of 0.4 and 2 mg/g of food. They also showed that the exposure to NPs with a diameter of 50 nm induced a higher level of DNA damage than in the case of particles with a diameter of 200 and 500 nm. The above mentioned results are consistent with other studies by Gopinath et al. (Citation2019), Shah et al. (Citation2020), and Brandts et al. (Citation2018) who showed that damage to the genetic material increased along with increasing doses of PS-NPs given to Allium cepa, Ctenopharyngodon Idella, and Mytilus galloprovincialis.
There are also studies showing the lack of genotoxic effect of plastic particles in various cell types. Studies by Cortés et al. (Citation2020) showed no significant effect of micro and nano PS at concentrations range from 25 to 200 μg/mL on DNA integrity in Caco-2 intestinal cells. These studies were confirmed by Domenech et al. (Citation2021) who did not notice any DNA damage in Caco-2 cells after long-term exposure to PS-NPs. Additionally, they used a modified version of the comet assay using the enzyme Fpg, which showed no tendency to increase oxidative DNA lesions. Similarly, Cole et al. (Citation2020) did not observe DNA damage in mussels (Mytilus spp.) exposed to PS-NPs with a diameter of 50 nm at 500 ng/mL.
In the last step of our study, the repair of damaged DNA in PBMCs after incubation with tested NPs was assessed. It was found that damage caused by PS-NPs of 44 and 72 nm was effectively repaired within 2 h (). In contrast, damage caused by the smallest NPs was not completely removed. Human DNA is constantly exposed to damage, which is why all living cells in the process of evolution developed various mechanisms of DNA repair, including base mismatch repair (MMR), base excision repair (BER), and nucleotide excision repair (NER). Repair by splitting non-homogeneous ends of DNA (NHEJ) and homologous recombination (HR) can also be distinguished (Carriere et al. Citation2017). Damage related to the presence of 8-oxodG is mostly repaired by the BER pathway. This modified DNA base is repaired by glycosylases, for example, OGG1 (Krokan, Standal, and Slupphaug Citation1997). The MMR pathway also plays an important role in the case of DNA damage caused by NPs. It can eliminate mutations caused by these particles, for example, repairs 8-oxodG formed as a result of guanine oxidation (Carriere et al. Citation2017). In the case of NER, DNA damage is repaired by the global genomic NER (GG-NER) and the NER is linked to transcription (Marteijn et al. Citation2014). DSBs can be repaired with NHEJ or HR, thanks to the presence of the p53 binding protein (Nakamura et al. Citation2006). NHEJ mechanism repairs double-stranded damage in a pathway dependent on POLµ and POLλ polymerases (Carriere et al. Citation2017).
In summary, tested PS-NPs caused SSBs and DSBs formation, induced oxidation of purines and pyrimidines and increased the level of 8-oxodG. The presence of the derivative 8-oxodG, and oxidation of purines and pyrimidines, as well as substantial ROS and hydroxyl radicals formation (Kik et al. Citation2021) indicate that radical mechanism is involved in the action of tested particles.
We have shown that among the non-functionalized PS-NPs, those with the smallest diameter exhibited the strongest genotoxicity, which was probably associated with their easiest penetration into tested cells, as reported by Xu et al. (Citation2019), Z. Liu et al. (Citation2021), and Hwang et al. (Citation2022).
Xu et al. (Citation2019) incubated the human alveolar epithelial A549 cell line with NPs in diameter of 25 and 70 nm and showed that smaller PS-NPs were rapidly internalized by these cells and caused greater changes in examined parameters. Similarly, studies by Z. Liu et al. (Citation2021) revealed that PS-NPs with a diameter of 50 and 500 nm entered the cells through active endocytosis due to hydrophobic interactions and van der Waals forces, as well as through passive penetration of the membrane resulting from the division of PS-NPs in the water-phospholipid system. It was proven that the smallest NPs (50 nm) were transported by RBL-2H3 cells (rat basophilic leukemia cells) via clathrin, caveolin, and macropinocytosis pathways, while larger particles (500 nm) were transported by macropinocytosis. Liu and coworkers also observed that PS-MPs with a size of 5000 nm were not adsorbed on the cell because of their too large size, which hindered their diffusion into the membrane surface. In addition, they showed that masses of the internalized PS50 inside the cells and the excreted PS50 outside the cells were both higher than the masses of PS500, indicating that the smaller particles more easily entered or leaved the cells than did their larger counterparts. According to the above mentioned study, the size of NPs may affect, among others on the kinetics and transport or the amount of particles that are taken up by the cells. Recently, Hwang et al. (Citation2022) observed that PS-NPs of 50 nm, unlike 100 nm ones, circulated in the blood vessels and accumulated in the brains of zebrafish larvae.
The value of zeta potential is very significant in PS-NPs toxicity. Musyanovych et al. (Citation2011) suggested that surface charge is very important for internalization particles by cells. They showed that PS-NPs with an anionic surfactant with a ζ-potential of −60 mV were internalized by HeLa cells more often than NPs stabilized with a nonionic surfactant having a ζ-potential of −5 mV. This was in agreement with the study of Ramsperger et al. (Citation2022) who observed that PS microplastic particles (3 μm) from Polysciences (P-MPP) with higher absolute value of negative potential zeta were more often covered by cellular membranes than PS microplastic particles (3 μm) from Micromod (MMPP), and therefore internalized by J774A.1 and ImKC macrophages. Additionally, it has been proven that particles showing a higher absolute value of negative ζ-potential induced a more significant metabolic response in a sensitive cell line and more strongly altered cell proliferation, especially at their higher concentrations.
These observations do not agree with our results, which indicate that if the particles are made of the same polymer, non-functionalized, but differ only in size, the effect is the opposite depending on the absolute value of the negative zeta potential. Increasing size of tested PS-NPs was correlated with increasing absolute value of negative zeta potential in RPMI medium, but it was not associated with higher particle cytotoxicity and genotoxicity (even weakened them). The smallest tested nanoparticles (26 nm), showed the strongest cytotoxicity and genotoxicity and had the lowest absolute value of negative zeta potential (−40.86 ± 2.77 mV), while the largest particles had the highest absolute value of negative zeta potential (−56 ± 2 mV) and exhibited the lowest cytotoxicity and genotoxicity. We observed an increase in absolute value of negative zeta potential of tested PS-NPs diluted in RPMI medium compared to those suspended in water. Probably, the observed changes in ζ-potential recorded after incubation of the particles in cell culture media may indicate on the formation of a protein corona (Partikel et al. Citation2019). Protein corona increases the diameter of the particle, but also changes the composition of the surface of the nanoparticles, and these changes affect biodistribution, efficacy, and toxicity of these substances (Breznica, Koliqi, and Daka Citation2020). The obtained diameter measured by DLS for the smallest particles (26 nm) in RPMI medium containing albumin was three times greater than that measured in water and declared by the manufacturer. The diameter of the largest 72 nm particles, measured by DLS, was the closest to that obtained in water, as well as declared by the manufacturer. Also, Gopinath et al. (Citation2019) showed that coronated-NPs with increased protein conformational changes caused higher genotoxic and cytotoxic effects in human blood cells than the virgin-NPs.
Summing up, we observed genotoxic changes in PBMCs incubated with PS-NPs (probably present in human organisms), which may raise concerns about the health of humans exposed to these substances. On the other hand, obtained data showed that tested PS-NPs caused mainly SSBs formation. Moreover, DNA damage, which was the most strongly induced by the smallest PS-NPs was repaired to a large extent, while DNA damage caused by tested NPs with a diameter of 44 and 72 nm was totally removed.
5. Conclusion
Non-functionalized PS-NPs induced SSBs and DSBs formation in PBMCs, caused oxidation of purines and pyrimidines and increased the level of 8-oxodG. The formation of the derivative 8-oxodG and oxidation of purines and pyrimidines indicate the radical mechanism of action of the tested PS-NPs. We have observed that PS-NPs with the smallest diameter and the lowest absolute value of negative zeta potentials exhibited the strongest cytotoxicity and genotoxicity, which was probably associated with their easiest penetration into tested cells.
Author contributions
K.M., B.B. and P.S. planned the experiments. K.M., P.S., E.Z., D.G., M.F., I.P. and A.K carried out the experimental part; K.M.; B.B., P.S., E.Z., D.G., M.F., I.P. and A.K analyzed the data; K.M., and P.S carried out the statistical analysis, K.M., P.S and I.P., prepared tables/figures/photos; K.M.; B.B., P.S., M.F., I.P. wrote the final manuscript.
Acknowledgment
The authors are thankful to Katarzyna Miłowska (Department of General Biophysics) that performed the measurements ζ-potential of the PS-NP using a Malvern Instruments Zetasizer Nano-ZS.
Disclosure Statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Alaraby, M., D. Abass, J. Domenech, A. Hernández, and R. Marcos. 2022. “Hazard Assessment of Ingested Polystyrene Nanoplastics in Drosophila Larvae.” Environmental Science: Nano 9 (5): 1845–1857. doi:10.1039/D1EN01199E
- Alimba, G. C., C. Faggio, S. Sivanesan, L. A. Ogunkanmi, and K. Krishnamurthi. 2021. “Micro(Nano)-Plastics in the Environment and Risk of Carcinogenesis: Insight into Possible Mechanisms.” Journal of Hazardous Materials. 416 (7): 126143. doi:10.1016/j.jhazmat.2021.126143
- Asgharian, B., W. Hofmann, and F. J. Miller. 2001. “Mucociliary Clearance of Insoluble Particles from the Tracheobronchial Airways of the Human Lung.” Journal of Aerosol Science 32 (6): 817–832. doi:10.1016/S0021-8502(00)00121-X
- Auta, H., Emenike, C, and Fauziah, S. 2017. “Distribution and Importance of Microplastics in the Marine Environment: A Review of the Sources, Fate, Effects, and Potential Solution.” Environment International 102: 165–176. doi:10.1016/j.envint.2017.02.013
- Avio, C. G., S. Gorbi, M. Milan, M. Benedetti, D. Fattorini, G. d‘Errico, M. Pauletto, L. Bargelloni, and F. Regoli. 2015. “Pollutants Bioavailability and Toxicological Risk from Microplastics to Marine Mussels.” Environmental Pollution (Barking, Essex : 1987) 198: 211–222. doi:10.1016/j.envpol.2014.12.021
- Ballesteros, S., J. Domenech, I. Barguilla, C. Cortés, R. Marcos, and A. Hernández. 2020. “Genotoxic and Immunomodulatory Effects in Human White Blood Cells after ex Vivo Exposure to Polystyrene Nanoplastics.” Environmental Science: Nano 7 (11): 3431–3446. doi:10.1039/D0EN00748J
- Brandts, I., M. Teles, A. P. Gonçalves, A. Barreto, L. Franco-Martinez, A. Tvarijonaviciute, M. A. Martins, A. M. V. M. Soares, L. Tort, and M. Oliveira. 2018. “Effects of Nanoplastics on Mytilus Galloprovincialis after Individual and Combined Exposure with Carbamazepine.” The Science of the Total Environment 643 (643): 775–784. doi:10.1016/j.scitotenv.2018.06.257
- Breznica, P., R. Koliqi, and A. Daka. 2020. “A Review of the Current Understanding of Nanoparticles Protein Corona Composition.” Medicine and Pharmacy Reports 93 (4): 342–350.
- Campanale, C., C. Massarelli, I. Savino, V. Locaputo, and V. F. Uricchio. 2020. “A Detailed Review Study on Potential Effects of Microplastics and Additives of Concern on Human Health.” International Journal of Environmental Research and Public Health 17 (4): 1212. doi:10.3390/ijerph17041212
- Carriere, M., S. Sauvaigo, T. Douki, and J.-L. Ravanat. 2017. “Impact of Nanoparticles on DNA Repair Processes: current Knowledge and Working Hypotheses.” Mutagenesis 32 (1): 203–213. doi:10.1093/mutage/gew052
- ChemicalSafetyFacts.org. 2020. [Accessed 3 Aug 2022]. https://www.chemicalsafetyfacts.org/polystyrene/
- Christmann, M, and B. Kaina. 2013. “Transcriptional Regulation of Human DNA Repair Genes following Genotoxic Stress: trigger Mechanisms, Inducible Responses and Genotoxic Adaptation.” Journal of Biological Chemistry 283: 1–5.
- Cole, M., C. Liddle, G. Consolandi, C. Drago, C. Hird, P. K. Lindeque, and T. S. Galloway. 2020. “Microplastics, Microfibres and Nanoplastics Cause Variable Sub-Lethal Responses in Mussels (Mytilus Spp.).” Marine Pollution Bulletin 160: 111552. doi:10.1016/j.marpolbul.2020.111552
- Cortés, C., J. Domenech, M. Salazar, S. Pastor, R. Marcos, and A. Hernandez. 2020. “Nanoplastics as a Potential Environmental Health Factor: Effects of Polystyrene Nanoparticles on Human Intestinal Epithelial Caco-2 Cells.” Environmental Science: Nano 7 (1): 272–285. doi:10.1039/C9EN00523D
- Cox, D. K., A. G. Covernton, L. H. Davies, F. J. Dower, F. Juanes, and S. E. Dudas. 2019. “Human Consumption of Microplastics.” Environmental Science & Technology 53 (12): 7068–7074. doi:10.1021/acs.est.9b01517
- Czarny, P., D. Kwiatkowski, D. Kacperska, D. Kawczyńska, M. Talarowska, A. Orzechowska, A. Bielecka-Kowalska, J. Szemraj, P. Gaandlstrokecki, and T. Śliwiński. 2015. “Elevated Level of DNA Damage and Impaired Repair of Oxidative DNA Damage in Patients with Recurrent Depressive Disorder.” Medical Science Monitor 6 (21): 412–418.
- Dizdaroglu, M., P. Jaruga, M. Birincioglu, and H. Rodriguez. 2002. “Free Radical-Induced Damage to DNA: Mechanisms and Measurement. FreeRadic.” Free Radical Biology & Medicine 32 (11): 1102–1115. doi:10.1016/S0891-5849(02)00826-2
- Domenech, J., M. de Britto, A. Velázquez, S. Pastor, A. Hernández, R. Marcos, and C. Cortés. 2021. “Long-Term Effects of Polystyrene Nanoplastics in Human Intestinal Caco-2 Cells.” Biomolecules 11 (10): 1442. doi:10.3390/biom11101442
- Enaud, R., R. Prevel, E. Ciarlo, F. Beaufils, G. Wieërs, B. Guery, and L. Delhaes. 2020. “The Gut-Lung Axis in Health and Respiratory Diseases: A Place for Inter-Organ and Inter-Kingdom Crosstalks.” Front Cell Infect Microbiol. Frontiers 10: 9.
- Enyoh, E. C., L. Shafea, W. A. Verla, N. E. Verla, W. Qingyue, T. Chowdhury, and M. Paredes. 2020. “Microplastics Exposure Routes and Toxicity Studies to Ecosystems: An Overview.” Environmental Analysis, Health and Toxicology 35 (1): e2020004. doi:10.5620/eaht.e2020004
- European Food Safety Authority (EFSA). 2016. “Presence of Microplastics and Nanoplastics in Food, with Particularfocus on Seafood”. EFSA Journal 14: 1–30.
- Facciolà, A., G. Visalli, P. M. Ciarello, and A. Di Pietro. 2021. “A Newly Emerging Airborne Pollutants: current Knowledge of Health Impact of Micro and Nanoplastics.” International Journal of Environmental Research and Public Health 18 (6): 2997. [Mismatch] doi:10.3390/ijerph18062997
- Farhat, S., C. Silva, M. Orione, L. Campos, A. Sallum, and A. Braga. 2011. “Air Pollution in Autoimmune Rheumatic Diseases: A Review.” Autoimmunity Reviews 11 (1): 14–21. doi:10.1016/j.autrev.2011.06.008
- Felgentreff, K., C. Schuetz, U. Baumann, C. Klemann, D. Viemann, S. Ursu, E.-M. Jacobsen, et al. 2021. “Differential DNA Damage Response of Peripheral Blood Lymphocyte Populations.” Frontiers in Immunology 12: 739675. doi:10.3389/fimmu.2021.739675
- Gackowski, D., M. Starczak, E. Zarakowska, M. Modrzejewska, A. Szpila, Z. Banaszkiewicz, and R. Olinski. 2016. “Accurate, Direct, and High-through Put Analyses of a Broad Spectrum of Endogenously Generated DNA Base Modifications with Isotope-Dilution Two-Dimensional Ultraperformance Liquid Chromatography with Tandem Mass Spectrometry: Possible Clinical Implication.” Analytical Chemistry 88 (24): 12128–12136. doi:10.1021/acs.analchem.6b02900
- Gigault, J., A. T. Halle, M. Baudrimont, P.-Y. Pascal, F. Gauffre, T.-L. Phi, H. El Hadri, B. Grassl, and S. Reynaud. 2018. “Current Opinion: What is a Nanoplastic?” Environmental Pollution (Barking, Essex : 1987) 235: 1030–1034. doi:10.1016/j.envpol.2018.01.024
- Gopinath, P. M., V. Saranya, S. Vijayakumar, M. Mythili Meera, S. Ruprekha, R. Kunal, A. Pranay, J. Thomas, A. Mukherjee, and N. Chandrasekaran. 2019. “Assessment on Interactive Prospectives of Nanoplastics with Plasma Proteins and the Toxicological Impacts of Virgin, Coronated and Environmentally Released-Nanoplastics.” Scientific Reports 9 (1): 15. doi:10.1038/s41598-019-45139-6
- Guimarães, B. T. A., Estrela, N. F., Pereira, S. P., de Andrade Vieira, E. J., de Lima Rodrigues, S. A., Silva, G. F, and Malafaia, G. 2021. “Toxicity of Polystyrene Nanoplastics in Ctenopharyngodonidella Juveniles: A Genotoxic, Mutagenic and Cytotoxic Perspective.” The Science of the Total Environment 752: 141937. doi:10.1016/j.scitotenv.2020.141937
- Hantoro, I., Löhr, A. J., Van Belleghem, F. G. A. J., Widianarko, B. and Ragas, A. M. J. 2019. “Microplastics in Coastal Areas and Seafood: implications for Food Safety.” Food Additives & Contaminants. Part A, Chemistry, Analysis, Control, Exposure & Risk Assessment 36 (5): 674–711. doi:10.1080/19440049.2019.1585581
- Hartmann, B. N., T. Hüffer, C. R. Thompson, M. Hassellöv, A. Verschoor, R. A. Daugaard, S. Rist, et al. 2019. “Are we Speaking the Same Language? Recommendations for a Definition and Categorization Framework for Plastic Debris.” Environmental Science & Technology 53 (3): 1039–1047. doi:10.1021/acs.est.8b05297
- Horie, M, and Y. Tabei. 2021. “Role of Oxidative Stress in Nanoparticles Toxicity.” Free Radical Research 55 (4): 331–342. doi:10.1080/10715762.2020.1859108
- Horton, A. A., Walton, A., Spurgeon, J. D., Lahive, E, and Svendsen, C. 2017. “Microplastics in Freshwater and Terrestrial Environments: evaluating the Current Understanding to Identify the Knowledge Gaps and Future Research Priorities.” The Science of the Total Environment 586: 127–141. doi:10.1016/j.scitotenv.2017.01.190
- Hu, M, and D. Palić. 2020. “Micro-and Nano-Plastics Activation of Oxidative and Inflammatory Adverse Outcome Pathways.” Redox Biology 37: 101620. doi:10.1016/j.redox.2020.101620
- Huff, J, and F. P. Infante. 2011. “Styrene Exposure and Risk Cancer.” Mutagenesis 26 (5): 583–584. doi:10.1093/mutage/ger033
- Hwang, K. S., Y. Son, S. S. Kim, D. S. Shin, S. H. Lim, J. Y. Yang, H. N. Jeong, B. H. Lee, and M. A. Bae. 2022. “Size-Dependent Effects of Polystyrene Nanoparticles (PS-NPs) on Behaviors and Endogenous Neurochemicals in Zebrafish Larvae.” International Journal of Molecular Sciences 23 (18): 10682. doi:10.3390/ijms231810682
- Kik, K., B. Bukowska, A. Krokosz, and P. Sicińska. 2021. “Oxidative Properties of Polystyrene Nanoparticles with Different Diameters in Human Peripheral Blood Mononuclear Cells (In Vitro Study).” International Journal of Molecular Sciences 22 (9): 4406. doi:10.3390/ijms22094406
- Klaude, M., S. Eriksson, J. Nygren, and G. Ahnstrom. 1996. “Polyribose Polymerase Deficient V79 Chinese Hamster, the Comet Assay: mechanisms and Technical Cell Line.” International Journal of Oncology 17: 955–962.
- Krokan, H. E., R. Standal, and G. Slupphaug. 1997. “DNA Glycosylases in the Base Excision Repair of DNA.” Biochemical Journal 325 (1): 1–16. doi:10.1042/bj3250001
- Kutralam-Muniasamy, G., F. Pérez-Guevara, I. Elizalde-Martínez, and V. Shruti. 2020. “Branded Milks- Are They Immune from Microplastics Contamination?” The Science of the Total Environment 714: 136823. doi:10.1016/j.scitotenv.2020.136823
- Leslie, A. H., van Velzen, M. J. M., Brandsma, H. S., Vethaak, D. A., Garcia-Vallejo, J. J. and Lamoree, M. H. 2022. “Discovery and Quantification of Plastic Particle Pollution in Human Blood.” Environment International 163: 107199. doi:10.1016/j.envint.2022.107199
- Li, B., Y. Ding, X. Cheng, D. Sheng, Z. Xu, Q. Rong, J. Wu, H. Zhao, X. Ji, and Y. Zhang. 2020. “Polyethylene Microplastics Affect the Distribution of Gut Microbiota and Inflammation Development in Mice.” Chemosphere 244: 125492. doi:10.1016/j.chemosphere.2019.125492
- Liu, Z., Y. Huang, Y. Jiao, Q. Chen, D. Wu, P. Yu, Y. Li, M. Cai, and Y. Zhao. 2020. “Polystyrene Nanoplastic Induces ROS Production and Affects the MAPK-HIF-1/NFkB-Mediated Antioxidant System in Daphnia pulex.” Aquatic Toxicology (Amsterdam, Netherlands) 220: 105420. doi:10.1016/j.aquatox.2020.105420
- Liu, L., K. Xu, B. Zhang, Y. Ye, Q. Zhang, and W. Jiang. 2021. “Cellular Internalization and Release of Polystyrene Microplastics and Nanoplastics.” The Science of the Total Environment 779: 146523. doi:10.1016/j.scitotenv.2021.146523
- Liu, Z., Y. Li, E. Pérez, Q. Jiang, Q. Chen, Y. Jiao, Y. Huang, Y. Yang, and Y. Zhao. 2021. “Polystyrene Nanoplastic Induces Oxidative Stress, Immune Defense, and Glycometabolism Change in Daphnia pulex. Application of Transcriptome Profiling in Risk Assessment of Nanoplastics.” Journal of Hazardous Materials 402: 123778. doi:10.1016/j.jhazmat.2020.123778
- Ma, B., M. Jing, P. Villalta, J. R. Kapphahn, R. S. Montezuma, A. D. Ferrington, and I. Stepanov. 2016. “Simultaneous Determination of 8-Oxo-2’-Deoxyguanosine and 8-Oxo-2’-Deoxyadenosine in Human Retinal DNA by Liquid Chromatography Nanoelectrospray-Tandem Mass Spectrometry.” Scientific Reports 6 (1): 22375. doi:10.1038/srep22375
- Maity, S., A. Chatterjee, R. Guchhait, S. De, and K. Pramanick. 2020. “Cytogenotoxic Potential of a Hazardous Material, Polystyrene Microparticles on Allium Cepa L.” Journal of Hazardous Materials 385: 121560. doi:10.1016/j.jhazmat.2019.121560
- Mangal, D., D. Vudathala, H.-J. Park, H. S. Lee, M. T. Penning, and I. A. Blair. 2009. “Analysis of 7,8-Dihydro-8-Oxo-2′-Deoxyguanosine in Cellular DNA during Oxidative Stress.” Chemical Research in Toxicology 22 (5): 788–797. 2009doi:10.1021/tx800343c
- Marteijn, J. A., H. Lans, W. Vermeulen, and J. H. Hoeijmakers. 2014. “Understanding Nucleotide Excision Repair and Its Roles in Cancer and Ageing.” Nature Reviews Molecular Cell Biology 15 (7): 465–481. doi:10.1038/nrm3822
- Mitrano, M., P. Wick, and B. Nowack. 2021. “Placing Nanoplastics in the Context of Global Plastic Pollution.” Nature Nanotechnology 16 (5): 491–500. doi:10.1038/s41565-021-00888-2
- Musyanovych, A., J. Dausend, M. Dass, P. Walther, V. Mail¨ander, and K. Landfester. 2011. “Criteria Impacting the Cellular Uptake of Nanoparticles: A Study Emphasizing Polymer Type and Surfactant Effects.” Acta Biomaterialia 7 (12): 4160–4168. doi:10.1016/j.actbio.2011.07.033
- Nakamura, K., W. Sakai, T. Kawamoto, R. T. Bree, N. F. Lowndes, S. Takeda, and Y. Taniguchi. 2006. “Genetic Dissection of Vertebrate 53BP1: A Major Role in Non-Homologous End Joining of DNA Double Strand Breaks.” DNA Repair 5 (6): 741–749. doi:10.1016/j.dnarep.2006.03.008
- Nemtsova, V., A. Shalimova, O. Kolesnikova, O. Vysotska, V. Zlatkina, and N. Zhelezniakova. 2022. “Prognostic Value of Plasma 8-Oxo-2'-Deoxyguanosine Levels in Cardio Vascular Complications Formation in Comorbidity of Arteria Hypertension and Type 2 Diabetes Mellitus.” Arterial Hyoertesion 19 (997): 1–12.
- Partikel, K., R. Korte, D. Mulac, H. U. Humpf, and K. Langer. 2019. “Serum Type and Concentration Both Affect the Protein-Corona Composition of PLGA Nanoparticles.” Beilstein Journal of Nanotechnology 10: 1002–1015. doi:10.3762/bjnano.10.101
- Poma, A., G. Vecchiotti, S. Colafarina, O. Zarivi, M. Aloisi, L. Arrizza, G. Chichiriccò, and P. Di Carlo. 2019. “In Vitro Genotoxicity of Polystyrene Nanoparticles on the Human Fibroblast Hs27 Cell Line.” Nanomaterials 9 (9): 1299–1213. doi:10.3390/nano9091299
- Prüst, M., J. Meijer, and R. H. S. Westerink. 2020. “The Plastic Brain: Neurotoxicity of Micro- and Nanoplastics. Part FibreToxicol.” BioMed Central 17: 1–16.
- Ragusa, A., A. Svelato, C. Santacroce, P. Catalano, V. Notarstefano, O. Carnevali, F. Papa, et al. 2021. “Plasticenta: First Evidence of Microplastics in Human Placenta.” Environment International 146: 106274. doi:10.1016/j.envint.2020.106274
- Rakowski, M, and A. Grzelak. 2020. “A New Occupational and Environmental Hazard- Nanoplastic.” Medycyna Pracy 71 (6): 743–756. doi:10.13075/mp.5893.00990
- Ramsperger, A. F. R. M., J. Jasinski, M. Völkl, T. Witzmann, M. Meinhart, V. Jérôme, W. P. Kretschmer, et al. 2022. “Supposedly Identical Microplastic Particles Substantially Differ in Their Material Properties Influencing Particle-Cell Interactions and Cellular Responses.” Journal of Hazardous Materials 425: 127961. doi:10.1016/j.jhazmat.2021.127961
- Ribeiro, F., R. A. Garcia, P. B. Pereira, M. Fonseca, C. N. Mestre, G. T. Fonseca, M. L. Ilharco, and M. J. Bebianno. 2017. “Microplastics Effects in Scrobicularia Plana.” Marine Pollution Bulletin 122 (1-2): 379–391. doi:10.1016/j.marpolbul.2017.06.078
- Rubio, L., I. Barguilla, J. Domenech, R. Marcos, and A. Hernandez. 2020. “Biological Effects, Including Oxidative Stress and Genotoxic Damage, of Polystyrene Nanoparticles in Different Human Hematopoietic Cell Lines.” Journal of Hazardous Materials 398: 122900. doi:10.1016/j.jhazmat.2020.122900
- Santovito, A., P. Cervella, and M. Delpero. 2014. “Chromosomal Damage in Peripheral Blood Lymphocytes from Nurses Occupationally Exposed to Chemicals.” Human & Experimental Toxicology 33 (9): 897–903. doi:10.1177/0960327113512338
- Sarasamma, S., G. Audira, P. Siregar, N. Malhotra, Y.-H. Lai, S.-T. Liang, J.-R. Chen, K. H.-C. Chen, and C.-D. Hsiao. 2020. “Nanoplastics Cause Neurobehavioral Impairments, Reproductive and Oxidative Damages and Biomarker Responses in Zebrafish: Throwing up Alarms of Wide Spread Health Risk of Exposure.” International Journal of Molecular Sciences 21 (4): 1410. doi:10.3390/ijms21041410
- Sarma, K. D., R. Dubey, M. R. Samarth, S. Shubham, P. Chowdhury, M. Kumawat, V. Verma, R. R. Tiwari, and K. Manoj. 2022. “The Biological Effects of Polystyrene Nanoplastics on Human Peripheral Blood Lymphocytes.” Nanomaterials 12 (10): 1632. doi:10.3390/nano12101632
- Shah, N., A. Khan, N. Habib Khan, and M. Khisroon. 2020. “Genotoxic Consequences in Common Grass Carp (Ctenopharyngodon Idella, (Valenciennes, 1844)) Exposed to Selected Toxic Metals.” Biological Trace Element Research 199: 305–314.
- Singh, N. P., McCoy, T.M., Tice, R. R. and Schneider, L. E. 1988. “Simple Technique for Quantitation of Low Levels of DNA Damage in Individual Cells.” Experimental Cell Research. 175 (1): 184–191. doi:10.1016/0014-4827(88)90265-0
- Singh, N. P, and E. R. Stephens. 1997. “Microgel Electrophoresis: Sensitivity, Mechanisms and DNA Electrostretching.” Mutation Research 383 (2): 167–175. doi:10.1016/S0921-8777(96)00056-0
- Smith, J. R. H., G. Etherington, A. L. Shutt, and M. J. Youngman. 2002. “ A Study of Aerosol Deposition and Clearance from the Human Nasal Passage.” Annals of Occupational Hygiene 46: 309–313.
- Sökmen, T. Ö., Sulukan, E., Türkoğlu, M., Baran, A., Özkaraca, M., Buğrahan Ceyhun, S. 2020. “Polystyrene nanoplastics (20 nm) are able to bioaccumulate and cause oxidative DNA damages in the brain tissue of zebrafish embryo (Danio rerio).” NeuroToxicology 77: 51–59,doi:10.1016/j.neuro.2019.12.010.
- Starczak, M., M. Gawronski, R. Olinski, and D. Gackowski. 2021. “Quantification of DNA Modifications Using Two-Dimensional Ultraperformance Liquid Chromatography Tandem Mass Spectrometry (2D-UPLC-MS/MS).” Methods in Molecular Biology (Clifton, N.J.) 2198: 91–108. doi:10.1007/978-1-0716-0876-0_8
- Sun, X., B. Chen, Q. Li, N. Liu, B. Xia, L. Zhu, and K. Qu. 2018. “Toxicities of Polystyrene Nano- and Microplastics toward Marine Bacterium Halomonas alkaliphila.” The Science of the Total Environment 642: 1378–1385. doi:10.1016/j.scitotenv.2018.06.141
- Sze, A., D. Erickson, L. Ren, and D. Li. 2003. “Zeta-Potential Measurement Using the Smoluchowski Equation and the Slope of the Current-Time Relationship in Electroosmotic Flow.” Journal of Colloid and Interface Science 261 (2): 402–410. doi:10.1016/S0021-9797(03)00142-5
- Thompson, J., J. Quigley, N. Halfpenny, D. Scott, and N. Hawkins. 2016. “Importance and Methods of Searching for e-Publications a Head of Print in Systematic Reviews.” Evidence-Based Medicine 21 (2): 55–59. doi:10.1136/ebmed-2015-110374
- Tice, R. R., E. Agurell, D. Anderson, B. Burlinson, A. Hartmann, H. Kobayashi, Y. Miyamae, E. Rojas, J.-C. Ryu, and F. Y. Sasaki. 2000. “Single Cell Gel/Comet Assay: Guidelines for in Vitro and in Vivo Genetic Toxicology Testing.” Environmental and Molecular Mutagenesis 35 (3): 206–221. doi:10.1002/(SICI)1098-2280(2000)35:3<206::AID-EM8>3.0.CO;2-J
- Toussaint, B., B. Raffael, A. Angers-Loustau, D. Gilliland, V. Kestens, M. Petrillo, M. I. Rio-Echevarria, and G. van den Eede. 2019. “Review of Micro- and Nanoplastic Contamination in the Food Chain.” Food Additives & Contaminants. Part A, Chemistry, Analysis, Control, Exposure & Risk Assessment 36 (5): 639–673. doi:10.1080/19440049.2019.1583381
- Toyokuni, S, and L.-J. Sagripanti. 1996. “Association between 8-Hydroxy-2′-Deoxyguanosine Formation and DNA Stand Break Mediated by Copper and Iron.” Free Radical Biology and Medicine 6: 859–864.
- Tubbs, A, and A. Nussenzweig. 2017. “Endogenous DNA Damage as a Source of Genomic Instability in Cancer.” Cell 168 (4): 644–656. doi:10.1016/j.cell.2017.01.002
- van Raamsdonk, L. W. D., M. van der Zande, A. A. Koelmans, R. L. A. P. Hoogenboom, R. J. B. Peters, M. J. Groot, M. C. A. A. Peijnenburg, and A. J. Y. Weesepoel. 2020. “Current Insights into Monitoring, Bioaccumulation, and Potential Health Effects of Microplastics Present in the Food Chain.” Foods 9 (1): 72. doi:10.3390/foods9010072
- Vecchiotti, G., S. Colafarina, M. Aloisi, O. Zarivi, P. Di Carlo, and A. Poma. 2021. “Genotoxicity and Oxidative Stress Induction by Polystyrene Nanoparticles in the Colorectal Cancer Cell Line HCT116.” PloS One 16 (7): e0255120. doi:10.1371/journal.pone.0255120
- Verster, C, and H. Bouwman. 2018. “Contextualization of Airborne Microplastic Pollution in the South African Environment.” In Proceedings of the 2018 Conference of the National Association for Clean Air. Riverside Sun, Vanderbijlpark, Gauteng.
- Woźniak, K., and J. Błasiak. 2003. “In Vitro Genotoxicity of Lead Acetate: induction of Single and Double DNA Strand Breaks and DNA-Protein Cross-Links.” Mutation Research 535 (2): 127–139. doi:10.1016/S1383-5718(02)00295-4
- Wright, L. S, and J. F. Kelly. 2017. “Plastic and Human Health: A Micro Issue?” Environmental Science & Technology 51 (12): 6634–6647. doi:10.1021/acs.est.7b00423
- Xia, T., M. Kovochich, M. Liong, J. Zink, and A. Nel. 2008. “Cationic Polystyrene Nanosphere Toxicity Depends on Cell-Specific Endocytic and Mitochondrial Injury Pathways.” ACS Nano 2 (1): 85–96. doi:10.1021/nn700256c
- Xu, M., G. Halimu, Q. Zhang, Y. Song, X. Fu, Y. Li, Y. Li, and H. Zhang. 2019. “Internalization and Toxicity: A Preliminary Study of Effects of Nanoplastic Particles on Human Lung Epithelial Cell.” The Science of the Total Environment 694: 133794. doi:10.1016/j.scitotenv.2019.133794
- Zheng, T., D. Yuan, and L. Chunguang. 2019. “Molecular Toxicity of Nanoplastics Involving in Oxidative Stress and Desoxyribonucleic Acid Damage.” Journal of Molecular Recognition : JMR 32 (11): e2804. doi:10.1002/jmr.2804