Abstract
Nanomaterials (NMs) offer plenty of novel functionalities. Moreover, their physicochemical properties can be fine-tuned to meet the needs of specific applications, leading to virtually unlimited numbers of NM variants. Hence, efficient hazard and risk assessment strategies building on New Approach Methodologies (NAMs) become indispensable. Indeed, the design, the development and implementation of NAMs has been a major topic in a substantial number of research projects. One of the promising strategies that can help to deal with the high number of NMs variants is grouping and read-across. Based on demonstrated structural and physicochemical similarity, NMs can be grouped and assessed together. Within an established NM group, read-across may be performed to fill in data gaps for data-poor variants using existing data for NMs within the group. Establishing a group requires a sound justification, usually based on a grouping hypothesis that links specific physicochemical properties to well-defined hazard endpoints. However, for NMs these interrelationships are only beginning to be understood. The aim of this review is to demonstrate the power of bioinformatics with a specific focus on Machine Learning (ML) approaches to unravel the NM Modes-of-Action (MoA) and identify the properties that are relevant to specific hazards, in support of grouping strategies. This review emphasizes the following messages: 1) ML supports identification of the most relevant properties contributing to specific hazards; 2) ML supports analysis of large omics datasets and identification of MoA patterns in support of hypothesis formulation in grouping approaches; 3) omics approaches are useful for shifting away from consideration of single endpoints towards a more mechanistic understanding across multiple endpoints gained from one experiment; and 4) approaches from other fields of Artificial Intelligence (AI) like Natural Language Processing or image analysis may support automated extraction and interlinkage of information related to NM toxicity. Here, existing ML models for predicting NM toxicity and for analyzing omics data in support of NM grouping are reviewed. Various challenges related to building robust models in the field of nanotoxicology exist and are also discussed.
1. Introduction
Engineering physicochemical properties of nanomaterials (NMs) such as size, morphology or surface chemistries has become common practice in order to meet the needs of specific applications. This has resulted in a large variety and a steadily increasing number of different NMs or nanoforms (NFs), as defined in specific regulatory frameworks (EC Citation2006). However, adjusting NM physicochemical properties does not only impact their desired functionalities but the fine-tuning also influences their original or expected behavior in a biological milieu including, their uptake by cells, biodistribution, dissolution rate and/or toxicity to humans or the environment, and more. To overcome the need to fully characterize each and every NM variant for all possible toxicological outcomes, the European chemicals legislation REACH (EC Citation2006) allows for grouping and read-across, to either justify waiving specific tests or to fill in data gaps.
For chemicals, grouping is well established as a ‘general approach for considering more than one chemical at the same time’ (OECD Citation2014a; ECHA Citation2008). The idea behind grouping approaches is that chemicals which are similar enough with respect to certain criteria (e.g. structural, physicochemical properties, etc.) can be considered as a group. Chemicals within one group are then expected to show similar (eco-)toxicological and/or environmental fate behavior. Within this group, data gaps on toxicological behavior for a certain member of the group can therefore be filled by read-across using information from the other members in the group. In general, grouping may support risk assessment as well as Safe(r)-and-Sustainable-by-Design (SSbD) approaches. Groups are established initially on the basis of structural similarity, which can be based on various principles such as common functional groups, precursors, breakdown products or a constant incremental change of the properties of interest across the group (OECD Citation2014a; ECHA Citation2017). However, it then has to be demonstrated that these structural similarities result in a similar fate and/or (eco-)toxicity. Thus, knowledge of a common toxic mechanism or Mode-of-Action (MoA) can strongly facilitate grouping, since grouping always requires a proper scientific justification which is mainly supported by establishing a link between specific properties and the toxicological endpoint of interest. In addition, grouping is endpoint-specific meaning that group membership may vary depending on which toxicity endpoint is considered.
In the last decade, several grouping frameworks have been developed for NMs, e.g. the MARINA grouping and read-across approach (Oomen et al. Citation2015) or the DF4nano Grouping Framework (Arts et al. Citation2015), which are comprehensively summarized in Oomen et al. (Oomen et al. Citation2018) and Giusti et al. (Giusti et al. Citation2019) The most recent and comprehensive framework is the GRACIOUS (Stone et al. Citation2020) grouping framework. Additional insights into NM grouping in the context of the EU chemicals legislation are detailed in Mech et al. (Mech et al. Citation2019) The most recent GRACIOUS framework is based on a hypothesis-driven approach. It proposes several grouping hypotheses, which link specific physicochemical properties with specific fates and/or toxicities, tested the hypotheses in case studies (Ag Seleci et al. Citation2022; Keller et al. Citation2021; Ruggiero et al. Citation2022; Jeliazkova et al. Citation2022a; Cross et al. Citation2022; Song et al. Citation2022) and lastly, offers guiding principles to support users to formulate their own grouping hypotheses (Murphy et al. Citation2023).
Nevertheless, grouping of NMs remains a challenge. In particular, unraveling relationships between specific physicochemical properties and toxicities is not trivial due to the large panel of partially interdependent physicochemical properties that are needed to describe a single NM (Lynch, Weiss, and Valsami-Jones Citation2014). The number-based particle size distribution, surface functionalization or treatment, shape or morphology as well as surface area are certainly the most central ones (Cosnier et al. Citation2021; Poulsen et al. Citation2016). Dissolution rate, state of agglomeration/aggregation and surface reactivity have also been shown to be of high relevance (EC Citation2018; Gutierrez et al. Citation2023). However, plenty of other NM properties exist that are not tested and, several of the NM properties are polydisperse already after production and additionally have the potential to change during the life cycle, depending on the environment/biological medium in which they are suspended or incorporated. This renders the physicochemical characterization of NMs both in their dry state and as applied, a complex task. Overall, identifying which physicochemical parameters are driving toxicity remains the key challenge in NM grouping (Ribeiro et al. Citation2017; Drasler et al. Citation2017).
Omics approaches are a very promising tool which have already frequently been applied for chemicals including NMs. Different omics layers can be investigated, e.g. levels of gene transcription (transcriptomics), protein abundances (proteomics) or levels of small molecule metabolites (metabolomics). Among them, transcriptomics is by far the most studied omics level for describing the molecular changes induced by NMs. This is mainly due to the fact that transcriptomics technologies are highly advanced, the evaluation is well standardized and interpretation of the results is relatively straightforward due to the well-studied and well-annotated state of this particular level of biological organization (Kinaret et al. Citation2020; Federico et al. Citation2020; Serra et al. Citation2020a) and well-established tools. On the other hand, while proteomics or metabolomics are closer to the actual phenotype, indicating the potential for more direct causal association with the adverse outcome (AO), i.e. endpoint of interest, they are more complex and do not at present have standardized protocols for the analysis and interpretation of data. The OECD reporting frameworks have been established for both transcriptomics (OECD Citation2021a) and metabolomics (OECD Citation2021b); however, standardized evaluation and interpretation for metabolomics and proteomics is still lagging behind. While for now, it is possible to individually assess and report the single omics endpoints (transcriptomics or metabolomics or proteomics), their combined evaluation, which may be important to obtain a holistic view of the biological response to an exposure remains difficult. Thus, developing models that are based on omics data is also consistent with the major shift away from pure consideration of single endpoints toward a more mechanistic understanding which can be observed within the field of toxicology. In general, omics approaches yield the advantage that multiple endpoints are considered at the same time as a whole panel of cellular changes is measured in one single experiment, allowing for investigation of dependencies between events and endpoints.
Within NM experiments, usually omics data for a few materials are obtained and analyzed subsequently (Ma et al. Citation2023; Nguyen and Falagan-Lotsch Citation2023). While this provides useful insights into various biological changes in that specific experiment, it is difficult to derive general patterns and to extract which changes are most relevant in terms of grouping NMs with respect to a certain endpoint. Especially, the high-dimensional setting of omics experiments with the number of parameters being much larger than the number of samples is a major challenge. Here, meta-analyses may be very useful to counterbalance this situation. Due to data sharing rules in the community and corresponding journals, huge databases of publicly available datasets exist for different omics levels like the National Center for Biotechnology Information Gene Expression Omnibus (NCBI GEO) (Edgar, Domrachev, and Lash Citation2002) for transcriptomics or PROteomic IDEntification (PRIDE) database (Perez-Riverol et al. Citation2021) for proteomics. While these datasets cover a wide range of studied effects such as those cause by drugs, other types of chemicals or diseases, omics datasets should theoretically allow integration across those effects. Such meta-analyses may be beneficial for the detection of broad biological patterns as they are derived from a greater knowledge base and noise occurring in single studies may be canceled out.
Machine learning (ML), a subfield of Artificial Intelligence (AI), may be highly valuable for addressing this task. The greatest advantage of ML is that such models are able to automatically learn patterns in large datasets and derive associated predictions. Generally, data to be modeled is described in a feature vector or matrix. This comprises values for measured input features like physicochemical properties or others. Additionally, a respective outcome variable, describing for instance a certain toxicity outcome can be linked to the input data. ML algorithms range from classical linear or logistic regression models to more complex ones such as Random Forests (RFs) or Support Vector Machines (SVMs) and Deep Learning models like Neural Networks (NNs). The choice of ML algorithm used in a study should be based on the amount and complexity of the available data as well as the specific goal in order to obtain robust trustworthy results. In an optimal dataset for the development of ML models, the number of samples should be much larger than the number of features describing these samples. However, in biological settings this is rarely ever the case. Therefore, the main goal for implementing useful ML models is to find the right balance between model complexity and generalizability – thus, avoiding overfitting. Model complexity means that a model is complex enough to be able to describe a set of so-called training data, but at the same time not too complex in order to be able to predict outcomes for previously unseen test data as well. Also, there is usually a tradeoff between flexibility/power and interpretability/inference capacity of a ML method (Gareth James, Hastie, and Tibshirani Citation2021; Gareth James et al. Citation2023). This means that simplest methods are also the most easily interpretable ones, such as linear regression (and variants such as elastic net regression) or decision trees. More complex methods such as RF and SVMs are significantly less interpretable, and the least interpretable models are NNs and the multilayered deep neural networks (deep learning models). Explainable AI (XAI) is a subset of AI and ML techniques focused on making AI systems more understandable, transparent, and interpretable for humans. This has implications also for regulatory toxicology, as models should be maximally interpretable for reasons of transparency and governance.
Again, ML is optimally suited to aid the analysis of omics data for several reasons. Omics datasets are high-dimensional and rich in describing many cellular alterations within each single measurement, in which patterns and relationships can be detected and that may not be visible using traditional statistical methods. However, the high dimensionality of the data can also be challenging. Here, moving from datasets limited to single experiment or materials toward integration of various datasets in a meta-analysis setting may be beneficial. In such cases, ML is well suited to handle the complexity of the datasets, unraveling the hidden interactions or dependencies between molecular components. ML can also be used to automatically select the most relevant features such as transcripts, proteins or metabolites related to specific molecular events or key event (KE), thereby reducing the dimensionality and improving the interpretability, resulting in identification of potential biomarkers. ML can also directly be used for omics-driven predictive modeling. Additionally, meta-analyses are great tools for handling noise in the data and distinguishing true from random signals (Cervantes-Gracia, Chahwan, and Husi Citation2022). ML can support integration of multiple omics layers. Finally, a great advantage of ML methods is that they can easily be adapted by re-training once new data become available. Thus, ML models are well suited for reducing the complexity of analysis of omics datasets, revealing the important biological traits perturbed after exposure. Moving forward, once NMs with similar MoAs have been identified, one may investigate corresponding similarities and differences in physicochemical properties and elucidate the most relevant properties for formulating a grouping hypothesis, and also to provide design principles for NMs with reduced hazards. Advances in high-throughput transcriptomics facilitate the creation of large and uniform datasets that are ideally suited for ML and grouping applications as the available technologies reduce the cost of transcriptome profiling by up to 10- to 20-fold (Ye et al. Citation2018). Coupling together omics and high-throughput screening technologies in a tiered approach increases the granularity and informativeness of the data even further (Grafström et al. Citation2015). XAI may also facilitates MoA discovery as it helps to interpret the decision made by ML models. In the context of transcriptomics-derived MoAs, tools need to be developed, in part, to conform to XAI principles while maximizing predictive ability (Boyadzhiev et al. Citation2021).
Some recent reviews have summarized ML models in the NM field (Stone et al. Citation2020; Lamon et al. Citation2019; Basei et al. Citation2019; Furxhi et al. Citation2020). However, they have not or only scarcely considered the potential of omics data in the context of NM grouping. The aim of this review is to provide an overview of existing ML models that enable complex data integration and analysis in support of NM grouping and to shed light on how omics data may be used in conjunction with ML models in the development of reliable grouping frameworks. The review focuses only on hazard endpoints which are considered relevant under REACH (see ) for human toxicity. ML models for other endpoints like cytotoxicity or ecotoxicity as well as those predicting NM uptake or protein corona formation are excluded from this review.
Table 1. Relevant endpoints for human health across multiple legislations (according to Jagiello et al. Citation2021, with modifications).
2. Relevant hazard endpoints
Grouping allows waiving of tests or filling the data gaps related to a target substance by using data from a previously tested source substance. In the European Union, the most important overarching regulation for chemicals is REACH. The Annexes VI to XI of the REACH regulation (EC Citation2006) specify which information the manufacturers need to provide when registering a substance. These requirements are dependent on the tonnage of production per year. Thus, higher tonnages lead to more extensive toxicological testing requirements. In order to tackle specific information requirements for NMs, the REACH annexes VI to X have been updated to specifically consider nano-specific information needs (EC Citation2018). In addition, work is underway to modify or adapt test guidelines in consideration of specific properties and property specific effects of NMs (Rasmussen et al. Citation2019).
Apart from REACH, other specific legislations exist for chemicals with specific applications, e.g. cosmetics (EC, Citation2009a), food contact materials (EC Citation2004) or pesticides (EC, Citation2009b). All these regulations consider (to a large extent) similar toxicological endpoints. The relevant endpoints for several legislations for NMs have also been collected in the European Union Observatory for NMs (EUON) report on New Approach Methodologies (NAMs, referring to a set of innovative techniques or testing strategies used in toxicology and risk assessment to evaluate the safety of chemicals) for NMs (Jagiello et al. Citation2022) as well as in a recent publication from Bleeker et al. (Bleeker Citation2023) which intends to support harmonization of the testing requirements for NMs across EU legislations. An overview on relevant hazard classes for human health under REACH or in at least two other legislations as described in these documents is given in .
While for some of the above-mentioned endpoints such as skin corrosion/irritation or serious eye damage/irritation, NAMs are well established and NAM data is accepted for regulatory decision making, for other more complex endpoints, NAM development is still on-going. Although a push for replacing animal tests with validated NAMs is in full swing internationally (Schmeisser et al. Citation2023), to date, toxicological testing for complex endpoints still largely relies on animal studies.
3. NAMs and NAM frameworks
For many chemicals, and especially for NMs, the data for complex endpoints is very scarce and an increasing number of new materials is entering the market every year. Filling the data gaps and testing new materials using animal studies raises not only ethical concerns but is also limited with respect to time and cost efficiency. In addition, human relevance of animal models is frequently questioned and the underlying toxicity mechanisms are often less obvious in animal models. Therefore, NAMs are becoming increasingly important for safety assessment in the light of the 3R principles for reducing, refining and replacing animal studies. Several NAMs exist and are based on in vitro, in chemico and in silico methods. This also includes high-throughput screening, allowing for testing of multiple chemicals at a time and high-content methods like omics approaches that enable comprehensive understanding of the underlying mechanisms (ECHA Citation2016).
While in vitro and in chemico methods are targeting the retrospective observation of NM toxicity, in silico models are able to predict potential hazards prior to development and testing and thus are well suited for SSbD approaches. Among the most frequently used in silico methods are Quantitative Structure-Activity Relationship (QSAR) models. QSAR models establish quantitative relationships between relevant physicochemical properties of NMs and their biological activity. These relationships are represented by mathematical functions in terms of regression models. Various QSAR models have been introduced for NMs and have been reviewed in multiple publications (Ciura et al. Citation2024; Li et al. Citation2022; Burello Citation2017). However, QSARs usually show certain limitations. The mathematical relationship usually holds true for few materials or material classes only. This is called the applicability domain (Roy, Kar, and Ambure Citation2015). In addition, most QSAR models are developed for simple toxicity endpoints like cytotoxicity due to the fact that modeling complex endpoints sufficiently well is usually not feasible. However, such endpoints are typically not of regulatory relevance. Also, mechanistic explanations on how the descriptors used for QSARs development are related to the outcome are usually missing and thus explainability of such approaches is limited which again leads to the fact that QSARs do not generalize well across different material classes on which they have not been developed. More complex ML models and omics approaches are well suited to overcome these limitations.
One major advantage of some NAMs is that they allow unraveling toxicity mechanisms which may greatly improve hazard and risk assessment in the future. However, single NAMs may not be sufficient to describe an AO. Instead, a battery of NAMs may be required to sufficiently assess an AO. Therefore, NAM frameworks combining multiple individual NAMs are needed.
To date, several NAM frameworks have been developed. Within REACH, one of the most important alternatives relying on NAM frameworks is the concept of grouping and read-across. In grouping, chemical similarity is assessed in terms of similarity of their physicochemical properties, their toxicity with respect to a certain endpoint as well as other properties which could be relevant in terms of their associated hazard. In case chemicals are similar with respect to all of these properties, they form a group. The main aim is to determine which physicochemical properties are most relevant with respect to the studied endpoint such that for new chemicals, for which no toxicity assessment has been performed so far, one may conclude whether this new chemical is likely to belong to the established group and thus induce similar toxicity or not. For chemicals in general, grouping is already well established and frequently used in regulatory decision making, with structural similarities or common functional groups being some of the key parameters defining similarity. In contrast, for NMs, the situation is much more complex and establishing reliable grouping approaches is not trivial. This stems from the facts that 1) the number of physicochemical properties needed to sufficiently describe a NM is much large and to date, no simple relationship between any single property and the toxicological outcomes has been consistently observed. This may also be due to the polydispersity of NM (or any other particle) in their properties, such that not all particles in one test preparation exhibit the exactly identical properties; 2) the NM physicochemical properties change during the lifecycle and/or in different environments; 3) other tasks such as exposure characterization and dose estimation are an issue; and 4) NMs interfere with some assay components, requiring optimization of existing methods or development of novel methods. All of this has resulted in inconsistencies in results and reporting, leading to an inability to generalize the observed results across NMs of similar properties.
For NM grouping approaches, one of the most critical questions is whether it is sufficient to establish them on the level of intrinsic physicochemical properties like chemical composition, primary particle size or morphology in combination with the toxicity data. Predictive models and grouping approaches based on intrinsic physicochemical properties describing the chemical and physical structures have two major advantages: 1) they can mostly be controlled directly during the production process and 2) many of the intrinsic properties can be measured more easily compared to extrinsic properties, which often require more complex methodologies, which are currently not well standardized. However, intrinsic physicochemical properties alone often seem to be insufficient for determining whether or not NMs behave similar with respect to a certain toxicity endpoint in a reliable manner. Instead, extrinsic properties like hydrodynamic diameter in relevant media, zeta potential or the formation of a protein corona, may be better suited for reflecting the actual biological activity of NMs. Thus, approaches based on such extrinsic properties may be superior compared to approaches based on simple physicochemical properties (Wohlleben, Mehling, and Landsiedel Citation2023) and they need to be derived to separate distinct NM hazard groups sufficiently well. However, existing grouping approaches of NMs are still not perfect and intrinsic as well as extrinsic physicochemical properties suffer from the fact that the applicability domain usually restricts models to very specific subsets of NMs. Due to all these reasons, NM grouping may be viewed as a complex endeavor and further efforts are needed to develop effective grouping strategies.
Instead of purely relying on overall potency read-outs from in vivo or in vitro testing, information on common MoAs or toxicity mechanisms may greatly advance NM grouping approaches and can help to justify an existing grouping hypothesis (OECD Citation2014a). Knowledge on MoAs may also aid some of the aforementioned challenges with implementing reliable NM grouping approaches. Especially, MoAs can provide qualitative information on whether two NMs induce the same kind of changes even if the actual quantitative values are not directly comparable due to challenges in dosimetry and dispersion. In addition, they drastically reduce the characterization efforts as instead of measuring a huge number of physicochemical properties, one only needs to perform very few omics measurements. These measurements are also directly related to induced changes while for the physicochemical properties if and how they are related to toxicity is not clear. Omics measurements may also be helpful to separate different MoAs and thus solve the difficulties for NM grouping that result from the fact that some AOs in vivo or in vitro may feature different MoAs and in other cases, different MOAs may lead to the same AO. The knowledge of underlying MoAs can then in turn greatly advance two important concepts: Adverse Outcome Pathways (AOPs) and Integrated Approaches to Testing and Assessment (IATAs).
AOPs (Ankley et al. Citation2010) are conceptual frameworks which aim to causally link certain biological events in a sequential manner, starting from a molecular initiating event (MIE), inducing multiple KEs and finally leading to an AO. The MIE thereby represents the interaction of a NM with a biomolecule or an event after its first interaction. That is followed by KEs at multiple levels of biological organization that are essential to the disease progression and can be measured, e.g. at the cellular, tissue or organ level. The AO may then be one of the endpoints mentioned in . Several AOPs have been proposed for chemical induced toxicity and may also be applicable or adaptable to NMs (Halappanavar et al. Citation2020; Gerloff et al. Citation2017; Gromelski et al. Citation2022; Nymark et al., Citation2021). One such example is AOP173 from the AOPwiki (https://aopwiki.org/aops/173) which describes the development of pulmonary fibrosis after substance interaction with pulmonary resident cell membrane components, which is relevant to NMs (Halappanavar Citation2023).
Often AOPs are used as a basis to establish IATAs, which are frameworks for evaluating complex hazard endpoints by integrating multiple sources of information for studying various aspects of the toxicity endpoint under consideration (Nymark et al., Citation2021; Halappanavar et al. Citation2021b). A proper understanding of the underlying MoA and related AOPs is important for developing reliable IATAs, as they enable identification of the right assays reflecting KEs to be tested. IATAs allow integration of the different assay outcomes and provide information on a potential hazard in a weight-of-evidence manner. Within an IATA, different kinds of NAMs can be combined.
In the following sections, an overview of existing approaches based on ML and omics supporting NM grouping is given.
4. ML And existing ML models for NMs
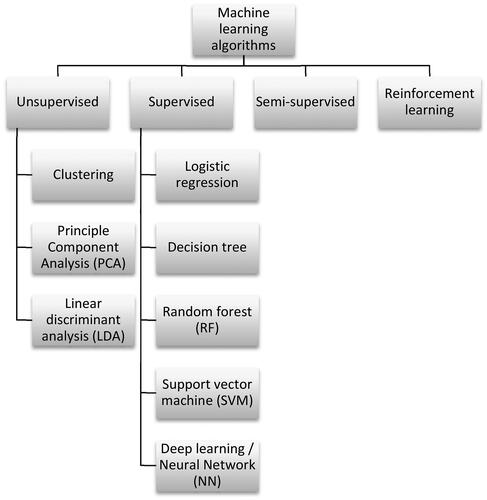
ML models are well suited for predicting outcomes with respect to NM toxicity and for selecting the most relevant descriptors influencing toxicity. In general, supervised and unsupervised ML models, as well as some mixed types exist. In supervised models, the goal is to map labels assigned to each sample to variances observed in the input data. In the case of NM grouping, these labels are usually representing a toxicity endpoint, e.g. the outcome of an in vivo study or an in vitro assay. Depending on the nature of this response variable, ML models can be divided into regression models in which the outcome variable is continuous and classification models with discrete outcome variables. Instead, unsupervised models use unlabeled data and thus, only rely on variances in the input data and seek to find patterns therein. Additionally, semi-supervised methods and reinforcement learning exist but are less frequently used. Semi-supervised ML methods combine labeled and unlabeled data thereby increasing the dataset size which can be used for training without the need for further labeling which might be time- or cost-intensive. In reinforcement learning an agent learns an optimal strategy for choosing actions in a way that the reward obtained for a sequence of actions is maximized. An overview of frequently used ML algorithms (Gareth James, Hastie, and Tibshirani Citation2021; Gareth James et al. Citation2023) is given in .
To identify relevant studies, a search in Scopus using the following search query: TITLE-ABS-KEY ((nanoparticle OR nanomaterial OR nanoparticles OR nanomaterials) AND (‘in silico’ OR computational OR ‘machine learning’ OR ‘case study’) AND toxicity), was conducted. This search matched 988 publications of which 657 publications were tagged as primary publications (Stand: 11/2023). From these publications, those using a ML model to predict one of the relevant endpoints in were identified. In addition, relevant studies from previous reviews on ML models in the NM field (Stone et al. Citation2020; Lamon et al. Citation2019; Basei et al. Citation2019; Furxhi et al. Citation2020) were also added. An overview of the relevant approaches is provided in .
Table 2. Predictive ML models for NM toxicity and grouping in mammals or mammalian cell models.
Except for one study, all identified approaches concentrate either on mutagenicity and genotoxicity or on STOT as the modeled endpoint. In addition, almost all models include supervised approaches. Most frequently, tree-based approaches, namely decision trees or RFs, are used to predict toxicity. Often, these are preceded by an unsupervised analysis using hierarchical clustering or Principle Component Analysis (PCA). Overall, the predictive performance of the models was found quite good (0.7 to 1.0). However, it is also easily visible from that the number of available datasets is very small in most cases and usually not sufficient to build robust ML models. In addition to predicting toxicity outcomes, many studies also perform feature selection to reduce the model to only the most relevant descriptors. From , it becomes obvious that the selected descriptors vary largely across studies. Thus, even though most models show relatively high predictive performance, it may be expected that their applicability domain is rather limited and that they may be expected to be overfitted for the training data and therefore not generalize well to new datasets. With respect to selected materials, there is a strong focus on metal oxide NMs and multi-walled carbon nanotubes (MWCNTs). Other materials are not sufficiently covered so far and thus, cannot easily be assessed with the available models. As properties by which different material classes and materials of different shapes can be described may differ, integrating various types of NMs in a common model is not straightforward.
5. Omics approaches revealing NM MoA
As described before, omics data have the potential to support NM grouping approaches by informing about underlying MoAs and induced AOPs. Thus, this review also focuses on predictive models for NM toxicity which include omics data as descriptors as well as other omics approaches which may potentially support NM grouping. Therefore, all omics-based tools and approaches were searched and below, important primary NM omics studies are described.
Several studies have been performed to describe the changes induced by NMs on the level of transcriptomics. Various in vitro (Bajak et al. Citation2015; Boyadzhiev et al. Citation2021; Snyder-Talkington et al. Citation2015) as well as in vivo (Halappanavar et al. Citation2015; Chézeau et al. Citation2018; Poulsen et al. Citation2021; Decan et al. Citation2016; Gosens et al. Citation2021; Solorio-Rodriguez et al. Citation2023) approaches have been described. Although scarcer, literature on the effects of NM treatment on other omics layers also exists. For proteomics, mass spectrometry gives the most comprehensive results and is therefore frequently used nowadays (Gallud et al. Citation2020; Torres et al. Citation2022; Billing et al. Citation2020; Miranda et al. Citation2022). Also, metabolomics changes are addressed in multiple studies (Bannuscher et al. Citation2020a; Cui et al. Citation2019; Chen et al. Citation2019). In addition, some approaches considered multiple omics layers at the same time (Bannuscher et al. Citation2020a; Karkossa et al. Citation2019; Nymark et al. Citation2015; Seidel et al. Citation2021; Aragoneses-Cazorla et al. Citation2022; Dekkers Citation2018). While this is not a comprehensive list of available studies, it is already clear, that these several omics datasets shed light on molecular changes induced by various NMs in vitro and in vivo from different perspectives, using different omics layers, techniques and methods, cell models, species and so on. The main question that remains is, how to use this existing information to support NM grouping. An overview of predictive models as well as other useful approaches will be given in the next section. In addition, the different obstacles rendering the development of omics-based models non-trivial are discussed.
In general, the biological model, the exposure pattern as well as the detection method for the respective omics layer may largely impact the result of a study. In vivo studies naturally represent the gold standard as they reflect the biological complexity present in animals and can inform about long-term effects. Depending on the use of the NM under study different exposure scenarios may be most relevant. For occupational exposure, the inhalation route is frequently considered to be most relevant. For medical applications, direct injection of the NMs may also be important. Within one exposure route, the actual exposure scenario might also impact the results. As an example, studies targeting the inhalative route of exposure may be performed using inhalation, instillation or pharyngeal aspiration exposure as well as single or repeated doses. While repeated doses in an inhalation setting may reflect the natural occupational exposure more closely, other methods like instillation have the advantage of better control of the actual delivered amounts of particles (Kinaret et al. Citation2017a).
While in vivo approaches may yield highly valuable results, they are time- and cost-intensive and require ethical approval for each study performed. Therefore, avoiding and replacing in vivo methods whenever possible has been a major endeavor in the past years. In vitro methods form one important pillar in this replacement. Naturally in vitro models can only reflect complex biological processes partially. However, as long as these models show high predictivity for the true toxicological impacts, they are still sufficient for testing purposes. This predictivity may critically depend on the chosen cell model under study. Here, cell types which are expected to be in direct contact with the NMs like lung macrophages may be expected to be most predictive. Also, the carcinogenic nature of most cell lines should be regarded as these cells might react substantially different from healthy cells in the organism.
Another advantage of in vitro omics studies is that they can be performed in a high-throughput manner and thus allow for testing of a large number of NMs. Here, the investigation of different omics layers may lead to information complementing each other. Optimally, the data on different omics layers should be derived from the very same cells in a multi-omics setting. Time points for the different omics layers should also be chosen carefully. Here, as an example transcriptomics changes are expected to happen much faster than proteomics changes and thus the time points for sample collection should differ. Also, the choice of analysis method and devices used may drastically change omics results. Here, more recent methods usually aim at untargeted assessment of all analytes available in a sample while older techniques focused on pre-defined analytes in a targeted manner.
As mentioned previously, one of the major advantages with respect to omics data is that almost all journals require study authors to make the raw datasets belonging to a publication available. Therefore, a large amount of data is available in public databases, e.g. NCBI GEO (https://www.ncbi.nlm.nih.gov/geo/) for transcriptomic data or PRIDE archive (https://www.ebi.ac.uk/pride/archive/) for proteomic data. As ML models require large datasets with respect to the number of samples, reuse of this data to train or test models is of great value for developing robust approaches and has frequently been applied. In addition, meta-analyses of several omics-based studies may also broaden the understanding of molecular mechanisms and MoAs of NMs. Predictive ML models and other useful tools developed in the field of omics-based NM grouping are summarized in and , respectively.
Table 3. Predictive ML models for NM toxicity and grouping based on omics data.
Table 4. Additional omics approaches that could support NM grouping.
From the literature review performed here, we identified a few models that aim to predict NM toxicity or grouping based on omics measurements (see ). The models use different ML algorithms to predict either in vitro or in vivo outcomes or to directly suggest grouping on NMs. All models yield high predictive performances of at least 0.7 for the validation set. To reduce the number of descriptors, two studies use feature selection (Yanamala et al. Citation2019; Fortino et al. Citation2022) for reducing the parameters included in the final model thereby improving explainability.
In addition, multiple other tools have been developed or used to support omics-based analysis of NM toxicity. Kohonen et al. (Kohonen et al. Citation2017) developed a ‘predictive toxicogenomics space’ (PTGS) tool which yields a predictive signature for drug-induced liver injury (DILI). This tool has also recently been applied successfully to NMs (Kohonen et al. Citation2019). Serra et al. (Serra et al. Citation2019) created the INSIdE NANO tools which contextualizes transcriptomic changes of NMs with those induced by drugs, other types of chemicals and diseases. A similar approach is followed by Bahl et al. (Bahl et al. Citation2023) who developed PROTEOMAS, a harmonized proteomic workflow which is applied to a case study in a very similar fashion. In addition, tools calculating benchmark doses (BMDs) based on omics data are also useful to support NM toxicity evaluation and grouping. Proposals for such tools have been made by Halappanavar et al. (Halappanavar et al. Citation2019), Gromelski et al. (Gromelski et al. Citation2022) and Serra et al. (Serra et al. Citation2020b). Others have attempted to link physicochemical properties of NMs to observations of changes in omics data. Kinaret et al. (Kinaret et al. Citation2017b) used coexpression networks and Bannuscher et al. (Bannuscher et al. Citation2020b) and Karkossa et al. (Karkossa et al. Citation2019) used Weighted Gene Correlation Network Analysis (WGCNA) (Langfelder and Horvath Citation2008). Jagiello et al. developed a QSAR models for predicting transcriptomic pathway level response (Jagiello et al. Citation2021). A complete list of identified approaches with more details is shown in . In addition, this table also lists several meta-analyses which may provide useful datasets for further model development or validation.
6. The value of AI for supporting NAMs
AI is a subfield of computer science in which algorithms and models are developed to mimic cognitive functions of human intelligence, such as learning and problem-solving. AI can aid risk assessment of NMs in various ways, especially by automating and improving commonly used processes. The major field of classical ML mainly dealing with pattern recognition and predictive modeling has already been introduced in detail above. However, this type of modeling and pattern recognition is only one part of existing AI methods. Other applications of AI may also be relevant for risk assessment of NMs and are briefly introduced below.
6.1. Automated (meta)data extraction and linked data
One important task in risk assessment is to gather all available data on the NM under study. This is a difficult and time-consuming task as data might be spread across various databases, tables or even be only present in scientific publications as unstructured texts. AI may support this task in terms of data and text mining using automated information retrieval and Natural Language Processing (NLP). Different approaches for mining chemical and biological data have been developed in the past. Swain and Cole developed the ChemDataExtractor for automated extraction of chemical information from scientific literature (Swain and Cole Citation2016). Also tmChem (Leaman, Wei, and Lu Citation2015) can perform chemical named entity recognition from texts. In addition, CD-REST (Xu et al. Citation2016) is able to extract chemical-induced disease relations from literature. This automated data extraction may support integration of individual findings across multiple publications which was not obvious previously. Also, relationships between diseases and genes (Lever et al. Citation2019) or proteins and drugs (Zheng et al. Citation2019) could be identified by text mining approaches. Similar approaches may also be used in the field of nanotoxicology. Especially, with recent developments in the field of Large Language Models (LLMs) this field is expected to be of major relevance in future research. Integration of such LLMs with knowledge graphs may further support data retrieval and storage in a structured way. This may also aid the curation of databases with respect to toxicological results as well as metadata. In addition, linking different databases holding certain information on NMs may also be facilitated by LLMs due to automated recognition of similar terms and alternative naming. As an example, AI may be useful for linking NM-specific information stored in databases like eNanoMapper (Jeliazkova et al. Citation2015) to omics databases like GEO or PRIDE in an efficient way even if the naming schema and underlying ontologies differ. In the broader context of risk assessment, toxicity information may be automatically integrated with other information such as their intended use or information based on different exposure scenarios. This may also be useful in terms of prioritization of NMs to be investigated and ML models may be developed specifically for this task. Additionally, if trained well, AI models can evaluate multiple risk factors and dependencies between them simultaneously. This may be of high value when evaluating complex mixtures of NMs or chemicals.
6.2. Data curation
Data curation is of utmost importance for developing reliable approaches and models for NM toxicity (Alves et al. Citation2021). However, if performed manually this is a highly time-consuming task with numerous challenges as shown in various projects before. AI may provide useful tools to facilitate this task (Martín et al. Citation2023). Here, anomaly detection is one of the most well-known information enrichment techniques which enables identification of outliers or patterns differing from the rest of the data in an automated way. Suitable models may significantly speed-up the identification of inconsistencies in the data. In addition, NLP may also improve the detection of data gaps in registration dossiers for NMs on the market in the context of regulatory process optimization by automated screening strategies. At the same time, recommendations based on historical data may automatically be generated. In addition to advantages in time consumption, AI methods are also less prone to errors compared to humans processing large amounts of data, given that the input data is of high quality. While not every published study can be considered as high quality, AI methods can directly support the identification of quality issues and assess the data quality. Anomaly detection is one very prominent example of data quality checks and is frequently supported by computational tools like Isolation Forests (Liu et al. Citation2008) or autoencoders (Shvetsova et al. Citation2021).
6.3. Image analysis
Another field which provides great opportunities for the application of AI is the field of image analysis. Automated image analysis may be enabled by deep learning approaches like convolutional neural networks. An example of such an approach has been provided by Aversa et al. (Aversa et al. Citation2018) who automatically identified NMs in Transmission Electron Microscopy/Scanning Electron Microscope images and derived their size and number. In addition, Karatzas et al. (Citation2020) used deep learning models to predict the effects of NMs on Daphnia magna. Similar approaches may also be derived from videos instead of images. However, the main challenge for image recognition is that large datasets need to be labeled before training the model.
6.4. Support during omics data analysis
Omics data are especially useful in the context of AI as usually many datasets are publicly available and, in addition, integration of NM-specific data with other chemicals or traits is comparatively easy. Thus, they allow to obtain more comprehensive insights into the underlying biological consequences of NM treatment. One of the most common applications of AI to omics data is the identification of potential biomarkers (Michelhaugh and Januzzi Citation2022). LLMs, in turn, could also support the interpretation of the identified biomarkers by quickly searching for existing literature and extracting relevant information and context. Similarly, one may also elucidate information on perturbed molecular pathways, affected targets or common patterns induced by treatment with different NMs. These developments are supported by the fact that LLMs like chatGPT can access databases such as GenBank, Ensembl and Gene Ontology, KEGG, Reactome, GEO or ArrayExpress thereby directly being able to connect the various information stored in these resources (Ng et al. Citation2023). The integration across different omics layers is another field which may be supported by AI. Commonly, ML and Deep Learning algorithms are applied in the context of multi-omics integration (Picard et al. Citation2021; Biswas and Chakrabarti Citation2020). In predictive ML models, omics data may also be used to infer links between physicochemical properties, molecular changes and toxicity which may support regulatory decision making or SSbD strategies. Especially, explainability is an important area of research allowing for new insights into outcomes of AI models thereby probably enhancing their acceptance in the field of toxicology (Santorsola and Lescai Citation2023).
Apart from all these advantages, AI models are highly dependent on the quality and amount of underlying data. Training AI models requires large datasets of high-quality data which are relevant for the studied question and are not subject to biases. One important limiting factor is data availability. Therefore, the implementation of the FAIR (findable, accessible, interoperable and reusable) principles is a key factor in the development of reliable AI models (Jeliazkova et al. Citation2021).
7. Remaining challenges and future requirements
7.1. Data availability
The first and most critical challenge in developing robust NM grouping frameworks is the limited availability of data. Any grouping approach can only be generalizable to different kinds of NMs if it was developed based on NMs belonging to different classes and property combinations. This requires many datasets to be available and to be comparable to each other in a way that they can be integrated. Especially, ML models can only make robust predictions if they have been trained on large sets of data containing as much variability as possible (Duan, Edwards, and Dwivedi Citation2019). Although high-throughput omics technologies are facilitating the generation of ever larger training data sets even by academic researchers, no single project will be able to generate sufficiently extensive datasets, and data reuse is the only option. In order to allow proper data reuse, the FAIR data principles (Jeliazkova et al. Citation2021; Wilkinson et al. Citation2016) should be considered when publishing any results. Briefly, the principles refer to e.g. clear and harmonized use of persistent identifiers, and that data as well as corresponding metadata can easily be found by both humans and machines at the same time. Data should also be accessible, either openly or through means of authorization or authentication depending on the need for restriction or not. Furthermore, data should be interoperable in order to allow for integration across highly diverse data sets. Overall, the principles guide in terms of making data reusable in the sense that descriptions of the data and metadata are sufficiently detailed to allow for reproducibility, automated assessments of, for example, data quality and large-scale integration allowing for novel interpretation opportunities.
7.2. Unique identification of NMs
Another central question in the case of NMs, which is directly related to the FAIR data principles, is how to uniquely identify a specific NM. Each NM has a multitude of different physicochemical properties which may also change during their lifecycle. Small changes in few of these properties may have large impacts on the toxicity outcome. Even for benchmark materials, variations may occur if they are obtained from different batches. Therefore, it is important to know exactly which NF was studied in a certain experiment in order to be able to interpret and reuse data. Due to the complexity of NMs, this is, however, not straightforward. Some attempts have been made to structure the nomenclature in such a way that each NM can be uniquely identified, e.g. the NInChI (Lynch et al. Citation2020) or labeled with a unique identifier even to the level of new batches, e.g. the European Materials Registry (van Rijn et al. Citation2022). These first approaches will have to be fine-tuned in the future and, importantly, also must be established within the community such that this information will be provided along with published data.
7.3. Reliability of data
Reliability of studies on NMs is another important factor. The main challenge for NMs is their handling during experiments. Especially, dispersion stability is a major issue as usually NMs are highly reactive and thus tend to agglomerate or interact with components in their surrounding environment like proteins from the medium they are dispersed in. The best way of dispersing NMs is still a matter of debate and thus is handled differently among researchers. In addition, there are no standardized ways on how to perform certain measurements of physicochemical properties or toxicity assays. Many of the OECD test guidelines are not yet adapted for NMs (Rasmussen et al. Citation2019; Bleeker Citation2023). For some of the toxicological assays, interferences of the assays with the tested NMs may occur and suitable replacements are not always available at the time being. All these uncertainties, missing adaptations to NMs and lacking standardizations lead to high variability of the resulting measurements and contradictory results in existing literature and renders integration of results from different studies a difficult task. In addition, only few NMs have been tested in in vivo studies and thus adapting NAMs such that they reflect the in vivo situation is not trivial.
In silico methods critically depend on data quality of the underlying experimental data. While especially ML models are capable of capturing very complex relations and interactions, they can, by definition, not be better than the underlying data. Thus, if the experimental results are not reliable, also developed models cannot be of high quality. Instead, data with low technical error levels allow for models which can reliably detect underlying patterns in the data. Quality criteria for NM experimental data have been introduced (Marchese Robinson et al. Citation2016; Comandella et al. Citation2020; Basei et al. Citation2022; Shao, Beronius, and Nymark Citation2023) and can guide scientists as well as modelers in judging the reliability of experimental data and thus allowing for subsequent robust model development.
7.4. Standardization and regulatory acceptance of omics data
Some of the uncertainties induced by grouping based on physicochemical properties and toxicity data alone can be overcome by adding omics data as these add an additional layer of mechanistic understanding. However, omics data often lack sufficient comparability. This is mainly because different measurement devices and analysis pipelines are in use. Therefore, also in the case of omics data harmonization and standardization of data reporting and analysis workflows are of utmost importance. The transcriptomic (OECD Citation2021a) and metabolomic (OECD Citation2021b) reporting frameworks developed by the OECD are highly valuable tools in this context. These frameworks define certain information that needs to be provided for transcriptomic and metabolomic experiments thereby supporting the implementation of the FAIR data principles.
7.5. Obtaining robust models
Finally, the question on how to combine and compare NM-related data in order to obtain robust models is another important aspect for grouping of NMs. Here, different aspects need to be taken into consideration: 1) Different measures of similarity exist and can be applied. A number of tools for quantifying similarity between NMs were recently compared by Jeliazkova et al. (Jeliazkova et al. Citation2022b) However, the different measures have different advantages and disadvantages and may eventually lead to different results. The use of high-dimensional omics datasets may even pose additional challenges in this regard; 2) Instead of focusing solely on NMs, integration with other more extensively studied effects like those from drugs, other types of chemicals or diseases may be supporting the acceptance of a certain grouping hypothesis as they may provide additional insights. While this is complicated with regards to physicochemical properties as those may be very different between NMs and other traits, on the level of omics data this approach seems to be reasonable. ML models like those based on transfer learning may be very valuable in this context; 3) AI in general is expected to form an important pillar in the generation of robust models as it will not be feasible to search, evaluate and integrate the huge and ever-increasing amount of data available manually; and 4) Once ML models have been developed on well-standardized data and methods, extensive validation is indispensable. The quality of a developed model may also be assessed based on certain criteria. For QSAR models, the OECD has published recommendations for determining the quality of developed models (OECD Citation2014b) as well as a corresponding QSAR assessment framework (OECD Citation2023). Such evaluation criteria are urgently needed in order to guarantee the use of high-quality models. These evaluation criteria may be used or adapted for more general ML and omics-based approaches to assess their robustness.
8. Conclusion
8.1. ML Models for NM grouping
ML is a valuable tool supporting NM grouping. Especially, the ability to extract the most important parameters describing toxicity in an automated, objective fashion is of great use. The extracted properties can then be used to determine the similarity of NMs. ML models can aid NM grouping approaches in several ways: 1) ML models are able to derive information and patterns from complex and high-dimensional datasets which cannot be easily detected by human inspection. Thus, they may capture more complex interactions between physical, chemical, and biological properties and can potentially enable more accurate classification and grouping of NMs; 2) ML models can predict the behavior of NMs under different conditions which may reduce time and cost when assessing safety, toxicity, and environmental impact of NMs and may guide SSbD approaches; 3) ML models are very time and cost efficient. Once a model is trained, incoming data for new NMs can usually be processed very quickly; and 4) ML can also aid the discovery of new properties or applications of NMs by identifying common patterns or correlations in large datasets. In addition, combining supervised and unsupervised approaches may also be important. While the prediction task of models is usually based on labeled data and supervised approaches, the advantage of unsupervised methods is that they can also use the large pool of unlabeled data to search for patterns or reduce dimensionality. As dataset size is of high relevance with respect to developing robust ML models with a large applicability domain, this is a very useful and recommended strategy.
8.2. Omics for NM grouping
Usually, NM grouping approaches are initially based on measured physicochemical properties or sometimes calculated descriptors. The advantage of directly linking physicochemical properties with toxicity is that one obtains information on how NMs need to be manipulated to make them safer, e.g. in SSbD approaches. However, limitations with respect to the generalizability and applicability domain have frequently been discussed and can also be observed from the different models presented in this review. This is because the importance of certain properties largely varies with the material types and shapes under study. Also, similar effects may be obtained from NMs with very different physicochemical properties. Omics approaches are very well suited to overcome these limitations as they yield additional mechanistic insights which can support NM grouping. Nevertheless, there are a few challenges which need to be overcome in order to successfully integrate omics results into NM grouping approaches. Especially, FAIRification of NM omics data, harmonization and standardization of measurements and analyses workflows, the definition of similarity and validation of findings are major fields in which improvement is still required in order to achieve robust models and regulatory acceptance. Once these hurdles are successfully tackled, omics approaches are a very promising source of information for supporting NM grouping. This is confirmed by the first approaches which obtained very good predictive performance when including omics data in the prediction models for NM toxicity. Thereby, omics approaches hold a number of benefits for NM grouping approaches: 1) Omics data provide detailed information about biological effects and interactions of NMs. This may aid the formulation and testing of a grouping hypothesis and thus support regulatory bodies in making informed decisions regarding the safety of NM; 2) While grouping approaches based on physicochemical properties suffer from the fact that these properties may change depending on their surrounding medium as well as over time, omics provide a more direct read-out of the actual state of the NM that was seen by the cells. In combination with ML, patterns and relationships not evident from traditional analysis methods may be detected; and 3) If well performed and analyzed, omics experiments are directly comparable among studies and thus well suited for meta-analyses. This does not only hold true within studies on different NMs but instead meta-analyses including other traits like drugs, other types of chemicals and so on are also possible. The fact that a huge number of such omics datasets is publicly available including raw data as well as metadata, is very promising for the implementation of robust ML models, whose performance largely depends on the amount of available data.
8.3. AI for NM grouping
AI in general will be unavoidable in the context of developing reliable grouping approaches to cope with the wealth of information available. The complexity of the task of finding suitable NAMs for NM toxicity assessment leads to a high number of potential applications of different AI methods. Especially NLP is expected to have a great influence on future data processing and retrieval. The following tasks have high potential for support by NLP: (1) automated review and data extraction from scientific literature which may also include automated generation of databases; (2) finding patterns, trends and inconsistencies in research papers or datasets; (3) standardization of terminologies; (4) compliance of research documents with regulatory standards; and (5) automated annotation and generation of metadata within databases. However, while AI may provide very helpful tools for obtaining information in an efficient and objective manner, human judgment will be of utmost importance and should not be underestimated. This may especially be important in terms of plausibility considerations, contextual interpretation, decision making, quality checks or results with high uncertainties.
8.4. Recommendations for future research investment
In order to implement ML and omics techniques successfully in the process of NM grouping, standardization of test methods including dispersion protocols and quality assessment followed by appropriate data curation of produced data are urgently needed in order to allow comparison between results from different studies. In line with that, raw data should be made available to the community once they are published and they should be accompanied by sufficient metadata. This may be supported by further refinement of file and model sharing formats, ontologies, terminologies and data quality assessment tools specific for the needs in the field of NMs. Along these lines, it is also of high relevance to find a solution for unambiguous naming of NMs which allows direct comparison of the data. New developments in AI, especially in the field of LLMs can aid the curation and linkage of existing databases. More reliable and comparable data will automatically support the implementation of more robust ML models with a larger applicability domain. At the same time, it will be necessary to enhance the explainability of ML models in order to derive a grouping hypothesis that can also be tested for validation purposes. Here, ML algorithms need to be explored and adjusted with respect to methods and insights that allow for interpretation of the outcome. While it is important to increase the explainability of the model itself, also unraveling the underlying MoA will support the hypothesis formulation. Here, more omics studies on NMs and meta-analyses will be highly useful for extracting predictive signatures that can be used in hazard assessment. Once predictive signatures and thus relevant transcripts, proteins or metabolites have been identified, targeted testing of only these entities will be possible allowing for high-throughput screening.
Disclosure statement
The authors report no conflict of interest.
Data availability statement
Data sharing not applicable – no new data generated.
Additional information
Funding
References
- Ag Seleci, Didem, Georgia Tsiliki, Kai Werle, Derek A. Elam, Omena Okpowe, Karsten Seidel, Xiangyu Bi, et al. 2022. “Determining Nanoform Similarity via Assessment of Surface Reactivity by Abiotic and in Vitro Assays.” NanoImpact 26: 100390. https://doi.org/10.1016/j.impact.2022.100390.
- Alves, Vinicius M., Scott S. Auerbach, Nicole Kleinstreuer, John P. Rooney, Eugene N. Muratov, Ivan Rusyn, Alexander Tropsha, et al. 2021. “Curated Data In - Trustworthy In Silico Models Out: The Impact of Data Quality on the Reliability of Artificial Intelligence Models as Alternatives to Animal Testing.” Alternatives to Laboratory Animals: ATLA 49 (3): 73–82. https://doi.org/10.1177/02611929211029635.
- Ambure, Pravin, Arantxa Ballesteros, Francisco Huertas, Pau Camilleri, Stephen J. Barigye, and Rafael Gozalbes. 2020. “Development of Generalized QSAR Models for Predicting Cytotoxicity and Genotoxicity of Metal Oxides Nanoparticles.” International Journal of Quantitative Structure-Property Relationships 5 (4): 83–100. https://doi.org/10.4018/IJQSPR.20201001.oa2.
- Ankley, Gerald T., Richard S. Bennett, Russell J. Erickson, Dale J. Hoff, Michael W. Hornung, Rodney D. Johnson, David R. Mount, et al. 2010. “Adverse Outcome Pathways: A Conceptual Framework to Support Ecotoxicology Research and Risk Assessment.” Environmental Toxicology and Chemistry 29 (3): 730–741. https://doi.org/10.1002/etc.34.
- Aragoneses-Cazorla, M. Pilar Buendia-Nacarino, Maria L. Mena and Jose L. Luque-Garcia. 2022. “A Multi-Omics Approach to Evaluate the Toxicity Mechanisms Associated with Silver Nanoparticles Exposure.” Nanomaterials 12 (10): 1762. https://doi.org/10.3390/nano12101762.
- Arts, Josje H. E., Mackenzie Hadi, Muhammad-Adeel Irfan, Athena M. Keene, Reinhard Kreiling, Delina Lyon, Monika Maier, et al. 2015. “A Decision-Making Framework for the Grouping and Testing of Nanomaterials (DF4nanoGrouping).” Regulatory Toxicology and Pharmacology: RTP 71 (2 Suppl): S1–S27. https://doi.org/10.1016/j.yrtph.2015.03.007.
- Aschberger, Karin, David Asturiol, Lara Lamon, Andrea Richarz, Kirsten Gerloff, and Andrew Worth. 2019. “Grouping of Multi-Walled Carbon Nanotubes to Read-across Genotoxicity: A Case Study to Evaluate the Applicability of Regulatory Guidance.” Computational Toxicology 9: 22–35. https://doi.org/10.1016/j.comtox.2018.10.001.
- Aversa, Rossella, Mohammad H. Modarres, Stefano Cozzini, Regina Ciancio, Alberto Chiusole. 2018. “The First Annotated Set of Scanning Electron Microscopy Images for Nanoscience.” Scientific Data 5 (1): 180172. https://doi.org/10.1038/sdata.2018.172.
- Bahl, Aileen, Bryan Hellack, Martin Wiemann, Anna Giusti, Kai Werle, Andrea Haase, Wendel Wohlleben, et al. 2020. “Nanomaterial Categorization by Surface Reactivity: A Case Study Comparing 35 Materials with Four Different Test Methods.” NanoImpact 19: 100234. https://doi.org/10.1016/j.impact.2020.100234.
- Bahl, Aileen, Bryan Hellack, Mihaela Balas, Anca Dinischiotu, Martin Wiemann, Joep Brinkmann, Andreas Luch, et al. 2019. “Recursive Feature Elimination in Random Forest Classification Supports Nanomaterial Grouping.” NanoImpact 15: 100179. https://doi.org/10.1016/j.impact.2019.100179.
- Bahl, Aileen, Celine Ibrahim, Kristina Plate, Andrea Haase, Jörn Dengjel, Penny Nymark, Verónica I. Dumit, et al. 2023. “PROTEOMAS: A Workflow Enabling Harmonized Proteomic Meta-Analysis and Proteomic Signature Mapping.” Journal of Cheminformatics 15 (1): 34. https://doi.org/10.1186/s13321-023-00710-2.
- Bajak, Edyta, Marco Fabbri, Jessica Ponti, Sabrina Gioria, Isaac Ojea-Jiménez, Angelo Collotta, Valentina Mariani, et al. 2015. “Changes in Caco-2 Cells Transcriptome Profiles upon Exposure to Gold Nanoparticles.” Toxicology Letters 233 (2): 187–199. https://doi.org/10.1016/j.toxlet.2014.12.008.
- Balfourier, Alice, Anne-Pia Marty, and Florence Gazeau. 2023. “Importance of Metal Biotransformation in Cell Response to Metallic Nanoparticles: A Transcriptomic Meta-Analysis Study.” ACS Nanoscience Au 3 (1): 46–57. https://doi.org/10.1021/acsnanoscienceau.2c00035.
- Ban, Zhan, Qixing Zhou, Anqi Sun, Li Mu, and Xiangang Hu. 2018. “Screening Priority Factors Determining and Predicting the Reproductive Toxicity of Various Nanoparticles.” Environmental Science & Technology 52 (17): 9666–9676. https://doi.org/10.1021/acs.est.8b02757.
- Bannuscher, Anne, Bryan Hellack, Aileen Bahl, Julie Laloy, Hildegard Herman, Miruna S. Stan, Anca Dinischiotu, et al. 2020a. “Metabolomics Profiling to Investigate Nanomaterial Toxicity in Vitro and in Vivo.” Nanotoxicology 14 (6): 807–826. https://doi.org/10.1080/17435390.2020.1764123.
- Bannuscher, Anne, Isabel Karkossa, Sophia Buhs, Peter Nollau, Katja Kettler, Mihaela Balas, Anca Dinischiotu, et al. 2020b. “A Multi-Omics Approach Reveals Mechanisms of Nanomaterial Toxicity and Structure-Activity Relationships in Alveolar Macrophages.” Nanotoxicology 14 (2): 181–195. https://doi.org/10.1080/17435390.2019.1684592.
- Basak, Subhash C., Marjan Vracko, and Frank A. Witzmann. 2016. “Mathematical Nanotoxicoproteomics: Quantitative Characterization of Effects of Multi-Walled Carbon Nanotubes (MWCNT) and TiO2 Nanobelts (TiO2-NB) on Protein Expression Patterns in Human Intestinal Cells.” Current Computer-Aided Drug Design 12 (4): 259–264. https://doi.org/10.2174/1573409912666160824145722.
- Basei, Gianpietro, Danail Hristozov, Lara Lamon, Alex Zabeo, Nina Jeliazkova, Georgia Tsiliki, Antonio Marcomini, et al. 2019. “Making Use of Available and Emerging data to predict the Hazards of Engineered Nanomaterials by Means of in Silico Tools: A Critical Review.” NanoImpact 13: 76–99. https://doi.org/10.1016/j.impact.2019.01.003.
- Basei, Gianpietro, Hubert Rauscher, Nina Jeliazkova, and Danail Hristozov. 2022. “A Methodology for the Automatic Evaluation of Data Quality and Completeness of Nanomaterials for Risk Assessment Purposes.” Nanotoxicology 16 (2): 195–216. https://doi.org/10.1080/17435390.2022.2065222.
- Billing, Anja M., Kristina B. Knudsen, Andrew J. Chetwynd, Laura-Jayne A. Ellis, Selina V. Y. Tang, Trine Berthing, Håkan Wallin, et al. 2020. “Fast and Robust Proteome Screening Platform Identifies Neutrophil Extracellular Trap Formation in the Lung in Response to Cobalt Ferrite Nanoparticles.” ACS Nano 14 (4): 4096–4110. https://doi.org/10.1021/acsnano.9b08818.
- Biswas, Nupur, and Saikat Chakrabarti. 2020. “Artificial Intelligence (AI)-Based Systems Biology Approaches in Multi-Omics Data Analysis of Cancer.” Frontiers in Oncology 10: 588221. https://doi.org/10.3389/fonc.2020.588221.
- Bleeker, Eric A. J., Elmer Swart, Hedwig Braakhuis, María L. Fernández Cruz, Steffi Friedrichs, Ilse Gosens, Frank Herzberg et al.. 2023. "Towards Harmonisation of Testing of Nanomaterials for EU Regulatory Requirements on Chemical safety - A Proposal for Further Actions." Regulatory Toxicology and Pharmacology 139: 105360. https://doi.org/10.1016/j.yrtph.2023.105360.
- Boyadzhiev, Andrey, Mary-Luyza Avramescu, Dongmei Wu, Andrew Williams, Pat Rasmussen, and Sabina Halappanavar. 2021. “Impact of Copper Oxide Particle Dissolution on Lung Epithelial Cell Toxicity: response Characterization Using Global Transcriptional Analysis.” Nanotoxicology 15 (3): 380–399. https://doi.org/10.1080/17435390.2021.1872114.
- Burello, Enrico 2017. “Review of (Q)SAR Models for Regulatory Assessment of Nanomaterials Risks.” NanoImpact 8: 48–58. https://doi.org/10.1016/j.impact.2017.07.002.
- Cai, Xiaoming, Jun Dong, Jing Liu, Huizhen Zheng, Chitrada Kaweeteerawat, Fangjun Wang, Zhaoxia Ji, et al. 2018. “Multi-Hierarchical Profiling the Structure-Activity Relationships of Engineered Nanomaterials at Nano-Bio Interfaces.” Nature Communications 9 (1): 4416. https://doi.org/10.1038/s41467-018-06869-9.
- Canzler, Sebastian, and Jörg Hackermüller. 2020. “multiGSEA: A GSEA-Based Pathway Enrichment Analysis for Multi-Omics Data.” BMC Bioinformatics 21 (1): 561. https://doi.org/10.1186/s12859-020-03910-x.
- Cervantes-Gracia, Karla, Richard Chahwan, and Holger Husi. 2022. “Integrative OMICS Data-Driven Procedure Using a Derivatized Meta-Analysis Approach.” Frontiers in Genetics 13: 828786. https://doi.org/10.3389/fgene.2022.828786.
- Chen, Zhangjian, Di Zhou, Shuo Han, Shupei Zhou, and Guang Jia. 2019. “Hepatotoxicity and the Role of the Gut-Liver Axis in Rats after Oral Administration of Titanium Dioxide Nanoparticles.” Particle and Fibre Toxicology 16 (1): 48. https://doi.org/10.1186/s12989-019-0332-2.
- Chézeau, Laëtitia, Sylvie Sébillaud, Ramia Safar, Carole Seidel, Doulaye Dembélé, Mylène Lorcin, Cristina Langlais, et al. 2018. “Short- and Long-Term Gene Expression Profiles Induced by Inhaled TiO(2) Nanostructured Aerosol in Rat Lung.” Toxicology and Applied Pharmacology 356: 54–64. https://doi.org/10.1016/j.taap.2018.07.013.
- Ciura, Krzesimir, Elisa Moschini, Maciej Stępnik, Tommaso Serchi, Arno Gutleb, Kamila Jarzyńska, Karolina Jagiello, et al. 2024. “Toward Nano-Specific In Silico NAMs: How to Adjust Nano-QSAR to the Recent Advancements of Nanotoxicology?” Small (Weinheim an Der Bergstrasse, Germany) 20 (6): e2305581. https://doi.org/10.1002/smll.202305581.
- Comandella, Daniele, Stefania Gottardo, Iria M. Rio-Echevarria, and Hubert Rauscher. 2020. “Quality of Physicochemical Data on Nanomaterials: An Assessment of Data Completeness and Variability.” Nanoscale 12 (7): 4695–4708. https://doi.org/10.1039/C9NR08323E.
- Cosnier, Frédéric, Carole Seidel, Sarah Valentino, Otmar Schmid, Sébastien Bau, Ulla Vogel, Jérôme Devoy, et al. 2021. “Retained Particle Surface Area Dose Drives Inflammation in Rat Lungs following Acute, Subacute, and Subchronic Inhalation of Nanomaterials.” Particle and Fibre Toxicology 18 (1): 29. https://doi.org/10.1186/s12989-021-00419-w.
- Cross, Richard, Marianne Matzke, Dave Spurgeon, María Diez, Veronica Gonzalez Andres, Elena Cerro Galvez, Maria Fernanda Esponda, et al. 2022. “Assessing the Similarity of Nanoforms Based on the Biodegradation of Organic Surface Treatment Chemicals.” NanoImpact 26: 100395. https://doi.org/10.1016/j.impact.2022.100395.
- Cui, Li, Xiang Wang, Bingbing Sun, Tian Xia, and Shen Hu. 2019. “Predictive Metabolomic Signatures for Safety Assessment of Metal Oxide Nanoparticles.” ACS Nano 13 (11): 13065–13082. https://doi.org/10.1021/acsnano.9b05793.
- Decan, Nathalie, Dongmei Wu, Andrew Williams, Stéphane Bernatchez, Michael Johnston, Myriam Hill, Sabina Halappanavar, et al. 2016. “Characterization of in Vitro Genotoxic, Cytotoxic and Transcriptomic Responses following Exposures to Amorphous Silica of Different Sizes.” Mutation Research. Genetic Toxicology and Environmental Mutagenesis 796: 8–22. https://doi.org/10.1016/j.mrgentox.2015.11.011.
- Dekkers, Susan, Tim D. Williams, Jinkang Zhang, Jiarui (Albert) Zhou, Rob J. Vandebriel, Liset J. J. De La Fonteyne, Eric R. Gremmer, et al. 2018. “Multi-Omics Approaches Confirm Metal Ions Mediate the Main Toxicological Pathways of Metal-Bearing Nanoparticles in Lung Epithelial A549 Cells. Environmental Science.” Nano 5: 1506–1517.
- Drasler, Barbara, Phil Sayre, Klaus G. Steinhäuser, Alke Petri-Fink, and Barbara Rothen-Rutishauser. 2017. “In Vitro Approaches to Assess the Hazard of Nanomaterials.” NanoImpact 8: 99–116. https://doi.org/10.1016/j.impact.2017.08.002.
- Drew, Nathan M., Eileen D. Kuempel, Ying Pei, and Feng Yang. 2017. “A Quantitative Framework to Group Nanoscale and Microscale Particles by Hazard Potency to Derive Occupational Exposure Limits: Proof of Concept Evaluation.” Regulatory Toxicology and Pharmacology: RTP 89: 253–267. https://doi.org/10.1016/j.yrtph.2017.08.003.
- Duan, Yanqing, John S. Edwards, and Yogesh K. Dwivedi. 2019. “Artificial Intelligence for Decision Making in the Era of Big Data – Evolution, Challenges and Research Agenda.” International Journal of Information Management 48: 63–71. https://doi.org/10.1016/j.ijinfomgt.2019.01.021.
- Dumit, Verónica I., Yuk-Chien Liu, Aileen Bahl, Pekka Kohonen, Roland C. Grafström, Penny Nymark, Christine Müller-Graf, et al. 2023. “Meta-Analysis of Integrated Proteomic and Transcriptomic Data Discerns Structure-Activity-Relationship of Carbon Materials with Different Morphologies.” Advanced Science 11 (9): e2306268. https://doi.org/10.1002/advs.202306268.
- EC. 2004. "Regulation (EC) No 1935/2004 of the European Parliament and of the Council of 27 October 2004 on Materials and Articles Intended to Come into Contact with Food and Repealing Directives 80/590/EEC and 89/109/EEC." Official Journal of the European Union L 338, 47: p. 4–17
- EC. 2006. "Regulation (EC) No 1907/2006 of the European Parliament and of the Council of 18 December 2006 concerning the Registration, Evaluation, Authorisation and Restriction of Chemicals (REACH)." Official Journal of t European Union L 396, 49: p. 1–849.
- EC. 2009a. "Regulation (EC) No 1107/2009 of the European Parliament and of the Council of 21 October 2009 Con-Cerning the Placing of Plant Protection Products on the Market." Official Journal of the European Union L 309, 52: p. 1–50.
- EC. 2009b. "Regulation (EC) No 1223/2009 of the European Parliament and of the Council of 30 November 2009 on Cosmetic Products." Official Journal of the European Union L 342, 52: p. 59–209.
- EC. 2018. "EU 2018/1881 Amending Regulation (EC) No 1907/2006 of the European Parliament and of the Council on the Registration, Evaluation, Authorisation and Restriction of Chemicals (REACH)." Official Journal of the European Union L 308, 04/12/2018, 61: p. 1–20.
- ECHA. 2008. "Guidance on Information Requirements and Chemical Safety Assessment Chapter R.6: QSARs and Grouping of Chemicals." Helsinki, Finland. Available at: https://echa.europa.eu/documents/10162/13632/information_requirements_r6_en.pdf
- ECHA. 2016. "New Approach Methodologies in Regulatory Science" Proceedings of a Scientific Workshop. Helsinki, Finland. Available at: https://echa.europa.eu/documents/10162/2174236/scientific_ws_proceedings_en.pdf/a2087434-0407-4705-9057-95d9c2c2cc57. https://doi.org/10.2823/543644.
- ECHA. 2017. “Read-Across Assessment Framework (RAAF).” Helsinki, Finland. Available at: https://echa.europa.eu/documents/10162/13628/raaf_en.pdf/614e5d61-891d-4154-8a47-87efebd1851a.
- Edgar, Ron, Michael Domrachev, and Alex E. Lash. 2002. “Gene Expression Omnibus: NCBI Gene Expression and Hybridization Array Data Repository.” Nucleic Acids Research 30 (1): 207–210. https://doi.org/10.1093/nar/30.1.207.
- El Yamani, Naouale, Espen Mariussen, Maciej Gromelski, Ewelina Wyrzykowska, Dawid Grabarek, Tomasz Puzyn, Speranta Tanasescu, et al. 2022. “Hazard Identification of Nanomaterials: In Silico Unraveling of Descriptors for Cytotoxicity and Genotoxicity.” Nano Today. 46: 101581. https://doi.org/10.1016/j.nantod.2022.101581.
- Enea, Maria, Ana Margarida Araújo, Miguel Peixoto de Almeida, Maria Elisa Soares, Salomé Gonçalves-Monteiro, Paula Guedes de Pinho, Eulália Pereira, et al. 2019. “A Metabolomic Approach for the In Vivo Study of Gold Nanospheres and Nanostars after a Single-Dose Intravenous Administration to Wistar Rats.” Nanomaterials (Basel, Switzerland) 9 (11): 1606. https://doi.org/10.3390/nano9111606.
- Federico, Antonio, Angela Serra, My Kieu Ha, Pekka Kohonen, Jang-Sik Choi, Irene Liampa, Penny Nymark, et al. 2020. “Transcriptomics in Toxicogenomics, Part II: Preprocessing and Differential Expression Analysis for High Quality Data.” Nanomaterials 10 (5): 903. https://doi.org/10.3390/nano10050903.
- Fortino, Vittorio, Pia Anneli Sofia Kinaret, Michele Fratello, Angela Serra, Laura Aliisa Saarimäki, Audrey Gallud, Govind Gupta, et al. 2022. “Biomarkers of Nanomaterials Hazard from Multi-Layer Data.” Nature Communications 13 (1): 3798. https://doi.org/10.1038/s41467-022-31609-5.
- Furxhi, Irini, and Finbarr Murphy. 2020. “Predicting In Vitro Neurotoxicity Induced by Nanoparticles Using Machine Learning.” International Journal of Molecular Sciences 21 (15): 5280. https://doi.org/10.3390/ijms21155280.
- Furxhi, Irini, Finbarr Murphy, Barry Sheehan, Martin Mullins, and Paride Mantecca. 2018. "Predicting Nanomaterials toxicity pathways based on genome-wide transcriptomics studies using Bayesian networks," IEEE 18th International Conference on Nanotechnology (IEEE-NANO) Cork, Ireland. pp. 1-4 https://doi.org/10.1109/NANO.2018.8626300.
- Furxhi, Irini, Finbarr Murphy, Martin Mullins, Athanasios Arvanitis, and Craig A. 2020. “Practices and Trends of Machine Learning Application in Nanotoxicology.” Nanomaterials 10 (1): 116. https://doi.org/10.3390/nano10010116.
- Gajewicz, Agnieszka, Tomasz Puzyn, Katarzyna Odziomek, Piotr Urbaszek, Andrea Haase, Christian Riebeling, Andreas Luch, et al. 2018. “Decision Tree Models to Classify Nanomaterials according to the DF4nanoGrouping Scheme.” Nanotoxicology 12 (1): 1–17. https://doi.org/10.1080/17435390.2017.1415388.
- Gallud, Audrey, Mathilde Delaval, Pia Kinaret, Veer Singh Marwah, Vittorio Fortino, Jimmy Ytterberg, Roman Zubarev, et al. 2020. “Multiparametric Profiling of Engineered Nanomaterials: Unmasking the Surface Coating Effect.” Advanced Science 7 (22): 2002221. https://doi.org/10.1002/advs.202002221.
- Gareth James, D. W., Trevor Hastie, and Robert Tibshirani. 2021. "An Introduction to Statistical Learning: With Applications in R." New York, United States of America: Springer Texts in Statistics.
- Gareth James, D. W., Trevor Hastie, Robert Tibshirani, and Jonathan Taylor. 2023. "An Introduction to Statistical Learning: With Applications in Python." New York, United States of America: Springer Texts in Statistics.
- Gerloff, Kirsten, Brigitte Landesmann, Andrew Worth, Sharon Munn, Taina Palosaari, and Maurice Whelan. 2017. “The Adverse Outcome Pathway Approach in Nanotoxicology.” Computational Toxicology 1: 3–11. https://doi.org/10.1016/j.comtox.2016.07.001.
- Gernand, Jeremy M., and Elizabeth A. Casman. 2014. “A Meta-Analysis of Carbon Nanotube Pulmonary Toxicity Studies–How Physical Dimensions and Impurities Affect the Toxicity of Carbon Nanotubes.” Risk Analysis: An Official Publication of the Society for Risk Analysis 34 (3): 583–597. https://doi.org/10.1111/risa.12109.
- Gernand, Jeremy M., and Elizabth A. Casman. 2016. “Nanoparticle Characteristic Interaction Effects on Pulmonary Toxicity: A Random Forest Modeling Framework to Compare Risks of Nanomaterial Variants.” ASCE-ASME Journal of Risk and Uncertainty in Engineering 2 (2): 021002. https://doi.org/10.1115/1.4031216.
- Ghojavand, Solmaz, Fatemeh Bagheri, and Hamzeh Mesrian Tanha. 2019. “Integrative Meta-Analysis of Publically Available Microarray Datasets of Several Epithelial Cell Lines Identifies Biological Processes Affected by Silver Nanoparticles Exposure.” Comparative Biochemistry and Physiology. Toxicology & Pharmacology: CBP 216: 67–74. https://doi.org/10.1016/j.cbpc.2018.11.003.
- Giusti, Anna, Rambabu Atluri, Rositsa Tsekovska, Agnieszka Gajewicz, Margarita D. Apostolova, Chiara L. Battistelli, Eric A. J. Bleeker, et al. 2019. “Nanomaterial Grouping: Existing Approaches and Future Recommendations.” NanoImpact 16: 100182. https://doi.org/10.1016/j.impact.2019.100182.
- González-Durruthy, Michael, Silvana Manske Nunes, Juliane Ventura-Lima, Marcos A. Gelesky, Humberto González-Díaz, José M. Monserrat, Riccardo Concu, et al. 2019. “MitoTarget Modeling Using ANN-Classification Models Based on Fractal SEM Nano-Descriptors: Carbon Nanotubes as Mitochondrial F0F1-ATPase Inhibitors.” Journal of Chemical Information and Modeling 59 (1): 86–97. https://doi.org/10.1021/acs.jcim.8b00631.
- Gosens, Ilse, Pedro M. Costa, Magnus Olsson, Vicki Stone, Anna L. Costa, Andrea Brunelli, Elena Badetti, et al. 2021. “Pulmonary Toxicity and Gene Expression Changes after Short-Term Inhalation Exposure to Surface-Modified Copper Oxide Nanoparticles.” NanoImpact 22: 100313. https://doi.org/10.1016/j.impact.2021.100313.
- Grafström, Roland C., Penny Nymark, Vesa Hongisto, Ola Spjuth, Rebecca Ceder, Egon Willighagen, Barry Hardy, et al. 2015. “Toward the Replacement of Animal Experiments through the Bioinformatics-Driven Analysis of ‘Omics’ Data from Human Cell Cultures.” Alternatives to Laboratory Animals: ATLA 43 (5): 325–332. https://doi.org/10.1177/026119291504300506.
- Gromelski, Maciej, Filip Stoliński, Karolina Jagiello, Anna Rybińska-Fryca, Andrew Williams, Sabina Halappanavar, Ulla Vogel, et al. 2022. “AOP173 Key Event Associated Pathway Predictor – Online Application for the Prediction of Benchmark Dose Lower Bound (BMDLs) of a Transcriptomic Pathway Involved in MWCNTs-Induced Lung Fibrosis.” Nanotoxicology 16 (2): 183–194. https://doi.org/10.1080/17435390.2022.2064250.
- Gutierrez, Claudia Torero, Charis Loizides, Iosif Hafez, Anders Brostrøm, Henrik Wolff, Józef Szarek, Trine Berthing, et al. 2023. “Acute Phase Response following Pulmonary Exposure to Soluble and Insoluble Metal Oxide Nanomaterials in Mice.” Particle and Fibre Toxicology 20 (1): 4. https://doi.org/10.1186/s12989-023-00514-0.
- Halappanavar, Sabina, Monita Sharma, Silvia Solorio-Rodriguez, Håkan Wallin, Ulla Vogel, Kristie Sullivan, Amy J. Clippinger. 2023. "Substance Interaction with the Pulmonary Resident Cell Membrane Components Leading to Pulmonary Fibrosis." OECD Series on Adverse Outcome Pathways 33, OECD Publishing. https://doi.org/10.1787/10372cb8-en.
- Halappanavar, Sabina, James D. Ede, Jo A. Shatkin, and Harald F. Krug. 2019. “A Systematic Process for Identifying Key Events for Advancing the Development of Nanomaterial Relevant Adverse Outcome Pathways.” NanoImpact 15: 100178. https://doi.org/10.1016/j.impact.2019.100178.
- Halappanavar, Sabina, Anne Thoustrup Saber, Nathalie Decan, Keld Alstrup Jensen, Dongmei Wu, Nicklas Raun Jacobsen, Charles Guo, et al. 2015. “Transcriptional Profiling Identifies Physicochemical Properties of Nanomaterials That Are Determinants of the in Vivo Pulmonary Response.” Environmental and Molecular Mutagenesis 56 (2): 245–264. https://doi.org/10.1002/em.21936.
- Halappanavar, Sabina, James D. Ede, Indrani Mahapatra, Harald F. Krug, Eileen D. Kuempel, Iseult Lynch, Rob J. Vandebriel, et al. 2021a. “A Methodology for Developing Key Events to Advance Nanomaterial-Relevant Adverse Outcome Pathways to Inform Risk Assessment.” Nanotoxicology 15 (3): 289–310. https://doi.org/10.1080/17435390.2020.1851419.
- Halappanavar, Sabina, Penny Nymark, Harald F. Krug, Martin J. D. Clift, Barbara Rothen-Rutishauser, and Ulla Vogel. 2021b. “Non-Animal Strategies for Toxicity Assessment of Nanoscale Materials: Role of Adverse Outcome Pathways in the Selection of Endpoints.” Small 17 (15): e2007628. https://doi.org/10.1002/smll.202007628.
- Halappanavar, Sabina, Sybille van den Brule, Penny Nymark, Laurent Gaté, Carole Seidel, Sarah Valentino, Vadim Zhernovkov, et al. 2020. “Adverse Outcome Pathways as a Tool for the Design of Testing Strategies to Support the Safety Assessment of Emerging Advanced Materials at the Nanoscale.” Particle and Fibre Toxicology 17 (1): 16. https://doi.org/10.1186/s12989-020-00344-4.
- Halder, Amit K., André. Melo, and M. Natália D. S. Cordeiro. 2020. “A Unified in Silico Model Based on Perturbation Theory for Assessing the Genotoxicity of Metal Oxide Nanoparticles.” Chemosphere 244: 125489. https://doi.org/10.1016/j.chemosphere.2019.125489.
- Jagiello, Karolina, Sabina Halappanavar., Anna Rybińska-Fryca, Andrew Willliams, Ulla Vogel, and Tomasz Puzyn. 2021. “Transcriptomics-Based and AOP-Informed Structure-Activity Relationshipsto Predict Pulmonary Pathology Induced by Multiwalled Carbon Nanotubes.” Small 17 (15): e2003465. https://doi.org/10.1002/smll.202003465.
- Jagiello, Karolina, Anita Sosnowska, Maciej Stępnik, Maciej Gromelski,Karolina Płonka.. 2022. “Nano-Specific Alternative Methods in Human Hazard/Safety Assessment under Different EU Regulations, considering the Animal Testing Bans Already in Place for Cosmetics and Their Ingredients.” EUON Report Helsinki, Finland. Available at: https://euon.echa.europa.eu/documents/2435000/3268573/ECHA-62-2022_final_report_published_02aug2023.pdf/4071837e-c4de-b496-b00f-4e5317646713?t=1691138948441.
- Jeliazkova, Nina, Lan Ma-Hock, Gemma Janer, Heidi Stratmann, and Wendel Wohlleben. 2022a. “Possibilities to Group Nanomaterials across Different Substances – A Case Study on Organic Pigments.” NanoImpact 26: 100391. https://doi.org/10.1016/j.impact.2022.100391.
- Jeliazkova, Nina, Charalampos Chomenidis, Philip Doganis, Bengt Fadeel, Roland Grafström, Barry Hardy, Janna Hastings, et al. 2015. “The eNanoMapper Database for Nanomaterial Safety Information.” Beilstein Journal of Nanotechnology 6: 1609–1634. https://doi.org/10.3762/bjnano.6.165.
- Jeliazkova, Nina, Eric Bleeker, Richard Cross, Andrea Haase, Gemma Janer, Willie Peijnenburg, Mario Pink, et al. 2022b. “How Can We Justify Grouping of Nanoforms for Hazard Assessment? Concepts and Tools to Quantify Similarity.” NanoImpact 25: 100366. https://doi.org/10.1016/j.impact.2021.100366.
- Jeliazkova, Nina, Margarita D. Apostolova, Cristina Andreoli, Flavia Barone, Andrew Barrick, Chiara Battistelli, Cecilia Bossa, et al. 2021. “Towards FAIR Nanosafety Data.” Nature Nanotechnology 16 (6): 644–654. https://doi.org/10.1038/s41565-021-00911-6.
- Karatzas, Pantelis , Georgia Melagraki, Laura-Jayne A. Ellis, Iseult Lynch, Dimitra-Danai Varsou, Antreas Afantitis, Andreas Tsoumanis, et al. 2020. "Development of Deep Learning Models for Predicting the Effects of Exposure to Engineered Nanomaterials on Daphnia magna." Small 16 (36): e2001080. https://doi.org/10.1002/smll.202001080.
- Karkossa, Isabel, Anne Bannuscher, Bryan Hellack, Aileen Bahl, Sophia Buhs, Peter Nollau, Andreas Luch, et al. 2019. “An in-Depth Multi-Omics Analysis in RLE-6TN Rat Alveolar Epithelial Cells Allows for Nanomaterial Categorization.” Particle and Fibre Toxicology 16 (1): 38. https://doi.org/10.1186/s12989-019-0321-5.
- Keller, Johannes G., Michael Persson, Philipp Müller, Lan Ma-Hock, Kai Werle, Josje Arts, Robert Landsiedel, et al. 2021. “Variation in Dissolution Behavior among Different Nanoforms and Its Implication for Grouping Approaches in Inhalation Toxicity.” NanoImpact 23: 100341. https://doi.org/10.1016/j.impact.2021.100341.
- Kinaret, Pia Anneli Sofia, Angela Serra, Antonio Federico, Pekka Kohonen, Penny Nymark, Irene Liampa, My Kieu Ha, et al. 2020. “Transcriptomics in Toxicogenomics, Part I: Experimental Design, Technologies, Publicly Available Data, and Regulatory Aspects.” Nanomaterials 10 (4): 750. https://doi.org/10.3390/nano10040750.
- Kinaret, Pia., Marit Ilves, Vittorio Fortino, Elina Rydman, Piia Karisola, Anna Lähde, Joonas Koivisto, et al. 2017a. “Inhalation and Oropharyngeal Aspiration Exposure to Rod-Like Carbon Nanotubes Induce Similar Airway Inflammation and Biological Responses in Mouse Lungs.” ACS Nano 11 (1): 291–303. https://doi.org/10.1021/acsnano.6b05652.
- Kinaret, Pia., Veer Marwah, Vittorio Fortino, Marit Ilves, Henrik Wolff, Lasse Ruokolainen, Petri Auvinen, et al. 2017b. “Network Analysis Reveals Similar Transcriptomic Responses to Intrinsic Properties of Carbon Nanomaterials in Vitro and in Vivo.” ACS Nano 11 (4): 3786–3796. https://doi.org/10.1021/acsnano.6b08650.
- Kohonen, Pekka, Juuso A. Parkkinen, Egon L. Willighagen, Rebecca Ceder, Krister Wennerberg, Samuel Kaski, Roland C. Grafström, et al. 2017. “A Transcriptomics Data-Driven Gene Space Accurately Predicts Liver Cytopathology and Drug-Induced Liver Injury.” Nature Communications 8 (1): 15932. https://doi.org/10.1038/ncomms15932.
- Kohonen Pekka J., Penny Nymark, Valtteri Hongisto, Roland Grafström. 2019. “Predictive toxicogenomics space modeling serves effectivelyto sensitive biomarker-based read across from capturing toxicmode-of-action of lowest-observable effect levels.” Abstracts of the 55th Congress of the European Societies of Toxicology (EUROTOX 2019), Toxicology Letters 314: S1–S309. https://doi.org/10.1016/j.toxlet.2019.09.002.
- Kotzabasaki, Marianna, Iason Sotiropoulos, Costas Charitidis, and Haralas Sarimveis. 2021. “Machine Learning Methods for Multi-Walled Carbon Nanotubes (MWCNT) Genotoxicity Prediction.” Nanoscale Advances 3 (11): 3167–3176. https://doi.org/10.1039/D0NA00600A.
- Labib, Sarah, Andrew Williams, Carole L. Yauk, Jake K. Nikota, Håkan Wallin, Ulla Vogel, Sabina Halappanavar, et al. 2016. “Nano-Risk Science: application of Toxicogenomics in an Adverse Outcome Pathway Framework for Risk Assessment of Multi-Walled Carbon Nanotubes.” Particle and Fibre Toxicology 13 (1): 15. https://doi.org/10.1186/s12989-016-0125-9.
- Lamon, Lara, David Asturiol, Andrea Richarz, Elisabeth Joossens, Rabea Graepel, Karin Aschberger, Andrew Worth, et al. 2018. “Grouping of Nanomaterials to Read-across Hazard Endpoints: From Data Collection to Assessment of the Grouping Hypothesis by Application of Chemoinformatic Techniques.” Particle and Fibre Toxicology 15 (1): 37. https://doi.org/10.1186/s12989-018-0273-1.
- Lamon, Lara, Karin Aschberger, David Asturiol, Andrea Richarz, and Andrew Worth. 2019. “Grouping of Nanomaterials to Read-across Hazard Endpoints: A Review.” Nanotoxicology 13 (1): 100–118. https://doi.org/10.1080/17435390.2018.1506060.
- Langfelder, Peter, and Steve Horvath. 2008. “WGCNA: An R Package for Weighted Correlation Network Analysis.” BMC Bioinformatics 9 (1): 559. https://doi.org/10.1186/1471-2105-9-559.
- Leaman, Robert, Chih-Hsuan Wei and Zhiyong Lu. 2015. “tmChem: A High Performance Approach for Chemical Named Entity Recognition and Normalization.” Journal of Cheminformatics 7 Supp 1: S3. https://doi.org/10.1186/1758-2946-7-S1-S3.
- Lever, Jake Eric Y. Zhao, Jasleen Grewal, Martinc R. Jones, and Seven J. Jones. 2019. "CancerMine: a Literature-Mined Resource for Drivers, Oncogenes and Tumor Suppressors in Cancer." Nature Methods 16, 505–507. https://doi.org/10.1038/s41592-019-0422-y
- Li, Jing, Chuanxi Wang, Le Yue, Feiran Chen, Xuesong Cao, and Zhenyu Wang. 2022. “Nano-QSAR Modeling for Predicting the Cytotoxicity of Metallic and Metal Oxide Nanoparticles: A Review.” Ecotoxicology and Environmental Safety 243: 113955. https://doi.org/10.1016/j.ecoenv.2022.113955.
- Liu, Fei T., Kai M. Ting, and Zhi-Hua Zhou. 2008. "Isolation Forest" Eighth IEEE International Conference on Data Mining Pisa, Italy, pp. 413-422, doi: 10.1109/ICDM.2008.17.
- Lynch, Iseult, Carsten Weiss, and Eugenia Valsami-Jones. 2014. “A Strategy for Grouping of Nanomaterials Based on Key Physico-Chemical Descriptors as a Basis for Safer-by-Design NMs.” Nano Today. 9 (3): 266–270. https://doi.org/10.1016/j.nantod.2014.05.001.
- Lynch, Iseult, Antreas Afantitis, Thomas Exner, Martin Himly, Vladimir Lobaskin, Philip Doganis, Dieter Maier, et al. 2020. “Can an InChI for Nano Address the Need for a Simplified Representation of Complex Nanomaterials across Experimental and Nanoinformatics Studies?” Nanomaterials 10 (12): 2493. https://doi.org/10.3390/nano10122493.
- Ma, Nyuk Ling, Nan Zhang, Wilson Thau Lym Yong, Suzana Misbah, Fatimah Hashim, Chin Fhong Soon, Gim Pao Lim, et al. 2023. “Use, Exposure and Omics Characterisation of Potential Hazard in Nanomaterials.” Materials Today Advances 17: 100341. https://doi.org/10.1016/j.mtadv.2023.100341.
- Marchese Robinson, Richard L., Iseult Lynch, Willie Peijnenburg, John Rumble, Fred Klaessig, Clarissa Marquardt, Hubert Rauscher, et al. 2016. “How Should the Completeness and Quality of Curated Nanomaterial Data Be Evaluated?” Nanoscale 8 (19): 9919–9943. https://doi.org/10.1039/C5NR08944A.
- Martens, Marvin, Tim Verbruggen, Penny Nymark, Roland Grafström, Lyle D. Burgoon, Hristo Aladjov, Fernando Torres Andón, et al. 2018. “Introducing WikiPathways as a Data-Source to Support Adverse Outcome Pathways for Regulatory Risk Assessment of Chemicals and Nanomaterials.” Frontiers in Genetics 9: 661. https://doi.org/10.3389/fgene.2018.00661.
- Martín, Laura, Luis Sánchez, Jorge Lanza, and Pablo Sotres. 2023. “Development and Evaluation of Artificial Intelligence Techniques for IoT Data Quality Assessment and Curation.” Internet of Things 22: 100779. https://doi.org/10.1016/j.iot.2023.100779.
- Marvin, Hans J. P., Yamine Bouzembrak, Esmée M. Janssen, Meike van der Zande, Finbarr Murphy, Barry Sheehan, Martin Mullins, et al. 2017. “Application of Bayesian Networks for Hazard Ranking of Nanomaterials to Support Human Health Risk Assessment.” Nanotoxicology 11 (1): 123–133. https://doi.org/10.1080/17435390.2016.1278481.
- Marwah, Veer Singh, Giovanni Scala, Pia Anneli Sofia Kinaret, Angela Serra, Harri Alenius, Vittorio Fortino, Dario Greco, et al. 2019. “eUTOPIA: solUTion for Omics Data PreprocessIng and Analysis.” Source Code for Biology and Medicine 14 (1): 1. https://doi.org/10.1186/s13029-019-0071-7.
- Mech, Agniezka, Kirsten Rasmussen, Paula Jantunen, Lothar Aicher, Maria Alessandrelli, Ulrike Bernauer, Eric A. J. Bleeker, et al. 2019. “Insights into Possibilities for Grouping and Read-across for Nanomaterials in EU Chemicals Legislation.” Nanotoxicology 13 (1): 119–141. https://doi.org/10.1080/17435390.2018.1513092.
- Michelhaugh, Sam A., and James L. Januzzi. Jr. 2022. “Using Artificial Intelligence to Better Predict and Develop Biomarkers.” Heart Failure Clinics 18 (2): 275–285. https://doi.org/10.1016/j.hfc.2021.11.004.
- Miranda, Renata R., Anny C. S. Oliveira, Lilian. Skytte, Kaare. L. Rasmussen, and Frank. Kjeldsen. 2022. “Proteome-Wide Analysis Reveals Molecular Pathways Affected by AgNP in a ROS-Dependent Manner.” Nanotoxicology 16 (1): 73–87. https://doi.org/10.1080/17435390.2022.2036844.
- Murphy, Fiona A., Helinor J. Johnston, Susan Dekkers, Eric A. J. Bleeker, Agnes G. Oomen, Teresa F. Fernandes, Kirsten Rasmussen, et al. 2023. “How to Formulate Hypotheses and IATAs to Support Grouping and Read-across of Nanoforms.” Altex 40 (1): 125–140. https://doi.org/10.14573/altex.2203241.
- Murugadoss, Sivakumar, Nilakash Das, Lode Godderis, Jan Mast, Peter H. Hoet, and Manosij Ghosh. 2021. “Identifying Nanodescriptors to Predict the Toxicity of Nanomaterials: A Case Study on Titanium Dioxide.” Environmental Science: Nano 8 (2): 580–590. https://doi.org/10.1039/D0EN01031F.
- Ng, Sandra, Sara Masarone, David Watson, and Michael R. Barnes. 2023. “The Benefits and Pitfalls of Machine Learning for Biomarker Discovery.” Cell and Tissue Research 394 (1): 17–31. https://doi.org/10.1007/s00441-023-03816-z.
- Nguyen, Nhung H. A., and Priscila. Falagan-Lotsch. 2023. “Mechanistic Insights into the Biological Effects of Engineered Nanomaterials: A Focus on Gold Nanoparticles.” International Journal of Molecular Sciences 24 (4): 4109. https://doi.org/10.3390/ijms24044109.
- Nikota, Jake, Andrew Williams, Carole L. Yauk, Håkan Wallin, Ulla Vogel, and Sabina Halappanavar. 2016. “Meta-Analysis of Transcriptomic Responses as a Means to Identify Pulmonary Disease Outcomes for Engineered Nanomaterials.” Particle and Fibre Toxicology 13 (1): 25. https://doi.org/10.1186/s12989-016-0137-5.
- Nymark, Penny, Hanna L. Karlsson, Sabina Halappanavar, and Ulla Vogel. 2021. “Adverse Outcome Pathway Development for Assessment of Lung Carcinogenicity by Nanoparticles.” Frontiers in Toxicology 3: 653386. https://doi.org/10.3389/ftox.2021.653386.
- Nymark, Penny, Peter Wijshoff, Rachel Cavill, Marcel van Herwijnen, Maarten L. J. Coonen, Sandra Claessen, Julia Catalán, et al. 2015. “Extensive Temporal Transcriptome and microRNA Analyses Identify Molecular Mechanisms Underlying Mitochondrial Dysfunction Induced by Multi-Walled Carbon Nanotubes in Human Lung Cells.” Nanotoxicology 9 (5): 624–635. https://doi.org/10.3109/17435390.2015.1017022.
- OECD. 2014a. “Guidance on Grouping of Chemicals. second Edition.” OECD Series on Testing & Assessment No. 194, OECD Publishing, Paris, France, https://doi.org/10.1787/9789264274679-en.
- OECD. 2014b. "Guidance Document on the Validation of (Quantitative) Structure-Activity Relationship [(Q)SAR] Models." OECD Series on Testing and Assessment, No. 69, OECD Publishing, Paris, France, https://doi.org/10.1787/9789264085442-en.
- OECD. 2021a. "Metabolomic Reporting Framework." Available at: https://web-archive.oecd.org/2021-09-30/592995-metabolomics-reporting-framework.pdf
- OECD. 2021b. "Transcriptomic Reporting Framework." Available at: https://web-archive.oecd.org/2021-09-30/592996-transcriptomic-reporting-framework.pdf
- OECD. 2023. "(Q)SAR Assessment Framework: Guidance for the Regulatory Assessment of (Quantitative) Structure − Activity Relationship Models, Predictions, and Results Based on Multiple Predictions." Available at: https://www.oecd.org/chemicalsafety/risk-assessment/qsar-assessment-framework.pdf
- Oomen, Agnes G., Eric A. J. Bleeker, Peter M. J. Bos, Fleur van Broekhuizen, Stefania Gottardo, Monique Groenewold, Danail Hristozov, et al. 2015. “Grouping and Read-Across Approaches for Risk Assessment of Nanomaterials.” International Journal of Environmental Research and Public Health 12 (10): 13415–13434. https://doi.org/10.3390/ijerph121013415.
- Oomen, Agnes G., Klaus G. Steinhäuser, Eric A. J. Bleeker, Fleur van Broekhuizen, Adriënne Sips, Susan Dekkers, Susan W. P. Wijnhoven, et al. 2018. “Risk Assessment Frameworks for Nanomaterials: Scope, Link to Regulations, Applicability, and Outline for Future Directions in View of Needed Increase in Efficiency.” NanoImpact 9: 1–13. https://doi.org/10.1016/j.impact.2017.09.001.
- Perez-Riverol, Yasset, Jingwen Bai, Chakradhar Bandla, David García-Seisdedos, Suresh Hewapathirana, Selvakumar Kamatchinathan, Deepti J. Kundu, et al. 2021. “The PRIDE Database Resources in 2022: A Hub for Mass Spectrometry-Based Proteomics Evidences.” Nucleic Acids Research 50 (D1): D543–D552. https://doi.org/10.1093/nar/gkab1038.
- Picard, Milan, Marie-Pier Scott-Boyer, Antoine Bodein, Olivier Périn, and Arnaud Droit. 2021. “Integration Strategies of Multi-Omics Data for Machine Learning Analysis.” Computational and Structural Biotechnology Journal 19: 3735–3746. https://doi.org/10.1016/j.csbj.2021.06.030.
- Poulsen, Sarah S., Petra Jackson, Kirsten Kling, Kristina B. Knudsen, Vidar Skaug, Zdenka O. Kyjovska, Birthe L. Thomsen, et al. 2016. “Multi-Walled Carbon Nanotube Physicochemical Properties Predict Pulmonary Inflammation and Genotoxicity.” Nanotoxicology 10 (9): 1263–1275. https://doi.org/10.1080/17435390.2016.1202351.
- Poulsen, Sarah S., Stefan Bengtson, Andrew Williams, Nicklas R. Jacobsen, Jesper T. Troelsen, Sabina Halappanavar, Ulla Vogel, et al. 2021. “A Transcriptomic Overview of Lung and Liver Changes One Day after Pulmonary Exposure to Graphene and Graphene Oxide.” Toxicology and Applied Pharmacology 410: 115343. https://doi.org/10.1016/j.taap.2020.115343.
- Rasmussen, Kirsten., Hubert Rauscher, Peter Kearns, Mar González, and Juan Riego Sintes. 2019. “Developing OECD Test Guidelines for Regulatory Testing of Nanomaterials to Ensure Mutual Acceptance of Test Data.” Regulatory Toxicology and Pharmacology: RTP 104: 74–83. https://doi.org/10.1016/j.yrtph.2019.02.008.
- Ribeiro, Ana R., Paulo E. Leite, Priscila Falagan-Lotsch, Federico Benetti, Christian Micheletti, Hans C. Budtz, Nicklas R. Jacobsen, et al. 2017. “Challenges on the Toxicological Predictions of Engineered Nanoparticles.” NanoImpact 8: 59–72. https://doi.org/10.1016/j.impact.2017.07.006.
- Roy, Kunal, Supratik Kar, and Pravin Ambure. 2015. “On a Simple Approach for Determining Applicability Domain of QSAR Models.” Chemometrics and Intelligent Laboratory Systems 145: 22–29. https://doi.org/10.1016/j.chemolab.2015.04.013.
- Ruggiero, Emmanuel, Katherine Y. Santizo, Michael Persson, Camilla Delpivo, and Wendel Wohlleben. 2022. “Food Contact of Paper and Plastic Products Containing SiO2, Cu-Phthalocyanine, Fe2O3, CaCO3: Ranking Factors That Control the Similarity of Form and Rate of Release.” NanoImpact 25: 100372. https://doi.org/10.1016/j.impact.2021.100372.
- Saarimäki, Laura Aliisa, Antonio Federico, Iseult Lynch, Anastasios G. Papadiamantis, Andreas Tsoumanis, Georgia Melagraki, Antreas Afantitis, et al. 2021. “Manually Curated Transcriptomics Data Collection for Toxicogenomic Assessment of Engineered Nanomaterials.” Scientific Data 8 (1): 49. https://doi.org/10.1038/s41597-021-00808-y.
- Saarimäki, Laura Aliisa, Jack Morikka, Alisa Pavel, Seela Korpilähde, Giusy Del Giudice, Antonio Federico, Michele Fratello, et al. 2023. “Toxicogenomics Data for Chemical Safety Assessment and Development of New Approach Methodologies: An Adverse Outcome Pathway-Based Approach.” Advanced Science (Weinheim, Baden-Wurttemberg, Germany) 10 (2): e2203984. https://doi.org/10.1002/advs.202203984.
- Santorsola, Mariangela, and Francesco Lescai. 2023. “The Promise of Explainable Deep Learning for Omics Data Analysis: Adding New Discovery Tools to AI.” New Biotechnology 77: 1–11. https://doi.org/10.1016/j.nbt.2023.06.002.
- Schmeisser, Sebastian, Andrea Miccoli, Martin von Bergen, Elisabet Berggren, Albert Braeuning, Wibke Busch, Christian Desaintes, et al. 2023. “New Approach Methodologies in Human Regulatory Toxicology – Not If, but How and When!.” Environment International 178: 108082. https://doi.org/10.1016/j.envint.2023.108082.
- Seidel, Carole, Vadim Zhernovkov, Hilary Cassidy, Boris Kholodenko, David Matallanas, Frédéric Cosnier, Laurent Gaté, et al. 2021. “Inhaled Multi-Walled Carbon Nanotubes Differently Modulate Global Gene and Protein Expression in Rat Lungs.” Nanotoxicology 15 (2): 238–256. https://doi.org/10.1080/17435390.2020.1851418.
- Serra, Angela, Laura A. Saarimäki, Michele Fratello, Veer S. Marwah, and Dario Greco. 2020a. “BMDx: A Graphical Shiny Application to Perform Benchmark Dose Analysis for Transcriptomics Data.” Bioinformatics (Oxford, England) 36 (9): 2932–2933. https://doi.org/10.1093/bioinformatics/btaa030.
- Serra, Angela, Ivica Letunic, Vittorio Fortino, Richard D. Handy, Bengt Fadeel, Roberto Tagliaferri, Dario Greco, et al. 2019. “INSIdE NANO: A Systems Biology Framework to Contextualize the Mechanism-of-Action of Engineered Nanomaterials.” Scientific Reports 9 (1): 179. https://doi.org/10.1038/s41598-018-37411-y.
- Serra, Angela, Michele Fratello, Giusy Del Giudice, Laura Aliisa Saarimäki, Michelangelo Paci, Antonio Federico, Dario Greco, et al. 2020b. “TinderMIX: Time-Dose Integrated Modelling of Toxicogenomics Data.” GigaScience 9 (5): giaa055. https://doi.org/10.1093/gigascience/giaa055.
- Serra, Angela, Michele Fratello, Luca Cattelani, Irene Liampa, Georgia Melagraki, Pekka Kohonen, Penny Nymark, et al. 2020c. “Transcriptomics in Toxicogenomics, Part III: Data Modelling for Risk Assessment.” Nanomaterials 10 (4): 708. https://doi.org/10.3390/nano10040708.
- Shao, Gen, Anna Beronius, and Penny Nymark. 2023. “SciRAPnano: A Pragmatic and Harmonized Approach for Quality Evaluation of in Vitro Toxicity Data to Support Risk Assessment of Nanomaterials.” Frontiers in Toxicology 5: 1319985. https://doi.org/10.3389/ftox.2023.1319985.
- Sheehan, Barry, Finbarr Murphy, Martin Mullins, Irini Furxhi, Anna L. Costa, Felice C. Simeone, Paride Mantecca, et al. 2018. “Hazard Screening Methods for Nanomaterials: A Comparative Study.” International Journal of Molecular Sciences 19 (3): 649. https://doi.org/10.3390/ijms19030649.
- Shin, Tae Hwan, Balachandran Manavalan, Da Yeon Lee, Shaherin Basith, Chan Seo, Man Jeong Paik, Sang-Wook Kim, et al. 2021. “Silica-Coated Magnetic-Nanoparticle-Induced Cytotoxicity is Reduced in Microglia by Glutathione and Citrate Identified Using Integrated Omics.” Particle and Fibre Toxicology 18 (1): 42. https://doi.org/10.1186/s12989-021-00433-y.
- Shvetsova, Nina, Bart Bakker, Irina Fedulova, Heinrich Schulz, and Dmitry. V. Dylov. 2021. “Anomaly Detection in Medical Imaging With Deep Perceptual Autoencoders.” IEEE Access. 9: 118571–118583. https://doi.org/10.1109/ACCESS.2021.3107163.
- Sizochenko, Natalia, Michael Syzochenko, Natalja Fjodorova, Bakhtiyor. Rasulev, and Jerzy Leszczynski. 2019. “Evaluating Genotoxicity of Metal Oxide Nanoparticles: Application of Advanced Supervised and Unsupervised Machine Learning Techniques.” Ecotoxicology and Environmental Safety 185: 109733. https://doi.org/10.1016/j.ecoenv.2019.109733.
- Snyder-Talkington, Brandi N., Chunlin Dong, Xiangyi Zhao, Julian Dymacek, Dale W. Porter, Michael G. Wolfarth, Vincent Castranova, et al. 2015. “Multi-Walled Carbon Nanotube-Induced Gene Expression in Vitro: concordance with in Vivo Studies.” Toxicology 328: 66–74. https://doi.org/10.1016/j.tox.2014.12.012.
- Solorio-Rodriguez, Silvia Aidee, Andrew Williams, Sarah Søs Poulsen, Kristina Bram Knudsen, Keld Alstrup Jensen, Per Axel Clausen, Pernille Høgh Danielsen, et al. 2023. “Single-Walled Vs. Multi-Walled Carbon Nanotubes: Influence of Physico-Chemical Properties on Toxicogenomics Responses in Mouse Lungs.” Nanomaterials 13 (6): 1059. https://doi.org/10.3390/nano13061059.
- Song, Yuchao., Eric Bleeker, Richard K. Cross, Martina G. Vijver, and Willie J. G. M. Peijnenburg. 2022. “Similarity Assessment of Metallic Nanoparticles within a Risk Assessment Framework: A Case Study on Metallic Nanoparticles and Lettuce.” NanoImpact 26: 100397. https://doi.org/10.1016/j.impact.2022.100397.
- Stone, Vicki, Stefania Gottardo, Eric A. J. Bleeker, Hedwig Braakhuis, Susan Dekkers, Teresa Fernandes, Andrea Haase, et al. 2020. “A Framework for Grouping and Read-across of Nanomaterials- Supporting Innovation and Risk Assessment.” Nano Today. 35: 100941. https://doi.org/10.1016/j.nantod.2020.100941.
- Swain, Matthew C., and Jaqueline M. Cole. 2016. “ChemDataExtractor: A Toolkit for Automated Extraction of Chemical Information from the Scientific Literature.” Journal of Chemical Information and Modeling 56 (10): 1894–1904. https://doi.org/10.1021/acs.jcim.6b00207.
- Torres, Anaelle, Véronique Collin-Faure, Hélène Diemer, Christine Moriscot, Daphna Fenel, Benoît Gallet, Sarah Cianférani, et al. 2022. “Repeated Exposure of Macrophages to Synthetic Amorphous Silica Induces Adaptive Proteome Changes and a Moderate Cell Activation.” Nanomaterials (Basel, Switzerland) 12 (9): 1424. https://doi.org/10.3390/nano12091424.
- van Rijn, Jeaphianne, Antreas Afantitis, Mustafa Culha, Maria Dusinska, Thomas E. Exner, Nina Jeliazkova, Eleonora Marta Longhin, et al. 2022. “European Registry of Materials: global, Unique Identifiers for (Undisclosed) Nanomaterials.” Journal of Cheminformatics 14 (1): 57. https://doi.org/10.1186/s13321-022-00614-7.
- Varsou, Dimitra-Danai, Georgia Tsiliki, Penny Nymark, Pekka Kohonen, Roland Grafström, and Haralambos Sarimveis. 2018. “toxFlow: A Web-Based Application for Read-Across Toxicity Prediction Using Omics and Physicochemical Data.” Journal of Chemical Information and Modeling 58 (3): 543–549. https://doi.org/10.1021/acs.jcim.7b00160.
- Wilkinson, Mark D., Michel Dumontier, I. Jsbrand Jan Aalbersberg, Gabrielle Appleton, Myles Axton, Arie Baak, Niklas Blomberg, et al. 2016. “The FAIR Guiding Principles for Scientific Data Management and Stewardship.” Scientific Data 3 (1): 160018. https://doi.org/10.1038/sdata.2016.18.
- Williams, Andrew, and Sabina Halappanavar. 2015. “Application of Biclustering of Gene Expression Data and Gene Set Enrichment Analysis Methods to Identify Potentially Disease Causing Nanomaterials.” Beilstein Journal of Nanotechnology 6: 2438–2448. https://doi.org/10.3762/bjnano.6.252.
- Wohlleben, Wendel, Annette Mehling, and Robert Landsiedel. 2023. “Lessons Learned from the Grouping of Chemicals to Assess Risks to Human Health.” Angewandte Chemie (International Ed. in English) 62 (22): e202210651. https://doi.org/10.1002/anie.202210651.
- Xu, Jun., Yonghui Wu, Yaoyun Zhang, Jingqi Wang, Hee-Jin Lee, and Hua Xu. 2016. “CD-REST: A System for Extracting Chemical-Induced Disease Relation in Literature.” Database: The Journal of Biological Databases and Curation 2016: baw036. https://doi.org/10.1093/database/baw036.
- Yanamala, Naveena, Ishika C. Desai, William Miller, Vamsi K. Kodali, Girija Syamlal, Jenny R. Roberts, Aaron D. Erdely, et al. 2019. “Grouping of Carbonaceous Nanomaterials Based on Association of Patterns of Inflammatory Markers in BAL Fluid with Adverse Outcomes in Lungs.” Nanotoxicology 13 (8): 1102–1116. https://doi.org/10.1080/17435390.2019.1640911.
- Ye, Chaoyang, Daniel J. Ho, Marilisa Neri, Chian Yang, Tripti Kulkarni, Ranjit Randhawa, Martin Henault, et al. 2018. “DRUG-Seq for Miniaturized High-Throughput Transcriptome Profiling in Drug Discovery.” Nature Communications 9 (1): 4307. https://doi.org/10.1038/s41467-018-06500-x.
- Zheng, Si, Shasia Dharssi, Meng Wu, Jiao Li, and Zhiyong Lu. 2019. "Text Mining for Drug Discovery." Methods in Molecular Biology 1939: 231–252. https://doi.org/10.1007/978-1-4939-9089-4_13.