1. Introduction: drug repositioning and current strategies
Drug research is a time-consuming and expensive process, where the main objective is to assess the impact of a specific drug in one or more biological targets for the treatment of certain disease. However, from all drugs evaluated in clinical development every year, only 15% of them are approved or even considered safe [Citation1]. In this respect, there are several limitations associated with drug discovery, such as the identification of a specific target as a relevant biomarker, adverse effects not previously identified in early stages of long-term treatments, and heterogeneity of patients and its implication in drug effectiveness; all these especially relevant in rheumatic diseases [Citation2]. In light of these issues, drug repositioning emerges as a new field in the drug industry. Drug repositioning is defined as the identification of new applications for existing drugs that have already passed development phases and were previously indicated to treat other diseases. Thus, this method allows saving time and resources providing new applications to existing drugs with known safety profiles [Citation3]. Traditionally, it has been centered on drug-based and disease-based approaches, focused on drugs and disease features, respectively. However, nowadays the signature-based strategy evaluates the transcriptomic profiles on the basis of drug or disease similarities, as mentioned in a recent review by Kingsmore and colleagues [Citation4]. In this review, we will focus on genomic methodologies for drug repositioning in systemic seropositive rheumatic diseases from a disease-based perspective.
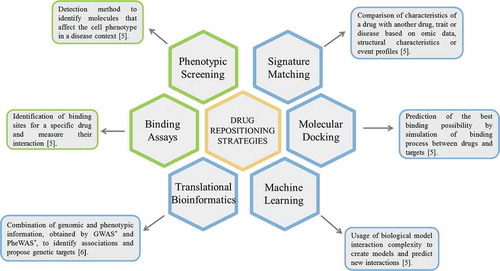
The first steps in drug repositioning starts with the selection of a drug directed to a specific indication. Then, the evaluation of the current knowledge of the drug based on previously described effects and its efficacy in phase II trials. This step is the most important and where diverse strategies are applied, mainly computational and experimental [Citation5,Citation6]. Due to the novelty of the field, there is still not a consensus on the classification strategies, although the computational techniques are the most used [Citation7]. These strategies are supported by clinical data and databases, and help to identify possible targets as well as different drug applications [Citation7]. Regarding the experimental approaches, there are several in vitro techniques that are very useful to test the effectiveness and safety of these drugs in the context of different diseases [Citation5]. The main strategies are summarized in .
2. Systemic seropositive rheumatic diseases and drug repositioning
Autoimmune diseases are a group of disorders characterized by the loss of immune self-tolerance of the organism, generating autoantibodies against its own tissues [Citation8]. In the cases where there is a systemic autoantibody response and the musculoskeletal system is affected, the disorders are known as ‘systemic seropositive rheumatic diseases’, which includes rheumatoid arthritis (RA), systemic lupus erythematosus (SLE), systemic sclerosis (SSc), idiopathic inflammatory myopathies (IIM), among others [Citation8,Citation9]. In general, these conditions are considered as seropositive, however it is worth mentioning that they can manifest without the presence of autoantibodies in a smaller proportion of patients.
In rheumatic diseases there have been several examples of drug repositioning, for instance, one of the most successful cases is the use of Methotrexate, initially developed for cancer treatment and repositioned in RA, SLE and primary Sjögren syndrome [Citation4]. Other important examples are the glucocorticoids, such as Prednisone, initially developed to treat inflammation in RA and later approved for SLE, among other rheumatic diseases. Current biologic treatments such as Tumor Necrosis Factor (TNF) inhibitors like Etanercept and Infliximab have been approved for RA, psoriatic arthritis (PsA), and ankylosing spondylitis (AS). Moreover, those biologics targeting Interleukin 6 (IL-6) inhibitors like Tocilizumab were initially developed to treat large-cell lung carcinoma, later for RA, and are currently used in other rheumatic diseases [Citation4].
3. Drug repositioning in the context of genomic studies
In spite of genetic similarities among humans, there is a small proportion of the genome that is crucial for explaining inter-individual differences in the risk of developing diseases, which together with external factors are involved in disease variability and treatment response. In this sense, genomic is an important tool in the identification of new relevant targets for drug repositioning and it has been demonstrated that drugs with genetic support doubles the chances to reach clinical approval than those without it [Citation10]. To provide this support, Genome-Wide Association Studies (GWAS) and Phenome-Wide Association Studies (PheWAS) are currently used for this prioritization. While GWAS have identified multiple susceptibility loci associated with a disease, PheWAS leverages electronic medical records and the literature to assess the association of a single genetic variant with many clinical manifestations [Citation4]. Nowadays, other genomic studies such as chromosomal interactions networks, are emerging strategies to highlight new pathways, molecules, and new genes associated with diseases [Citation11]. Thus, the combination of multi-omic studies allows providing important insights into multifactorial conditions, such as the rheumatic diseases [Citation12].
Several genomic studies have been carried out in systemic seropositive rheumatic diseases, allowing the identification of novel genetic loci as targets for existing drugs. In the largest genetic study conducted to date in RA, Okada and colleagues performed a trans-ethnic meta-analysis identifying novel loci associated with the disease. Using these results, they performed a drug target gene enrichment analysis, proving for the first time the utility of genetic data in the drug discovery process in RA [Citation13]. In the same way, the largest genetic study in SSc performed to date, combines GWAS data together with chromatin interaction and enrichment in epigenetic marks. Using these approaches, the authors were able to nominate 78 target genes that were used in a drug enrichment analysis. From these, seven loci overlapped with active drug targets and two of them, CD80 molecule and BLK proto-oncogene, Src family tyrosine kinase (BLK), were already targets for SSc drugs, highlighting the utility of this approach in the drug repositioning field [Citation14].
Considering the involvement of the immune system and the clinical manifestations in seropositive rheumatic diseases, a shared genetic component is proposed, suggesting that genetically related disorders may be treated with the same drugs [Citation15]. Consequently, cross-disease association studies combining different affections have become relevant in the identification of shared loci and propose them for drug repositioning. A recent report by Acosta-Herrera et al. summarizes the main findings in the cross-disease meta-analysis in systemic seropositive rheumatic diseases to date [Citation16]. An interesting study to highlight is the one combining four of these conditions, namely SSc, SLE, IIM and RA, where the authors evaluate drug targets indicated for prevalent diseases such as RA, and provide possibilities of being repurposed in genetically related, less prevalent or rare diseases such as SSc [Citation8]. The main results of these studies are summarized in .
Table 1. Main drugs proposed for repositioning in systemic seropositive rheumatic diseases.
However, most of genetic variants associated with rheumatic diseases map in non-coding regulatory regions of the genome, and may be affecting the expression of remote genes. For this reason, the knowledge of epigenetic modifications as well as structural changes represents a great opportunity to contextualize genetic variations in the pathways where they are involved [Citation17]. For instance, several methylation analyses in SSc considered DNA-methyltransferases (DNMT) as good targets for inhibitory drugs, such as 2-deoxy-5-azacytidine (5-aza) and 5-azacytidine (5-azaC) that were developed as treatment for leukemia and myelodysplastic syndrome [Citation18]. A recent comparative analysis carried out by Carnero-Montoro and colleagues identified an interferon-related epigenetic signature in Mixed Connective Tissue Disease (MCTD), common with patients diagnosed with SLE, or Sjögren’s syndrome, but significantly different to the ones observed in RA and SSc patients. In addition, the authors observed methylation changes associated with treatment, especially in interferon (IFN) related genes, suggesting these loci as potential targets to revert the disease [Citation19]. Within epigenetic modifications, microRNA (miRNAs) analyses have been performed in several IIMs, such as dermatomyositis and polymyositis, proposing them as potential targets for diagnosis and treatment [Citation20]. On the other hand, Kroef and colleagues demonstrated the influence of H3K27ac and H3K27ac histone modifications in the expression of genes related to IFN and cytokines signaling pathways in monocytes of SSc patients, highlighting these as promising for the treatment of SSc [Citation21].
Moreover, given the complex structure of the genome it becomes necessary to investigate the correlation of associated genetic variants with chromatin interactions to assess their functionality and causal genes and mechanisms [Citation17]. Jeng and colleagues assessed the linkage of noncoding variants associated with SLE, dermatomyositis, Sjögren’s syndrome, and SSc in active enhancers to identify potential causal genes applying HiChIP high-resolution contact maps. The authors reported an enrichment of enhancers at the major histocompatibility complex (HLA) locus and key players at the JAK-STAT signaling pathways, a common pathway among many of the inflammatory conditions under study. Authors highlighted how the enhancer connectome will assist in the identification of therapeutically relevant targets from genomic interactions [Citation17]. Another interesting approach utilized in rheumatic diseases, is the Capture Hi-C (CHi-C) strategy. In this case, Martin and colleagues successfully identified causal genes and drug targets that were existing treatments and that could be repositioned for RA, Juvenile idiopathic arthritis, PsA, and Type 1 diabetes, highlighting the potential of CHi-C for the identification of functionally relevant genes [Citation9].
4. Future perspectives
Genomic studies have been useful tools to identify associated genes that may serve as targets for drug discovery and/or repositioning in rheumatic diseases. However, given the novelty of the field, additional studies are required that assess the full frequency spectrum of genomic variation by using whole genome and whole-exome sequencing. This will eventually allow uncovering new genes associated with these conditions in a comprehensive fashion [Citation22].
Keeping in mind the opportunities offered in the drug repositioning field, there are remaining limitations to overcome. First, the establishment of novel safety evaluations taking into account the new potential interactions between the repurposed drug and its new application; second, the development of new platforms to integrate different ‘omics’ data; and third, the novel use of an existing therapeutic agent may need new patent considerations, which has a great economic impact for the industry and might be considered nowadays as the biggest limitation in the repositioning process [Citation5].
Key issues will be essential to approach as the field develops, including the engagement of different stakeholders, such as regulatory agencies, pharmaceutical industry and patients’ groups, in order to bridge the gap between basic research and therapy development. Global collaborations will be essential to boost the progress of the field and make it a health priority [Citation23].
Declaration of interest
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
Reviewer disclosures
Peer reviewers on this manuscript have no relevant financial or other relationships to disclose.
Additional information
Funding
References
- Corsello SM, Bittker JA, Liu Z, et al. The Drug Repurposing Hub: a next-generation drug library and information resource. Nat Med. 2017 Apr 7;23(4):405–408.
- Mina-Osorio P. Review: basics of drug development in rheumatology. Arthritis Rheumatol. 2015 Oct;67(10):2581–2590.
- David T, Ling SF, Barton A. Genetics of immune-mediated inflammatory diseases. Clin Exp Immunol. 2018 Jul;193(1):3–12.
- Kingsmore KM, Grammer AC, Lipsky PE. Drug repurposing to improve treatment of rheumatic autoimmune inflammatory diseases. Nat Rev Rheumatol. 2020 Jan;16(1):32–52.
- Pushpakom S, Iorio F, Eyers PA, et al. Drug repurposing: progress, challenges and recommendations. Nat Rev Drug Discov. 2019 Jan;18(1):41–58.
- Grammer AC, Lipsky PE. Drug repositioning strategies for the identification of novel therapies for rheumatic autoimmune inflammatory diseases. Rheum Dis Clin North Am. 2017 Aug;43(3):467–480.
- Masoudi-Sobhanzadeh Y, Omidi Y, Amanlou M, et al. Drug databases and their contributions to drug repurposing. Genomics. 2020 Mar;112(2):1087–1095.
- Acosta-Herrera M, Kerick M, Gonzalez-Serna D, et al. Genome-wide meta-analysis reveals shared new loci in systemic seropositive rheumatic diseases. Ann Rheum Dis. 2019 Mar;78(3):311–319.
- Martin P, Ding J, Duffus K, et al. Chromatin interactions reveal novel gene targets for drug repositioning in rheumatic diseases. Ann Rheum Dis. 2019 Aug;78(8):1127–1134.
- Nelson MR, Tipney H, Painter JL, et al. The support of human genetic evidence for approved drug indications. Nat Genet. 2015 Aug;47(8):856–860.
- Ding J, Orozco G. Identification of rheumatoid arthritis causal genes using functional genomics. Scand J Immunol. 2019 May;89(5):e12753.
- Prachayasittikul V, Prathipati P, Pratiwi R, et al. Exploring the epigenetic drug discovery landscape. Expert Opin Drug Discov. 2017 Apr;12(4):345–362.
- Okada Y, Wu D, Trynka G, et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature. 2014 Feb 20;506(7488):376–381.
- Lopez-Isac E, Acosta-Herrera M, Kerick M, et al. GWAS for systemic sclerosis identifies multiple risk loci and highlights fibrotic and vasculopathy pathways. Nat Commun. 2019 Oct 31;10(1):4955.
- Kirino Y, Remmers EF. Genetic architectures of seropositive and seronegative rheumatic diseases. Nat Rev Rheumatol. 2015 Jul;11(7):401–414.
- Acosta-Herrera M, Lopez-Isac E, Martin J. Towards a better classification and novel therapies based on the genetics of systemic sclerosis. Curr Rheumatol Rep. 2019 Jul 15;21(9):44.
- Jeng MY, Mumbach MR, Granja JM, et al. Enhancer connectome nominates target genes of inherited risk variants from inflammatory skin disorders. J Invest Dermatol. 2019 Mar;139(3):605–614.
- Angiolilli C, Marut W, van der Kroef M, et al. New insights into the genetics and epigenetics of systemic sclerosis. Nat Rev Rheumatol. 2018 Nov;14(11):657–673.
- Carnero-Montoro E, Barturen G, Povedano E, et al. Epigenome-wide comparative study reveals key differences between mixed connective tissue disease and related systemic autoimmune diseases. Front Immunol. 2019;10:1880.
- Gao S, Zhang H, Zuo X, et al. Integrated comparison of the miRNAome and mRNAome in muscles of dermatomyositis and polymyositis reveals common and specific miRNA-mRNAs. Epigenomics. 2019 Jan;11(1):23–33.
- van der Kroef M, Castellucci M, Mokry M, et al. Histone modifications underlie monocyte dysregulation in patients with systemic sclerosis, underlining the treatment potential of epigenetic targeting. Ann Rheum Dis. 2019 Apr;78(4):529–538.
- Donlin LT, Park SH, Giannopoulou E, et al. Insights into rheumatic diseases from next-generation sequencing. Nat Rev Rheumatol. 2019 Jun;15(6):327–339.
- Tambuyzer E, Vandendriessche B, Austin CP, et al. Therapies for rare diseases: therapeutic modalities, progress and challenges ahead. Nat Rev Drug Discov. 2020 Feb;19(2):93–111.