Abstract
A new goatfish, Upeneus heemstra sp. nov. (Mullidae), from the Western Indian Ocean and SE India is described from initial DNA barcoding and quantitative morphological screening, followed by a taxonomic analysis featuring the comparison of 56 meristic, morphometric and colour characters compiled from 340 specimens of 10 phenotypically similar species. The new species differs clearly from U. oligospilus (Persian Gulf), U. tragula (Eastern Indian Ocean and West Pacific) and U. niebuhri – resurrected here – from the Gulf of Suez (Red Sea) in the combination of the following characters: caudal peduncle, head, snout, postorbital, barbel and caudal-fin length, anal-fin and second dorsal-fin height, and the number of oblique bars on the caudal fin. These four species can be distinguished from the six other species of the so-called tragula group primarily by colour pattern and appear to represent a distinct ‘dark-freckled’ species complex. An updated identification key for the tragula species group is provided and remarks on size-related and population differences are made.
Introduction
Ocean habitats are mostly connected and spacious, thus allowing highly mobile organisms such as fishes to distribute widely. Determining the spatial delimitations of fish populations or species is therefore often difficult, an additional obstacle being the still insufficiently known diversity. Detailed genetic and phenotypic studies, at best in combination, are a promising tool to uncover ‘cryptic’ diversity that is not immediately evident, thus providing new insights for both basic and applied marine biological science.
The family of the goatfishes (Mullidae), ecologically and commercially important inhabitants of sand-associated, shallow habitats (Uiblein Citation2007), is a clade that requires enhanced biodiversity-related research. For instance, in the Western Indian Ocean, 12 new species of the genera Mulloidichthys, Upeneus and Parupeneus have been described only recently (Randall & Heemstra Citation2009; Randall & King Citation2009; Uiblein & Heemstra Citation2010, Citation2011a,Citationb; Uiblein Citation2011; Uiblein & Lisher Citation2013) and preliminary evidence of population differentiation at various geographic scales has been documented for distinct species (e.g. Uiblein Citation2011; Uiblein & Heemstra Citation2011a,Citationb).
In their regional revision of the Upeneus species from the Western Indian Ocean, Uiblein & Heemstra (Citation2010) assembled phenotypically similar species into groups to simplify comparisons among the many species of this genus. This approach was followed in several recent taxonomic accounts (Uiblein & Heemstra Citation2011a,Citationb; Yamashita et al. Citation2011; Motomura et al. Citation2012; Uiblein & McGrouther Citation2012; Uiblein & Causse Citation2013; Uiblein & Lisher Citation2013) with additional new species being added and species groups updated. Accordingly, one very distinct species, U. filifer (Ogilby, 1910), and five phenotypically separable species groups, each containing between four and nine species, have been distinguished (Uiblein & Heemstra Citation2010; Uiblein & Causse Citation2013; Uiblein & Lisher Citation2013): the japonicus group with U. asymmetricus Lachner, 1954, U. australiae Kim & Nakaya, 2002, U. francisi Randall & Guézé, 1992, U. guttatus (Day, 1868), U. itoui Yamashita et al., 2011, U. japonicus (Houttuyn, 1782), U. pori Ben-Tuvia & Golani, 1989, U. saiab Uiblein & Lisher, 2013, and U. seychellensis Uiblein & Heemstra, 2011; the moluccensis group with U. doriae (Günther, 1869), U. moluccensis (Bleeker, 1855), U. quadrilineatus Cheng & Wang in Chu et al., 1963, and U. sulphureus (Cuvier, 1829); the stenopsis group with U. davidaromi Golani, 2001, U. mascareinsis Fourmanoir & Guézé, 1967, U. stenopsis Uiblein & McGrouther, 2012, U. subvittatus (Temminck & Schlegel, 1843), and U. vanuatu Uiblein & Causse, 2013; the vittatus group with U. indicus Uiblein & Heemstra, 2010, U. parvus Poey, 1852, U. suahelicus Uiblein & Heemstra, 2010, U. supravittatus Uiblein & Heemstra, 2010, and U. vittatus (Forsskål, 1775); and the tragula group with U. luzonius Jordan & Seale, Citation1907, U. margarethae Uiblein & Heemstra, 2010, U. mouthami Randall & Kulbicki, 2006, U. oligospilus Lachner, 1954, U. randalli Uiblein & Heemstra, 2011, U. sundaicus (Bleeker, 1855), U. taeniopterus Cuvier in Cuvier & Valenciennes, 1829, and U. tragula Richardson, 1846.
Species from the tragula group can be distinguished from all other congeners by the following combination of characteristics: VIII dorsal-fin spines, total gill rakers 18–26, pectoral-fin rays 12–15, pelvic-fin length 0.8–1.1 times in pectoral fins, bars on caudal fin in fresh fish of all species, and bars retained or not retained in preserved fish. The name-bearing species for this group, the freckled goatfish Upeneus tragula, has been documented to be particularly widely distributed in the Indo-Pacific from South Africa to Japan (e.g. Uiblein et al. Citation1998; Randall & Kulbicki Citation2006; Uiblein & Heemstra Citation2010) and one may be tempted to hypothesize the existence of a yet insufficiently explored diversity. In fact, Lachner (Citation1954) described an additional species from the Persian Gulf, showing dark freckles on the body and fins like U. tragula and named it U. oligospilus. Assumptions were made that U. tragula may overlap with this second species in distribution (Thomas Citation1969). Another goatfish species, U. niebuhri Guézé, Citation1976, was described more recently from the Gulf of Suez. Both U. niebuhri and U. oligospilus were later synonymized with U. tragula by Bauchot et al. (Citation1985) and Randall & Kulbicki (Citation2006), respectively.
In their regional taxonomic review, Uiblein & Heemstra (Citation2010) resurrected U. oligospilus based on its clear distinction from U. tragula in colour and morphometric characters. After this review was published, a large number of Upeneus specimens from off Australia, Indonesia, Vietnam and several other areas were examined and compared with the Western Indian Ocean material. In parallel with this work, tissue samples mostly taken from collected or photographed voucher specimens from various localities in the Indo-West Pacific became available for molecular studies.
Here, we analyse a large number of genetic and phenotypic data gathered from specimens originally identified as U. tragula and from several other Upeneus species. Based on initial screening of three independent data sets, involving DNA sequencing and quantitative morphometric and meristic comparisons, we provide cumulative evidence for the existence of a still undescribed species that occurs in the Western Indian Ocean and SE India. Based on comparative studies of 56 meristic, morphometric and colour characters compiled from a total of 340 specimens representing all species of the tragula group, we describe Upeneus heemstra sp. nov., a species that was previously confused with U. tragula. We also confirm the validity of the species U. oligospilus, resurrect the species U. niebuhri, and present a key for the Indian Ocean species of the tragula group. Remarks on differences among size groups and populations are also made.
Materials and methods
Molecular approach
Representatives of four Upeneus species were included in the genetic analysis. These included U. heemstra sp. nov. and two representatives (U. tragula and U. margarethae) of the tragula species group (Uiblein & Heemstra Citation2010). Representatives of U. tragula were included from the West Pacific and Eastern Indian Ocean. These included specimens collected from Guangdong, China, near the type locality of the species (Richardson Citation1846). Representatives of U. heemstra sp. nov. were from the Western Indian Ocean and Sri Lanka. Three specimens of U. suahelicus, collected from Tanzania (Zanzibar) and Madagascar, served as more distantly related outgroups for the analysis. Representatives of the three other Upeneus species groups were not included in the present analysis, as the relationships among and constitution of these groups are the focus of ongoing research by the authors. Details of the specimens included and the data sources are provided in .
Table I. Details of the representative specimens of the four Upeneus species (U. heemstra sp. nov., U. margarethae, U. suahelicus and U. tragula) included in the genetic study. These include the GenBank number (if available) or, alternatively, the BOLD Process ID, the collection locality, the accession details of the corresponding voucher specimen or unvouchered image, and details on the original reference for the data and other significant information. Voucher specimens included in the phenotypic studies are emphasized in bold.
Sequence data of the ‘DNA barcoding’ (sensu Hebert et al. Citation2003) fragment of the mitochondrial cytochrome oxidase c subunit I (COI) gene were downloaded from public repositories, including GenBank and BOLD (http://www.boldsystems.org (accessed 21 October 2013)) () or generated specifically for this study. For the latter, DNA was extracted from ethanol-preserved tissues using the Wizard Genomic DNA Purification (Promega, Madison, Wisconsin) kit. The COI gene fragment was amplified for each sample by polymerase chain reaction (PCR) using the primers (dgLCO-1490 and dgHCO-2198) of Meyer (Citation2003). Reactions contained 1 × buffer, 2.5 mM MgCl2, 0.2 µM of each primer, 0.2 mM of each dNTP, 0.5 U Taq-polymerase (Southern Cross Biotechnology, South Africa) and 3 µl of template DNA, and were made to the final 25 µl volume with ultrapure water. The thermocycling regime included an initial denaturing step of 95°C for 2 min, followed by 35 cycles of denaturing (94°C for 30 s), annealing (48°C for 30 s) and extension (72°C for 50 s). A final extension step (72°C) of 7 min followed. To determine whether amplification was successful, PCR products were visualized on a UV-transilluminator, following electrophoresis in 1% agarose gels stained with ethidium bromide. PCR products were purified using Millipore (Billerica, Massachusetts) Montáge PCR filter units, sequenced using BigDye v3.1 (Applied Biosystems, Austin, Texas) terminator chemistry and analysed on an ABI 3730XL (Applied Biosystems) automated sequencer by a commercial sequencing facility (Macrogen, Seoul, South Korea).
The resulting sequences were checked against their chromatograms for misreads and sequencing errors using ChromasLITE (Technylesium). Sequences were aligned and edited further using Lasergene SeqMan Pro 9 (DNASTAR, Madison, Wisconsin). The final alignment of the data set was performed using ClustalX2 (Larkin et al. Citation2007). Data were analysed under maximum likelihood (ML) and unweighted parsimony (UP) phylogenetic frameworks in PAUP*4b10 (Swofford Citation2002). In each case, heuristic searches were conducted to determine the most likely or parsimonious tree(s), using TBR-branch swapping of a starting tree obtained by a random addition of taxa. The likelihood and parsimony analyses employed 100 and 1000 such iterations, respectively. Prior to the ML analysis, ModelTest3.7 (Posada & Crandall Citation1998) was used to determine the optimal model of nucleotide evolution for the data set. The Akaike (Citation1974) Information Criterion was used to select among competing models and the parameters of the most likely model were then implemented in the analysis. Nodal support for clades and relationships was determined using non-parametric bootstrapping (Felsenstein Citation1985) of the data set under both approaches, with 1000 bootstrapping pseudo-replicates being used in each case. Relationships were also considered using a distance-based, minimum-evolution approach, through the construction of a neighbour-joining (NJ: Saitou & Nei Citation1987) tree. In this case, nodal support was determined through 10000 bootstrapping replicates.
Sequence divergences among individuals and taxa were calculated using PAUP. These included uncorrected and model-corrected divergences. A Kimura (Citation1980) 2-parameter (K2P) model was employed here, rather than the ML-model selected above, to enable comparison to other DNA barcoding surveys.
Phenotypic approach
A total of 205 specimens from various museum collections were identified as Upeneus tragula using the available keys of Uiblein & Heemstra (Citation2010, Citation2011b) and further examined based on a large set of characters including SL, 41 additional morphometric (all in mm, to the nearest second decimal) and 12 meristic characters following earlier published work (e.g. Uiblein & Heemstra Citation2010, Citation2011a,Citationb; see for descriptions, abbreviations, and use of these characters). In addition, three qualitative colour characters, the number and colour of lateral body stripes and the presence of a dark first dorsal-fin tip, were used for detailed comparisons. Methods for measuring and counting as well as descriptions of colour based on preserved specimens and photographs of fresh fish follow Uiblein & Heemstra (Citation2010). The holotype of U. niebuhri and another more recently collected specimen from the Gulf of Suez (Red Sea), originally identified as U. cf. tragula by the senior author, were also studied. For interspecific comparisons, 124 specimens of 5 other phenotypically similar species from the Indian Ocean (U. margarethae, U. oligospilus, U. randalli, U. sundaicus and U. taeniopterus) and 9 specimens of 2 Pacific species (U. luzonius, U. mouthami) were studied. A complete list of the comparative material examined is provided at the end of the Material and Methods section.
Table II. Abbreviations and descriptions of morphometric and meristic characters examined in the current study.
Quantitative morphological screening
The morphological screening was performed in subsequent steps, starting with Principal Components Analysis (PCA; SYSTAT software) that allows detection of possible groupings a posteriori using initially ungrouped data. The data were gathered from a subsample of 143 adult specimens (> 70 mm SL) originally identified as Upeneus tragula for which a complete matrix from measurements of 37 morphometric characters was available. The PCA was then followed by one-way ANOVA comparisons of groups previously identified by PCA for the characters with the highest loadings from those components that allowed the detection of the groupings. All morphometric data used in these analyses were size-adjusted using the residuals derived from log-log regressions against SL (Uiblein & Winkler Citation1994). All regressions were highly correlated and significant.
As a further step, 204 specimens from the same areas as the 2 groups identified by PCA were analysed regarding possible group differences in 5 selected meristic characters using frequency tables and Chi2 test for trends (GraphPad Prism 5 software).
In both analysis steps geographical subgroups were identified to allow additional comparisons.
Taxonomic analysis
In the taxonomic study, particular attention was paid to elaborate the most important diagnostic characters for distinction among species in the subsequent comparisons, either singly or in combination, taking sample size and intraspecific variation into account. During the preparation of the identification key, special emphasis was given to the combined use of two or three different character types and to include also diagnostically important information on colour patterns of fresh and preserved fish. Types were chosen from the available material according to their condition and the collection area being relatively close to the type locality (E and SE African coast).
Attention was paid to detect possible geographical variation in the new species, Upeneus heemstra sp. nov. To account for allometric changes, the morphometric and caudal-fin bar data obtained from subadults of U. heemstra sp. nov., U. oligospilus and U. tragula were studied separately from adults (> 70 mm SL).
Comparative material examined
Institutional abbreviations follow Sabaj Pérez (Citation2012). HIFIRE is the abbreviation for the scientific fish reference collection at the Institute of Marine Research, Bergen, Norway. Measurements of specimens examined refer to SL.
Upeneus luzonius (n = 5): W Pacific, Philippines: SU 9244, 95 mm, holotype, Cavite, Luzon; SU 20101, 2: 68–70 mm, paratypes, same locality as previous lot; USNM 53067, 2: 65–71 mm, paratypes, Manila.
Upeneus margarethae (n = 50): W Indian Ocean: Kenya: SAIAB 82825, 2: 65–95 mm, off Kipini, 02°38′S 40°28′E, 12 m; Tanzania: SAIAB 87108, 112 mm, Zanzibar, Mazizini, landing site adjacent to Zanzibar Beach Resort; Mozambique: SAIAB 81741, 128 mm, 26.16°S 32.98°E, 45 m; SAIAB 86466, 4: 75–84 mm, 19°47.44′S 35°30.72′E, 28–29 m; Red Sea, Sudan: BMNH 1960.3.15.841, 63 mm, Ibn Abbas Island, 5 km south of island; Madagascar: MNHN 1966–881, 90 mm, no locality; E Indian Ocean: Thailand: ZMUC P49560, 85 mm, Phuket; NW Australia: AMS 22831-021, 3 (of 6): 81–97 mm, Northwest Shelf, 140 km west of Port Hedland, 20°00′S 117°16′E, 50 m; CSIRO-CA 3052, 98 mm, off Port Hedland, 20°01′S 117°13′E, 46–49 m; WAM 23785-6, 100 mm, Exmouth Gulf; WAM 25397.004, 3 (of 7): 94–102 mm, Rowley Shoals, 17°29′S 121°52′E, 42 m; WAM 32680-002, 93 mm, Exmouth Gulf, Bundegi Reef, 21°50.879′S 114°15.530′E, 24 m; and 30 specimens (including 21 types) from the Western Indian Ocean studied by Uiblein & Heemstra (Citation2010, Citation2011b).
Upeneus mouthami (n = 4): SW Pacific, E Australia: MNHN 2004-1571, 73 mm, Chesterfield Islands, Chesterfield Bank; and three specimens from same area studied by Uiblein & Heemstra (Citation2010).
Upeneus niebuhri (n = 2): Holotype: MNHN 1977-174, 105 mm, Red Sea, Egypt, Gulf of Suez. Non-type: SAIAB 88873, 74 mm, Gulf of Suez.
Upeneus oligospilus (n = 21): Persian Gulf: BMNH 2000.4.19.1190, 73 mm, United Arab Emirates, Abu Dhabi, south side of Ras Ghurab, 24°35′48″N 54°31′11″E; BPBM 29499, 4: 72–118 mm, Bahrain; and 16 specimens from same area studied by Uiblein & Heemstra (Citation2010).
Upeneus randalli (n = 8): 8 types from the Persian Gulf studied by Uiblein & Heemstra (Citation2011b).
Upeneus sundaicus (n = 28): W Indian Ocean: Persian Gulf: Iran: SMF26056, 119 mm, 28°57′49″N 49°43′81″E; SMF 26057, 103 mm, 29°11′51″N 49°13′04″E; ZMUC P49121, 132 mm, Bushehr; ZMUC P49122, 110 mm, same locality as previous lot; ZMUC P49123, 105 mm; ZMUC P49124, 109 mm, off Kangan; Kuwait: BPBM 33212, 81 mm, Kuwait Bay, 8–10 m; E Indian Ocean: Myanmar: USNM 379299, 6: 118–141 mm, 15°32′00″N 94°53′00″E, 16–29 m; Thailand: ZMUC P49402, 119 mm, East of Phuket, 07°53′N 98°50′E; ZMUC P49403, 115 mm, same locality as previous lot; ZMUC P49562, 127 mm, Phuket; W Indonesia: RMNH 5735, 6: 89–125 mm, Bleeker types, Java, Madoera, Nias; NW Australia: AMS 3006, 109 mm, Exmouth Gulf, 22°S 114°E; AMS 21638-001, 158 mm, Timor Sea, 13°41′S 129°15′E, 35 m; CSIRO-CA 2028, 130 mm, SE of Barrow Island; CSIRO-CA 2036, 137 mm, west of Bynoe Harbour, 12°30′S 130°05′E, 38–44 m; RMNH 24737, 119 mm, Exmouth Gulf; WAM 25095.049, 100 mm, Exmouth Gulf, 22°05′S 114°15′E, 12 m.
Upeneus taeniopterus (n = 17): all specimens from the Indian Ocean studied by Uiblein & Heemstra (Citation2010).
Upeneus tragula (n = 171): E Indian Ocean: Myanmar to Northwestern Australia (Timor Sea): Myanmar: USNM 360813, 78 mm, Rakhine, Sandoway, Gwa, Sar Chet Chaung, 17°47′N 94°30′E; Thailand: FRLM 30610, 82 mm, Libong Island, Trang, 7°15′00″N, 99°15′00″E; FRLM 30611, 66 mm, same station data as previous lot; ZMUC 49406-07, 2: 160–174 mm, Thai-Danish Expedition st. 1079, 07°50′N 98°48′E; ZMUC P49589, 98 mm, Phuket; ZMUC P49611, 147 mm, Phuket; ZMUC P49613, 141 mm, Phuket; Singapore: SU 31233; 4: 98–122 mm, Sutton Shoal, AWH III-34; SU 33356, 124 mm, AWH 7-V-37; W Indonesia: RMNH 480, 2: 140–158 mm, Java; USNM 396085, 147 mm, Bali, Kedonganan; Lombok, Tanjung Luar: CSIRO LM383, 3: 81–115 mm; CSIRO LM607, 118 mm; CSIRO LM608, 81 mm; CSIRO LM609, 91 mm; CSIRO LM728, 103 mm; CSIRO LM770, 181 mm; CSIRO LM771, 144 mm; CSIRO LM772, 132 mm; CSIRO LM846, 120 mm; CSIRO LM847, 132 mm; CSIRO LM848, 133 mm; CSIRO LM960, 126 mm; CSIRO LM961, 122 mm; Lombok, Gerupuk: FRLM 16109, 83 mm; FRLM 16110, 58 mm; Northwestern Australia: BMNH 1983.5.5.23-26, 4: 131–51 mm, 20°10′0″S 118°25′00″E; CSIRO C2582, 142 mm, off Carnavon, Shark Bay, Lancelin; CSIRO H1451-3, 165 mm, North West Shelf, FRV Soela, 20°03′S 118°14′E, 20–21 m; WAM 4402.001, 112 mm, Exmouth Gulf, 22°05′S 114°15′E; WAM 5485.001, 115 mm, Exmouth Gulf, 22°05′S 113°44′E; WAM 25108.006, 2: 141–169 mm, Dampier Archipelago, Kendrew Island, 20°29′S 116°32′E, 6–10 m; WAM 25397.004, 4: 123–137 mm, Rowley Shoals, 17°29′S 121°52′E, 42 m; WAM 28883.002, 159 mm, Carnarvon, 24°55′S 113°30′E; WAM 30084.007, 114 mm, Shark Bay, Slope Island, 26°07′S 113°14′E, 1–4 m; WAM 30320.029, 4: 64–113 mm, Buccaneer Archipelago, Powerful Island, 16°05′00″S 123°27′00″E; WAM 30321.019, 82 mm, Sunday Island, 16°26′00″S 123°11′00″E, 1.5 m; WAM 31097.032, 2: 63–83 mm, Cape Londonderry, 13°45′S 126°58′E, 1–2 m; WAM 31240.017, 1 (of 4): 75 mm, Berthier Island, 14°30′S 125°00′E, 1–2 m; WAM 32277.001, 178 mm, Shark Bay, Cape Peron North, 25°22.958′S 113°37.141′E, 16 m; WAM 32667.008, 120 mm, Exmouth Gulf, Bundegi Reef, 21°51.815′S 114°11.902′E, 22–23 m; W Pacific: E Indonesia: RMNH 459, 190 mm, Celebes, Macassar; Moluccas, Ambon: CSIRO KD898, 155 mm, Teluk Baguala, off Suli, 3°38′00″S 128°17′30″E, 0–1.5 m; FRLM 24034, 71 mm, Maluku, Baguala Bay; FRLM 24763, 63 mm, same locality; N Australia (Arafura Sea to Torres Strait, 142.5°E): CSIRO H 3349-01, 139 mm, Torres Strait, NW of Prince of Wales Island, FRV Southern Surveyor, 10°23′S 141°47′E, 8 m; CSIRO H 3637-09, 92 mm, W of Thursday Island, FRV Southern Surveyor, 10°34′S 141°59′E, 12 m; CSIRO H 6558-02, 152 mm, Torres Strait, Endeavour Strait, RV Gwendoline May, 10°49′S 142°09′E, 15 m; CSIRO H 6558-03, 139 mm, same station data as previous lot; CSIRO H 6795-02, 83 mm, Torres Strait, W of Prince of Wales Island, 10°41.65′S, 141°53.61′E, 14 m; CSIRO H 6895-03, 138 mm, same station data as CSIRO H 6558-02; CSIRO H 6920-02, 2: 152–182 mm, Torres Strait, N of Cape York Peninsula, RV Gwendoline May, 10°24′S 142°41′E, 20 m; CSIRO-H 7203-01, 126 mm, Torres Strait, W of Saibai Island, 9°27.90′S 142°26.74′E, 7m; WAM 28943.001, 141 mm, Gulf of Carpentaria, 14°00′S 140°00′E, 12 m; Eastern Australia (Torres Strait (142.5°E) to Moreton Bay): AMS 20753-003, 16: 54–128 mm, Lizard Island area, two miles off Nymph Island, 14°36′S 145°14′E, 15 m; AMS 34397-028, 2 (of 5): 130–160 mm, Port Clinton, adjacent to West Flat, South Arm channel, SWB 93-98, 22°34′08″S 150°44′34″E, 10–11 m; CSIRO-DGQ0414, 2: 95–112 mm, NE of Yeppoon, 22°39.88′S 151°16.40′E, 13 m; CSIRO H 6717-03 (DGQO-133), 146 mm, N of Rockingham Bay, site 896, 17°38.82′S 146°22.28′E, 26 m; WAM 28824.003, 79 mm, Moreton Bay, 27°05′S 153°09′E, 1–2 m; Western Thailand: CAS 206431, 10: 59–105 mm, Thailand Bay, Rayong, southeast Ban Phe Fisheries Training Center, 12°35′40″N 101°25′43″E, 0–1 m; CAS 206433, 10: 50–93 mm, Gulf of Thailand, west shore of Ko Samet; Cambodia: MNHN 1963-580, 2: 79–84 mm, 10°21′ N 104°18′ E; Vietnam: Ha Long: HIFIRE F58133, 119 mm, st. 5201; HIFIRE F58134, 137 mm, st. 5258; HIFIRE F58135, 130 mm, st. 5257; Nha Trang: FRLM 31773, 107 mm; HIFIRE F58100, 111 mm; HIFIRE F58101, 7: 116–174 mm; ZMUC 49481, 165 mm, Luong Son, north of Nha Trang; ZMUC 49489, 100 mm; Vung Tau: USNM 305043, 191 mm, Cap St. Jacques, Vung Tau fish market, 10°21′N 107°15′E; no locality information: MNHN 1905-223, 121 mm. Philippines: CAS 52572, 80 mm, Mindanao Island, Zamboanga, JD VI-48ZAM; SU 9686, 2: 74–98 mm, Luzon Island, Manila, GAL 1901; SU 20882, 108 mm, Luzon Island, Sorsogon, Bacon, CJP; SU 38939, 114 mm, Palawan, Coron, Busuanga, AWH 29-VI-40; SU 38941, 87 mm, Panay Island, Iloilo, AWH 27-VII-40; Palau: CAS 206389, 2: 90–104 mm, Btilawel District, Koror Island, GVF 1378, 7°20′39″N 134°30′44″E, 0–2 ft; CAS 206435, 4: 82–107 mm, Madalai District, west end of Koror Island, GVF 0509, 7°20′22″N 134°28′5″E, 0–7 ft; CAS 206464, 118 mm, Kossol Reef, GVF 1968, 7°54′30″N 134°40′50″E, 1–3 fa; Papua New Guinea: CSIRO A4, 60 mm, New Britain, Matupi Island, Simpson Harbour, FRV Fairwind; CSIRO C1772, 71 mm, New Britain, Matupi Isl., Rabaul Hbr., FRV Fairwind; ZMUC 49479-80, 2: 112–122 mm, Bay of Port Moresby, Galathea Expedition st. 527, 3 m; New Caledonia: AMS 25756-001, 159 mm, St Vincent; S China: SU 61120, 2: 101–120 mm, Hong Kong Spec. Admin. Reg., Kau Sai Chau, GVF 1717, 22°22′35″N 114°18′42″E, 0–2 ft; Taiwan: SAIAB 35138, 135 mm, Penghu Islands, southern end of island, 23°30.6′N 119°34.3′E; SAIAB 35638, 3: 125–140 mm, Penghu Islands, Makung market; SU 21003, 3: 72–165 mm, Kaohsiung (Takao); Japan: FRLM 16819, 98 mm, Shima City, Mie Pref, Ago Bay; FRLM 22176, 105 mm, Iriomote Island, Kuira River; FRLM 22177, 118 mm, same locality as previous lot; FRLM 29923, 140 mm, same locality as FRLM 16819; SU 23506, 2: 87–92 mm, Honshu Island, Shizuoka Province, Shizuoka market.
Results
Molecular approach
Once sequences were aligned and trimmed to equal length, the COI data set provided 626 nucleotides for analysis. The alignment included 107 variable characters, of which 86 were parsimony-informative. A transversional model with a gamma-distribution of rate variation (TvM + Γ: Posada & Crandall Citation1998) was selected as being the most appropriate among those models available in Modeltest for the data set. Implementing the parameters of this model in a heuristic search in the ML analysis provided a topology with a log-likelihood (ln L) of –1426.733 ().
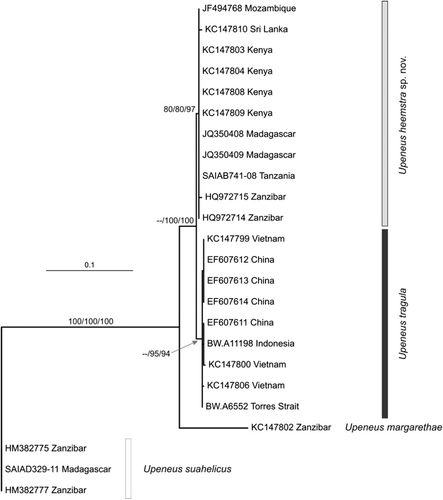
The parsimony analysis yielded two equally parsimonious trees, each 102 steps in length (CI = 0.971, RI = 0.987, Rescaled CI = 0.958). Strict and majority-rule consensus trees of these equally parsimonious trees (not shown) were in agreement with both the ML-phylogram (above) and the NJ tree (not shown), with the retrieval of two reciprocally monophyletic clades corresponding to Upeneus heemstra sp. nov. and U. tragula, respectively (). Both clades were well supported, with bootstraps ≥ 80%, with the exception of the ML analysis, which provided only fair support (64%, not shown) for U. tragula. These two clades were well supported by UP and NJ (bootstraps of 100%) and weakly supported (55%, not shown) by ML as sister taxa. Upeneus margarethae was sister to both, and this larger U. tragula species group was well supported (100% bootstrap) as a monophyletic group with respect to the included outgroup (U. suahelicus).
Sequence divergences among representatives of the included taxa are presented in . Sequence divergences among conspecific individuals ranged from zero to 0.6% (irrespective of whether distances were corrected according to the K2P model), with mean intraspecific values between zero and 0.2% for the three taxa with multiple individuals included. Uncorrected sequence divergences among individuals belonging to different Upeneus species ranged from 0.8 to 14.1%, while the corresponding K2P model-corrected values ranged from 0.8 to 15.8%. The mean interspecific divergences obtained between U. heemstra sp. nov. and U. tragula (1.1% uncorrected, 1.2% corrected) were the lowest observed, with mean uncorrected and corrected divergences among the remaining species being greater than 7.4 and 7.9%, respectively. Regardless, distances obtained in comparisons among conspecific individuals were always lower and showed no overlap with the range of values obtained in interspecific comparisons.
Table III. Means and ranges (in parentheses) of COI sequence divergences among representatives of the four included Upeneus species. Values below the diagonal represent uncorrected sequence divergences (presented as percentages), while those above the diagonal represent divergences corrected according to the Kimura (Citation1980) two-parameter model of nucleotide substitution. Values on the diagonal (in bold) represent the intraspecific sequence divergences for those species where multiple individuals were included. Uncorrected and corrected mean divergences and ranges were identical for intraspecific comparisons.
Quantitative morphological screening
The results of the PCA are shown in and . The third and fourth principal components (PCs) in combination resulted in a clear separation of two geographical groups, one from the Western Indian Ocean (WIO), including Sri Lanka and SE India, and the other from the Eastern Indian Ocean (Myanmar southwards to eastern Indonesia and eastern Australia) and the Pacific. The latter group is here ascribed to Upeneus tragula. In the NE Indian Ocean, the specimens of U. tragula occurring from Myanmar to Singapore are particularly well separated from the WIO and SE Indian form, described as Upeneus heemstra sp. nov. in the following section. The characters showing the highest loadings for these two PCs are caudal-fin length, anal-fin height, first and second dorsal-fin height, caudal-peduncle length, length of first dorsal-fin base and pectoral-fin length ().
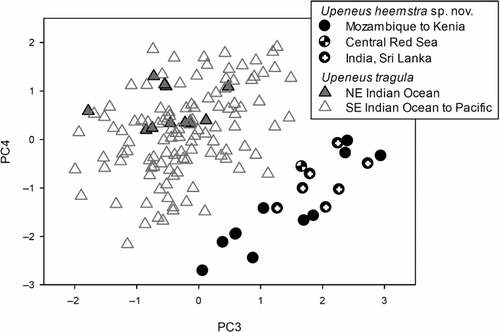
Table IV. Results of multi- and univariate analysis of 37 morphometric characters from 19 Upeneus heemstra sp. nov. and 124 U. tragula individuals, with character loadings, Eigenvalues and variance explained (PCA; highest loadings for components 3 and 4 emphasized), means of residuals (log-log regressions with SL), and F-values (one-way ANOVA; values reflecting highly significant differences (P < 0.01) are emphasized); see for explanation of abbreviations.
When compared using one-way ANOVA, the two groups differed significantly in seven characters: caudal-fin length, caudal-peduncle length, first and second dorsal-fin height, anal-fin height, barbel length and pectoral-fin length. Barbel length is the only character that was not among the most important contributing characters in the PCA, while first dorsal-fin base did not show significant differences.
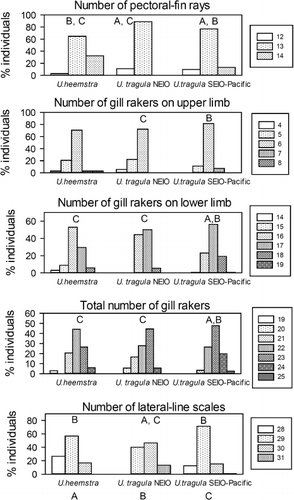
Upeneus heemstra sp. nov. could also be distinguished from U. tragula in meristic characters (). In addition, there were some differences between two geographic subgroups of the latter species, one from the NE Indian Ocean and the other from the SE Indian Ocean and Pacific. Regarding pectoral rays and number of lateral-line scales, the differences between the NEIO population and U. heemstra sp. nov. were stronger than with the other subgroup of U. tragula.
Taxonomy
Family Mullidae
Genus Upeneus Cuvier, 1829
Upeneus Cuvier, 1829: 157. Type species Mullus vittatus (Forsskål, 1775) by subsequent designation of Desmarest (1856).
Diagnosis
Dorsal fins VII or VIII + 9; anal fin I, 6; pectoral fins 12–17; pelvic fins I, 5; principal caudal-fin rays 7 + 8 (median 13 branched); gill rakers 4–9 + 13–24 = 18–33; lateral-line scales 28–39, lateral line complete; small scales present basally on second dorsal, anal and caudal fins; small teeth present on vomer, palatines and jaws, multiserial and villiform on jaws; body oblong, slightly compressed; barbel length in adults 4–7 times in SL; snout length 7–11 times in SL, subequal to postorbital length (6–10 times in SL); in fresh fish lateral body stripes and/or caudal-fin bars of differing colours, dark caudal-fin bars frequently retained on preserved fish.
Distribution
In all major oceans, tropical to subtropical, only a single species in the Atlantic (Upeneus parvus) and two in the Mediterranean (U. moluccensis and U. pori), both immigrants from the Red Sea (Ben-Tuvia Citation1966).
Remarks
We recognize 34 species as valid, Upeneus heemstra sp. nov. and U. niebuhri included. The Eastern Pacific Upeneus xanthogrammus Gilbert, 1892 is probably a synonym of Mulloidichthys dentatus (Gill, 1862) (FU, unpublished data including type material from CAS).
Key to Indian Ocean species of the tragula group
1a. No dark dots, spots or blotches on body and paired fins in adult fish (except for area of lateral line in fresh U. randalli); no dark blotch around or on first dorsal-fin tip; 0–2 yellowish or pale brown lateral body stripes on fresh fish, mostly not retained in preserved fish; barbels white or yellow in fresh fish2
1b. Dark dots, spots or blotches on body and paired fins; dark blotch close to or on first dorsal-fin tip; one dark (red, brown or black) mid-lateral body stripe in fresh and preserved fish; barbels yellow or pale brown in fresh fish3
2a. Lateral-line scales 35–39; pectoral-fin length 5.0–5.8 in SL; second-dorsal fin height 6.3–7.2 times in SL; at least two lateral body stripes in fresh fish, one pale brown mid-lateral body stripe and a weaker, more yellowish stripe below; caudal-fin with 7–13 dark bars in adult fish, well retained in preserved fish (Indo-Pacific) U. taeniopterus
2b. Lateral-line scales 28–34; pectoral-fin length 4.2–5.3 in SL; second-dorsal fin height 5.1–6.7 times in SL; no or only one yellow or pale brown mid-lateral body stripe in fresh fish; caudal-fin lobe with 4–13 red or grey bars in adult fish, not or only weakly retained on preserved fish…………………6
3a. Bars on lower caudal-fin lobe 3–5 (3 in subadults ≤ 7 cm SL); postorbital length in anal-fin height 1.2–1.4 times; caudal-fin length 3.5–4.1 times in SL and 1.0–1.3 times in head length; pelvic-fin length 4.6–6.0 times in SL (Persian Gulf) …………U. oligospilus
3b. Bars on lower caudal-fin lobe 4–10 (4–5 in subadults ≤ 7 cm SL); postorbital length in anal-fin height 1.4–2.0 times; caudal-fin length 2.9–3.7 times in SL and 0.8–1.1 times in head length; pelvic-fin length 4.2–5.4 times in SL…………………………. 4
4a. Bars on lower caudal-fin lobe 4–5; barbel length 4.5–4.7 in SL; upper-jaw length 9.6–10 in SL; first-dorsal fin height 5.3 in SL (Gulf of Suez) U. niebuhri
4b. Bars on lower caudal-fin lobe 4–10; barbel length 4.8–7.5 in SL; upper-jaw length 7.2–10 in SL; first-dorsal fin height 3.9–5.3 in SL 5
5a. Caudal-fin length 3.5–3.7 in SL, caudal-peduncle length 4.3–4.7; first dorsal-fin height 4.4–5.3; second-dorsal fin height 5.4–6.0 in SL (Western Indian Ocean, incl. SE India) U. heemstra sp. nov.
5b. Caudal-fin length 2.9–3.5 in SL, caudal-peduncle length 3.7–4.6; first dorsal-fin height 3.9–5.1; second dorsal-fin height 4.7–6.0 in SL (Eastern Indian Ocean, W Pacific)U. tragula
6a. Total gill rakers 18–22; lateral-line scales 31–34; first dorsal-fin height 3.7–4.6 times in SL; caudal-peduncle depth 7.7–9.1 times in SL; barbels frequently yellow in fresh fish (Indo-Pacific) U. sundaicus
6b. Total gill rakers 21–25; lateral-line scales 28–30; first dorsal-fin height 4.3–5.3 times in SL; caudal-peduncle depth 8.7–11 times in SL; barbels white 7
7a. Total gill rakers 21–24; caudal-peduncle width 17–28 times in SL and 4.9–7.9 in head length; anal-fin base 7.2–10 times in SL; pectoral-fin width 19–24 times in SL and 5.2–7.1 in head length; lower caudal-fin lobe with broad red band, covering up to 5 or 6 red bars, the latter only partly visible along ventral fin margin in fresh fish (Western Indian Ocean); mid-lateral body stripe running through eye, red from snout tip to eye and yellow from behind eye to caudal-fin base, stripe absent in preserved fish (Indian Ocean and Arafura Sea)U. margarethae
7b. Total gill rakers 23–25; caudal-peduncle width 27–34 times in SL and 7.5–10 in head length; anal-fin base 8.9–11 times in SL; pectoral-fin width 23–26 times in SL and 6.5–7.6 in head length; lower caudal-fin lobe with 6–8 dark red bars, bars not covered by a band; mid-lateral body stripe only vaguely visible in fresh fish, stripe absent in preserved fish (Persian Gulf) U. randalli
Upeneus heemstra sp. nov.
Heemstra goatfish
(, VI; –)
Table V. Comparative data for the Indian Ocean Upeneus species of the tragula group; (a) meristic and colour characters, (b) morphometric characters as %SL and (c) morphometric characters as ratios of SL and postorbital length in anal-fin height (last column); morphometric and caudal-fin bar data taken from adult fishes only (nd = no data).
Table VI. Morphometric, meristic and caudal-fin colour (preserved specimens only) characters in adult Upeneus heemstra sp. nov., U. niebuhri, U. oligospilus, and U. tragula; see for explanation of abbreviations; HT: holotype.
Upeneus tragula Richardson, 1846: Ben-Tuvia Citation1986: 613, plate 70; Uiblein & Heemstra Citation2010: 55–56, plates 2–3 (in part); Uiblein & Heemstra Citation2011a: 639; Uiblein & Heemstra Citation2011b: 588.
Material examined
Holotype: SAIAB 119042, 124 mm, Kenya, off Kipini, 02°38′S, 40°28′E, RV Dr. F. Nansen, station 889, shrimp trawl, 12 m, 15 Dec. 1980, collector Phil C. Heemstra.
Paratypes (n = 19): W Indian Ocean: Kenya: SAIAB 13763, 7: 52–121 mm, same data as holotype; Tanzania: SAIAB 13894, 3: 94–150 mm, Dar es Salaam; SAIAB 13898, 79 mm, Zanzibar, Zanzibar market; SAIAB 80384, 142 mm, Nyama reef (paragenetype: BOLD sequence SAIAB741-08); SAIAB 188307, 2: 173–177 mm, Shimoni (paragenetypes: GenBank KC147808 and KC147809); Mozambique: SAIAB 3955, 98 mm, Ibo Island; SAIAB 88453, 2: 118–128 mm, Pomene (paragenetype: GenBank JF494768); SAIAB 186009, 2: 117–147 mm, Pemba.
Non-types (n = 16): W Indian Ocean: Mozambique: SAIAB 13899, 3: 90–101 mm, Delagoa Bay; SAIAB 13911, 104 mm, same locality; Seychelles: SAIAB 13896, 113 mm, Mahé; SAIAB 13908, 100 mm, Aldabra; SW India: BPBM 27721, 2: 92–99 mm, Vizhinjam, Kerala; Sri Lanka: SAIAB 187361, 133 mm, Negombo; USNM 396086, 3: 48–131, Jaffna area, Kakaithivu, about 3 miles south of Vaddukkodai, 2–3 m; USNM 396087, 110 mm, Trincomalee, south side of second bay north of harbour, 0–8 m; ZMUC P4946, 138 mm, Sri Lanka or India; Red Sea: Saudi Arabia: BMNH 1982.7.27.8, 113 mm, coast of Jeddah; E Indian Ocean: SW India: BPBM 20656, 128 mm, Chennai (Madras).
Diagnosis
Dorsal fins VIII + 9; pectoral-fin rays 12–14; gill rakers 4–8 + 14–18 = 19–24; lateral-line scales 28–30; measurements in %SL: body depth at first dorsal-fin origin 22–26; body depth at anal-fin origin 19–23; caudal-peduncle depth 9.5–11; caudal-peduncle width 2.9–4.6; maximum head depth 19–22; head depth through eye 14–17; head length 27–31; postorbital length 10–13; orbit length 6.3–8.3; upper jaw length 9.8–13; barbel length 15–20; caudal-peduncle length 21–23; caudal-fin length 27–30; anal-fin height 16–19; pelvic-fin length 20–22; pectoral-fin length 19–21; pectoral-fin width 3.7–4.9; first dorsal-fin height 19–23; second dorsal-fin height 17–18; postorbital length 1.4–1.8 times in anal-fin height; all fins with red, brown or black stripes, bars or blotches; caudal fin with 9–12 oblique bars, 4–6 bars on upper caudal-fin lobe and 5–6 bars on lower caudal-fin lobe; first dorsal fin with a large blotch around tip; one pale- or dark-brown mid-lateral body stripe from tip of snout through eye to caudal base; body and head ground colour white or beige, slightly darker above lateral line, with irregular red, brown or black spots and/or blotches; barbels yellow or pale brown in fresh fish; fin and body pigmentation mostly retained in preserved fish.
Description
For measurements in %SL, a single ratio, and counts of meristic and colour characters see ; morphometric data as ratios of SL for holotype (with ranges for the 17 paratypes in brackets): body elongate, body depth at first dorsal-fin origin 4.3 [3.8–4.4] exceeding length of pectoral fin (5.0 [4.4–5.1]), body depth at anal-fin origin 4.8 [4.3–5.1], caudal-peduncle depth 9.5 [8.9–10], much larger than orbit length (13 [12–16]), maximum head depth 4.8 [4.5–5.2], head depth across eye 6.6 [5.8–6.4], head length 3.4 [3.1–3.6], larger than maximum depth of body and subequal to caudal-fin length (3.6 [3.1–3.7]), pelvic-fin length 4.6 [4.2–4.9], pectoral-fin width 23 [21–27], first dorsal-fin height 4.6 [4.2–5.0], second dorsal-fin height 5.4 [4.8–5.8], greater than barbel length (6.2 [5.5–6.5]).
Fresh colour (based on types and field photographs of specimens from Mozambique; ): head and body ground colouration white below and beige or brown above lateral line, with red or brown spots associated with individual scales creating a speckled pattern and with large red or brown blotches differing in size occurring in various body regions; one brown mid-lateral body stripe from tip of snout through eye to caudal-fin base, positioned slightly below lateral line on anterior half of body and closely associated with it posteriorly; up to five vertical rows of brown bands of differing width and colour intensity extending from dorsal edge of body to or beyond mid-lateral body stripe, the most prominent band forming a dark-brown ‘saddle’ behind second dorsal fin; first dorsal fin showing a large red, pale-brown or dark-brown blotch that covers or surrounds the fin tip, indistinctly separated from or closely connected to a second smaller and paler blotch that covers mostly the central part of the fin base; second dorsal fin with one or two weaker stripes closer to fin base and one dark stripe covering fin tip or close to white fin tip; pectoral fins hyaline with mostly indistinct pigmentation, pelvic and anal fins hyaline with red or brown stripes or blotches; caudal fin hyaline or whitish pigmented, overlain with up to 12 oblique red, brown or dark-brown bars which mostly traverse entire fin lobes, being usually wider than unpigmented or white interspaces and in some instances interconnected with each other by a perpendicular dark stripe at mid of lobes; 4–6 bars on upper and 5–6 bars on lower lobe, distal-most bars close to or covering lobe tips; barbels pale-yellow, yellow, or pale brown.
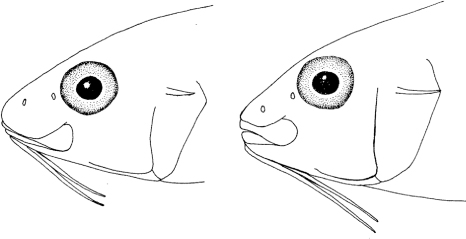
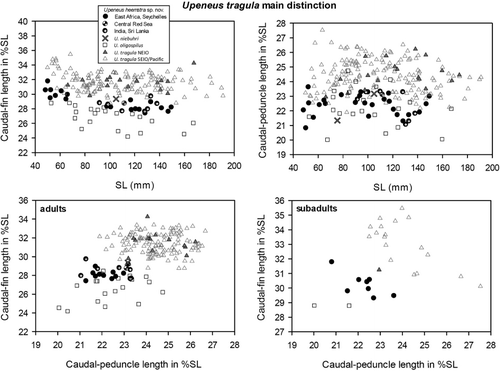
Table VII. Morphometric, meristic and caudal-fin colour (preserved specimens only) characters in adults of Upeneus margarethae, U. randalli, U. sundaicus, and U. taeniopterus from the Indian Ocean, and U. luzonius and U. mouthami from the W Pacific; see for explanation of abbreviations.
Preserved colour: head and body pale or dark brown or grey, dark blotches sometimes visible; body of holotype rather uniformly brown, becoming darker dorsally, with broad dark-brown mid-lateral body stripe from snout to caudal-fin base; holotype and several paratypes show dark saddle behind second dorsal fin that reaches to lateral line; all fins hyaline with pigment mostly retained, especially on first dorsal and anal fins, and on lower caudal-fin lobe in holotype; first dorsal-fin blotch around tip and caudal fin bars mostly fully retained.
Distribution and size
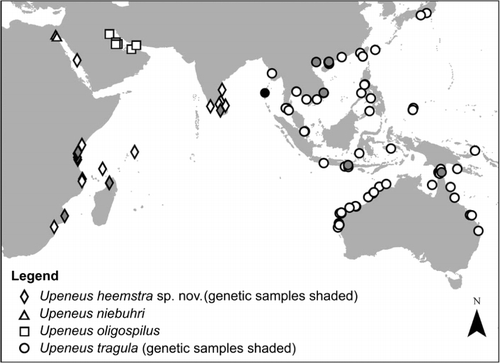
Western Indian Ocean proper, Mozambique to Central Red Sea, Seychelles, W India and Sri Lanka, and SE India (); 0–12 m depth; Upeneus heemstra sp. nov. attains 15 cm SL.
Etymology
The name of this species ‘heemstra’ is in honour of the esteemed ichthyologist Phil C. Heemstra, who collected and photographed the holotype on a cruise of the RV Dr. F. Nansen and his wife Elaine Heemstra, who provided the head drawings and assistance throughout this study.
Upeneus niebuhri Guézé, 1976
Niebuhr's goatfish
(, VI; –)
Upeneus niebuhri Guézé, 1976: 596 (type locality: Gulf of Suez)
Upeneus tragula Richardson, 1846; Bauchot et al. Citation1985: 8; Ben-Tuvia & Golani Citation1989: 110.
Diagnosis
Dorsal fins VIII + 9; pectoral-fin rays 13–14; gill rakers 5–6 + 16–17 = 22–23; lateral-line scales 29–30; measurements in %SL: body depth at first dorsal-fin origin 23–26; body depth at anal-fin origin 19–22; caudal-peduncle depth 9.7–9.9; caudal-peduncle width 3.2–3.7; maximum head depth 19–20; head depth through eye 16–17; head length 29–31; postorbital length 11–12; orbit length 6.9–7.3; upper jaw length 10; barbel length 21–22; caudal-peduncle length 21–23; caudal-fin length 29; pelvic-fin length 21; pectoral-fin length 21; pectoral-fin width 4.1–4.7; first dorsal-fin height 19; second dorsal-fin height 17; all fins with brown or black stripes, bars or blotches; caudal fin with 8 oblique bars, 3–4 bars on upper caudal-fin lobe and 4–5 bars on lower caudal-fin lobe; first and second dorsal fin tips with dark-brown blotches; one brown mid-lateral body stripe from behind opercle to caudal base; body and head ground colour beige, darker above lateral line, with irregular red, brown or black spots and blotches; barbels pale brown in fresh fish; fin and body pigmentation retained in preserved fish.
Distribution and size
Gulf of Suez (); depth: shallow; Upeneus niebuhri attains at least 11 cm SL.
Upeneus oligospilus Lachner, 1954
Short-fin goatfish
(, ; , –)
Upeneus oligospilus Lachner, 1954: 525, plate 14 (type locality: Persian Gulf); Uiblein & Heemstra Citation2010: 48, plate 1; Uiblein & Heemstra Citation2011a: 639; Uiblein & Heemstra Citation2011b: 588.
Diagnosis
Dorsal fins VIII + 9; pectoral-fin rays 13–14; gill rakers 4–7 + 15–18 = 20–24; lateral-line scales 29–31; measurements in %SL: body depth at first dorsal-fin origin 22–26; body depth at anal-fin origin 19–22; caudal-peduncle depth 9.6–11; caudal-peduncle width 2.2–4.1; maximum head depth 19–23; head depth through eye 15–19; head length 29–33; postorbital length 12–13; orbit length 5.5–8.1; upper-jaw length 11–14; barbel length 16–22; caudal-peduncle length 20–25; caudal-fin length 24–28; anal-fin height 15–17; pelvic-fin length 17–22; pectoral-fin length 18–22; pectoral-fin width 3.6–5.4; first dorsal-fin height 18–22; second dorsal-fin height 16–18; postorbital length 1.2–1.4 times in anal-fin height; all fins with dark brown or black stripes, bars or blotches; caudal fin with 6–10 oblique bars, 3–5 bars on upper caudal-fin lobe and 3–5 bars on lower caudal-fin lobe; first dorsal fin with a large blotch around tip; one dark-brown or black mid-lateral body stripe from tip of snout through eye to caudal base; body and head ground colour white or beige, slightly darker above lateral line, with irregular dark brown or black spots, dots and blotches; barbels yellow or pale brown; fin and body pigmentation mostly retained in preserved fish.
Distribution and size
Persian Gulf (); depth: 0–13 m; Upeneus oligospilus attains 13 cm SL.
Upeneus tragula Richardson, 1846
Freckled goatfish
(, ; , –)
Upeneus tragula Richardson, 1846: 220 (type locality: Guangzhou, China); Okamura & Amaoka Citation1997: 372; Randall et al. Citation1997: 212; Uiblein et al. Citation1998: 124–130, ; Randall & Kulbicki Citation2006: 305–306, ; Imamura et al. Citation2009: 176; Uiblein & Heemstra Citation2010: 55–56, plates 2–3 (in part); Allen & Erdmann Citation2012: 510.
Upeneus luzonius non Jordan & Seale, 1907; Gloerfelt-Tarp & Kailola Citation1984: 214–215; Sainsbury et al. Citation1985: 238–239.
Diagnosis
Dorsal fins VIII + 9; pectoral-fin rays 12–14; gill rakers 4–7 + 15–19 = 20–25; lateral-line scales 28–31; measurements in %SL: body depth at first dorsal-fin origin 21–28; body depth at anal-fin origin 18–24; caudal-peduncle depth 9.5–12; caudal-peduncle width 3.1–4.9; maximum head depth 18–24; head depth through eye 14–20; head length 26–32; postorbital length 9.9–13; orbit length 5.3–8.9; upper jaw length 9.8–14; barbel length 13–21; caudal-peduncle length 22–27; caudal-fin length 29–34; anal-fin height 16–20; pelvic-fin length 19–24; pectoral-fin length 17–22; pectoral-fin width 3.5–5.3; first dorsal-fin height 20–25; second dorsal-fin height 17–21; postorbital length 1.4–2.0 times in anal-fin height; all fins with red, brown or black stripes, bars or blotches; caudal fin with 7–19 oblique bars, 3–9 bars on upper caudal-fin lobe and 4–10 bars on lower caudal-fin lobe; first dorsal fin with a large blotch around tip; one red, brown or black mid-lateral body stripe from tip of snout through eye to caudal base; body and head ground colour white or beige, slightly darker above lateral line, with irregular red, brown or black spots and/or blotches; barbels yellow, pale brown or orange in fresh fish; fin and body pigmentation mostly retained in preserved fish.
Distribution and size
Indo-Pacific from Andaman Islands to E Australia and to Japan and New Caledonia; depth: 0–42 m; Upeneus tragula attains 19 cm SL.
Comparisons among species of the tragula group
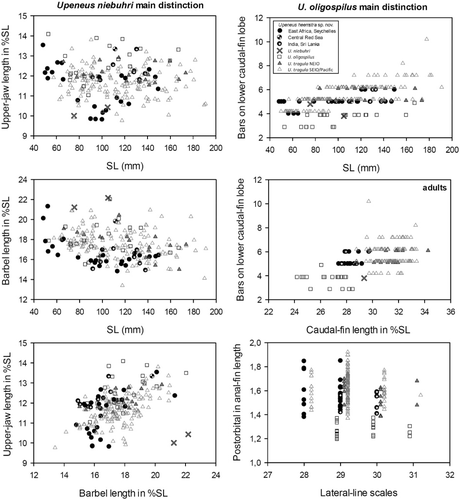
The four dark-freckled species Upeneus heemstra sp. nov., U. niebuhri, U. oligospilus and U. tragula differ from each other as follows (morphometric values in %SL, if not otherwise indicated; , , –, ): Upeneus heemstra sp. nov. differs from U. niebuhri in deeper caudal peduncle (9.5–11 vs. 9.7–9.9), shorter barbels (15–20 vs. 21–22) and more bars on caudal fin in total (9–12 vs. 8); from U. oligospilus it differs in shorter head (27–31 vs. 29–33), shorter snout (10–13 vs. 12–14), shorter postorbital (10–13 vs. 12–13), shorter distance from snout to first dorsal-fin origin (33–38 vs. 36–41), longer caudal-fin length (27–30 vs. 24–28), larger anal-fin height (16–19 vs. 15–17), larger postorbital length in anal-fin height ratio (1.4–1.8 vs. 1.2–1.4 times) and more caudal-fin bars in total (9–12 vs. 6–10); and from U. tragula it differs in shorter caudal peduncle (21–23 vs. 22–27), shorter caudal fin (27–30 vs. 29–34) and shallower second dorsal fin (17–18 vs. 17–21). Upeneus niebuhri differs from U. oligospilus in shorter jaws (upper-jaw length 10 vs. 11–14; lower-jaw length 9.7 vs. 10–13), longer barbels (21–22 vs. 16–22) and longer caudal fin (29 vs. 24–28); and from U. tragula it differs in longer barbels (21–22 vs. 13–21), shorter caudal peduncle (21–23 vs. 22–27), shorter caudal-fin length (29 vs. 29–34) and shallower first dorsal fin (19 vs. 20–25). Upeneus oligospilus differs from U. tragula in shorter caudal fin (24–28 vs. 29–34), shallower anal fin (15–17 vs. 16–20), shallower dorsal fins (first dorsal-fin height 18–22 vs. 20–25; second dorsal-fin height 16–18 vs. 17–21), smaller postorbital length in anal-fin height ratio (1.2–1.4 vs. 1.4–2.0 times) and fewer bars in total on caudal fin (6–10 vs. 7–19).
The four dark-freckled species differ from other congenerics as follows (–, ; Uiblein & Heemstra Citation2010, Citation2011a,Citationb; Yamashita et al. Citation2011; Uiblein & McGrouther Citation2012): from all others by presence of dark-freckled colour patterns consisting of dark blotches or spots on the body, anal and paired fins, and the retention of those colour patterns in preserved fish. They differ from all other species of the tragula group by a dark blotch close to the tip of the first dorsal fin and by the presence of a single, dark mid-lateral body stripe that is retained in preserved fish. In addition, the four dark-freckled species differ from U. margarethae and U. randalli in yellow to pale brown vs. white barbels in fresh fish and caudal-fin bars fully retained in preserved fish, from U. sundaicus in shallower first dorsal fin (18–25 vs. 22–27 %SL) and presence of dark bars on lower caudal-fin lobe both in fresh and preserved fish, and from U. taeniopterus in fewer lateral line scales (18–31 vs. 35–39), higher second dorsal fin (16–21 vs. 14–16 %SL), one dark mid-lateral body stripe vs. two (or three) yellow to pale brown lateral body stripes in fresh fish, and the stripes not retained in preserved fish of the latter species. The main morphometric differences between the four dark-freckled species and the two Pacific species of the tragula group are the shallower first dorsal fin compared to U. luzonius (18–25 vs. 24–29 %SL) and the longer first dorsal-fin base compared to U. mouthami (14–17 vs. 11–13 %SL).
Additional differences between the four dark-freckled species and the other species of the tragula group are (–): Upeneus heemstra differs from U. margarethae in shallower head through eye, shorter caudal peduncle and shorter pectoral length; from U. randalli in fewer total gill rakers, shorter caudal peduncle, and fewer bars on lower caudal-fin lobe; from U. sundaicus in fewer lateral-line scales, shallower and shorter caudal peduncle, shallower head through eye and suborbital distance, and shallower first dorsal fin; and from U. taeniopterus in longer head, larger eyes, and longer paired fins; from the Pacific U. luzonius in fewer lateral-line scales, shorter barbels, shorter caudal peduncle, shallower first dorsal fin, and more caudal-fin bars; and from the Pacific U. mouthami it differs in fewer total gill rakers, greater maximum body depth, deeper head through eye, longer first dorsal-fin base, and shorter pectoral fins.
Upeneus niebuhri differs from U. margarethae in narrower and shorter caudal peduncle and longer barbels; from U randalli in fewer gill rakers, longer barbels, shorter caudal peduncle and shorter first dorsal-fin base; from U. sundaicus in more total gill rakers, fewer lateral-line scales, shallower caudal peduncle, longer barbels and shallower first dorsal fin; it differs from U. taeniopterus in longer head, larger eyes, shorter jaws, longer barbels, longer paired fins and shallower first dorsal fin; from U. luzonius in more total gill rakers, fewer lateral-line scales, shorter barbels, shorter caudal peduncle, and shallower first dorsal fin; and from U. mouthami it differs in more pectoral rays and shorter paired fins.
Upeneus oligospilus differs from U. margarethae in narrower caudal peduncle, longer postorbital distance, longer jaws, and shorter caudal and paired fins; from U randalli in fewer gill rakers, longer head and postorbital distance, shorter caudal fin, smaller postorbital length in anal-fin height ratio and fewer bars on caudal fin; from U. sundaicus more total gill rakers, fewer lateral-line scales, shallower body at anal-fin origin and caudal peduncle, longer head and postorbital distance, and smaller postorbital length in anal-fin height ratio; from U. taeniopterus in longer head and postorbital distance, shorter caudal fin, smaller postorbital length in anal-fin height ratio and fewer bars on upper caudal-fin lobe; from U. luzonius it differs in more total gill rakers, fewer lateral-line scales, in longer head and postorbital distance, shorter caudal peduncle, shorter caudal fin and smaller postorbital length in anal-fin height ratio; and from U. mouthami it differs in fewer total gill rakers, longer snout and postorbital distance, longer jaws and in shorter paired fins.
Upeneus tragula differs from U. margarethae in fewer pectoral-fin rays, longer caudal fin, higher anal fin, shorter pectoral fins and higher second dorsal fin; from U. randalli in fewer pectoral-fin rays, longer caudal fin and higher second dorsal fin; from U. sundaicus in fewer pectoral rays, more gill rakers, fewer lateral-line scales, shallower caudal peduncle, longer caudal fin, higher anal fin and longer second dorsal fin; from U. taeniopterus in larger eyes, longer pelvic fin, higher second dorsal fin and larger postorbital length in anal-fin height ratio; from U. luzonius it differs in fewer pectoral-fin rays, fewer lateral-line scales, longer caudal fin, higher anal fin and higher second dorsal fin; and from U. mouthami it differs in shorter barbels, longer caudal peduncle, longer caudal fin and higher first dorsal fin.
Remarks
For three of the four dark-freckled species, subadults were available for studies of allometric changes (). When compared with the adults (> 70 mm SL; ), subadult Upeneus heemstra sp. nov. and U. oligospilus showed longer heads, longer and higher fins, and fewer caudal-fin bars, while subadult U. tragula differ from adults in slightly longer caudal fins and fewer caudal-fin bars. Species differences in morphometric characters among the subadults mostly follow those of adults. Subadult U. heemstra sp. nov. differ from U. oligospilus in smaller mouth width, longer caudal peduncle, longer caudal fin, longer anal fin and more caudal-fin bars and they differ from U. tragula in longer heads, shorter caudal peduncle, shorter caudal fin and longer pectoral fins. Subadult Upeneus oligospilus differ from U. tragula in longer head, snout and postorbital, longer jaws and wider mouth, shorter caudal peduncle, shorter caudal fin, longer anal-fin base, shallower anal fin, longer pectoral fins and fewer caudal-fin bars.
Table VIII. Morphometric, meristic and caudal-fin colour (preserved specimens only) characters of subadults of Upeneus heemstra sp. nov., U. oligospilus, and U. tragula (no subadult specimen for U. niebuhri was available for study).
There are only minor differences among geographic groups of adult U. heemstra sp. nov. (), with the two specimens from Aldabra (Seychelles) having shorter heads, snouts and jaws and the central Red Sea specimen having a shallower half body depth at anal-fin origin, longer postorbital and longer barbels than all other conspecifics, respectively. Among subadults, the two specimens from Sri Lanka differ from those of the East African coast population in having a deeper and longer head, a wider snout, longer barbels and shallower dorsal fins ().
Upeneus heemstra sp. nov. and U. tragula show considerable variation in colour patterns, based on direct observations in the field and freshly collected fish (), and distinction among the four dark-freckled species based on colour characters alone is difficult. In fact, the only reliable diagnostic colour characters are the oblique bars on the caudal fin, which seem to undergo less variation and are also mostly retained in preserved fish. However, because also the number of bars overlaps among species and shows size-dependency (), caudal-fin bars can only be used in combination with morphometric characters for a reliable diagnosis.
Discussion
Based on the cumulative evidence from molecular and quantitative morphological screening, a new goatfish species has been discovered and subsequently described.
The genetic approach, using DNA sequence data from the ‘barcoding’ fragment of the mitochondrial COI gene, provides important supporting evidence for the recognition of Upeneus heemstra sp. nov. as an independent taxon, distinct from U. tragula. The extent of sequence divergence observed between these two taxa was relatively low (0.8–1.6%), but exceeded the range of divergences observed among individuals within each of these species (≤ 0.6%). This lower range of intraspecific values and the disjuncture between these and the higher interspecific values among the two species – which occur adjacently and are separated by distances (i.e. the U. heemstra sp. nov. specimens from Sri Lanka (WIO) and the Vietnamese or Indonesian U. tragula specimens) comparable to the geographic extent of sampling within each species – suggests taxonomic differentiation rather than geographic genetic structure or population differentiation within a widespread species.
Several DNA barcoding surveys of sequence variation at the COI fragment of both known and cryptic marine fish species have attempted to typify divergence values characteristic of conspecific and congeneric comparisons. Intraspecific divergence values in the current study were broadly comparable to those obtained in these studies (Ward et al. Citation2005; Mabragaña et al. Citation2011; Zhang Citation2011; Zhang & Hanner Citation2012) and were typical of those characterizing widespread species (Hubert et al. Citation2012). Similarly, divergences among the species included in the present study (with the exception of the comparison among U. heemstra sp. nov. and U. tragula) were comparable to documented mean interspecific values (Ward et al. Citation2005; Zhang Citation2011; Hubert et al. Citation2012; Weigt et al. Citation2012; Zhang & Hanner Citation2012). These intra- and interspecific divergences were also comparable to those calculated for the Mullidae specifically (Zhang Citation2011; Zhang & Hanner Citation2012). Against these, the divergence between U. heemstra sp. nov. and U. tragula appears slight. However, the extent of interspecific divergence varies widely by taxon (e.g. Ward et al. Citation2005; Zhang Citation2011) and mean values calculated for various groups (used comparatively here) are dependent on taxonomic coverage. Importantly, DNA barcoding has revealed low levels of divergence among valid species in certain genera (e.g. Ward et al. Citation2005). Species have also been recognized and designated on the basis of cumulative evidence despite low levels of sequence divergence in even more variable gene regions (e.g. Randall et al. Citation2008). More importantly, individuals of each species clustered together and formed tight units; Upeneus heemstra sp. nov. and U. tragula both formed well-supported, reciprocally monophyletic clades. Such patterns of cohesive units are typically retrieved for individual species through the barcoding approach (e.g. Mabragaña et al. Citation2011; Zhang & Hanner Citation2012).
Given the criticisms and limitations of single (mtDNA) marker sets and of the use of mtDNA and DNA barcoding in species delineation, it is important that results and conclusions are subjected to thorough examination through additional or extended sampling and the examination and rigorous, appropriate analyses of additional data sets or additional molecular – particularly nuclear – markers (Brower Citation2006, Citation2010; Elias et al. Citation2007; Ilves et al. Citation2010).
The multivariate morphometric comparisons support the barcoding results concerning the occurrence of a distinct new species. Detailed comparisons in the subsequent alpha-taxonomic approach further revealed that U. heemstra sp. nov. can be clearly distinguished from the most similar species U. niebuhri, U. oligospilus and U. tragula by the combination of a relatively small set of mostly morphometric characters: caudal peduncle, head, snout, postorbital, barbel and caudal-fin length, anal-fin and second dorsal-fin height, and the number of oblique bars on the caudal fin. These four species represent a ‘dark-freckled’ species complex that can be primarily distinguished from all other species of the tragula group by a typical colour pattern consisting of dark dots, spots or blotches on the body and paired fins in both fresh and preserved fish. The occurrence of species complexes rooted within phenotypically similar species groups of Upeneus may not be uncommon, requiring an integration of alpha taxonomy with phylogenetic analyses (Uiblein & Causse Citation2013).
Preliminary evidence for intraspecific geographic differentiation has been found in the Red Sea and Seychelles populations of U. heemstra sp. nov. in morphometric characters. Also, the NE Indian Ocean U. tragula deviate from those of other areas in meristic characters and there are some indications of character displacement (in two meristic and a combination of several morphometric characters) with U. heemstra sp. nov. in areas of possible overlap. However, for the NEIO, where both species occur (), only small sample sizes (12 U. tragula from Myanmar to Singapore and a single U. heemstra sp. nov. from SE India) were available for detailed examination, apart from an unvouchered photograph of Upeneus tragula from the Andaman Islands (, ). To further examine our preliminary evidence of population differentiation, large samples from geographically separated areas need be studied, genetically and phenotypically. Such an approach is currently underway towards thoroughly exploring the phenotypic and phylogeographic diversity among W Pacific and SE Indian Ocean U. tragula populations.
Editorial responsibility: Peter R. Møller
Acknowledgements
We thank the following colleagues for hospitality and assistance during visits to collections or for providing other collection-related favours: Mark McGrouther, Amanda Hay, Sally Reader and John Paxton (AMS); James Maclaine (BMNH); Arnold Suzumoto, Jack Randall and Lori O'Hara (BPBM); Dave Catania, Mysi Hoang and Tomio Iwamoto (CAS); Alastair Graham, Will White, Daniel Gledhill, Peter Last, John Pogonoski, and Robert Ward (CSIRO); Seishi Kimura (FRLM); Rupert Wienerroither (HIFIRE); Daniel Golani (HUJ); Hiroyuki Motomura (KAUM); Christine Thacker and Rick Feeney (LACM); Romain Causse and Patrice Pruvost (MNHN); Martin Gomon and Dianne Bray (NMV); Sven Kullander and Erik Åhlander (NRM); Ronald de Reuter and Martien van Oijen (RMNH); Roger Bills, Mark Lisher and the SAIAB National Fish Collection staff, Elaine and Phil Heemstra, Alan Whitfield, Wouter Holleman, Monica Mwale (all SAIAB); Friedhelm Krupp and Horst Zetzsche (SMF); Jeff Williams, Jerry Finan and David Smith (USNM); Sue Morrison (WAM); and Peter Møller, Jørgen Nielsen, Marcus Krag, and Tammes Menne (ZMUC); Sergey Bogorodsky, Dimitri Pavlov and Pako Uiblein. For photographs of fresh fish and/or additional information we thank Phil and Elaine Heemstra, Alan Connell, Mike and Valda Fraser, Dennis King, Sergey Bogorodsky, Dimitri Pavlov, Jack Randall, William White and Gordon Yearsley. The first author thanks the South African Institute for Aquatic Biodiversity, the Nansen Programme of the Center for Developmental Fisheries at the Institute of Marine Research (IMR), Bergen, and CSIRO Hobart for travel support. This research was supported, and the genetic component specifically funded, by the National Research Foundation (NRF) of South Africa. We gratefully acknowledge CSIRO Australia for the use of their barcode data and thank Bob Ward, Will White and Alastair Graham for their assistance in this regard. We thank Willem Coetzer (SAIAB) for the drafting of the map.
References
- Akaike H. 1974. A new look at the statistical model identification. IEEE Transactions on Automatic Control 19:716–23. doi:10.1109/TAC.1974.1100705
- Allen GR, Erdmann MV. 2012. Reef Fishes of the East Indies. Volume 2. Renon, Bali, Indonesia: Conservation International, p 425–856.
- Bauchot ML, Desoutter M, Guézé P, Randall JE. 1985. Catalogue critique des types de poissons du Muséum national d'Histoire naturelle. Bulletin du Muséum national d'Histoire naturelle 4 (7), sect. A 2, supplement:1–125.
- Ben-Tuvia A. 1966. Red Sea fishes recently found in the Mediterranean. Copeia 1966:254–75. doi:10.2307/1441133
- Ben-Tuvia A. 1986. Mullidae. Family account 196 in: Smith MM, Heemstra PC, editors. Smith's Sea Fishes. Johannesburg: Macmillan South Africa, p 610–13.
- Ben-Tuvia A, Golani D. 1989. A new species of goatfish (Mullidae) of the genus Upeneus from the Red Sea and the Eastern Mediterranean. Israel Journal of Zoology 36:103–12.
- Brower AVZ. 2006. Problems with DNA barcodes for species delimitation: ‘Ten species’ of Astraptes fulgerator reassessed (Lepidoptera: Hesperiidae). Systematics and Biodiversity 4:127–32. doi:10.1017/S147720000500191X
- Brower AVZ. 2010. Alleviating the taxonomic impediment of DNA barcoding and setting a bad precedent: Names for ten species of ‘Astraptes fulgerator’ (Lepidoptera: Hesperiidae: Eudaminae) with DNA-based diagnoses. Systematics and Biodiversity 8:485–91. doi:10.1080/14772000.2010.534512
- Elias M, Hill RI, Willmott KR, Dasmahapatra KK, Brower AVZ, Mallet J, et al. 2007. Limited performance of DNA barcoding in a diverse community of tropical butterflies. Proceedings of the Royal Society B 274:2881–89. doi:10.1098/rspb.2007.1035
- Felsenstein J. 1985. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39:783–91. doi:10.2307/2408678
- Gloerfelt-Tarp T, Kailola PJ. 1984. Trawled Fishes of Southern Indonesia and Northwestern Australia. Canberra, Australia: Australian Development Assistance Bureau. 406 pages.
- Guézé P. 1976. Upeneus niebuhri, espèce nouvelle de Mullidae de la Mer Rouge (Pisces, Perciformes). Revue des Travaux de l'Institut des Pêsches Maritimes 40:596.
- Hebert PDN, Cywinska A, Ball SL, DeWaart JR. 2003. Biological identifications through DNA barcodes. Proceedings of the Royal Society B 270:313–21. doi:10.1098/rspb.2002.2218
- Hubert N, Meyer CP, Bruggemann HJ, Guérin F, Komeno RJL, Espiau B, et al. 2012. Cryptic diversity in Indo-Pacific coral-reef fishes revealed by DNA-barcoding provides new support to the Centre-of-Overlap hypothesis. PLoS One 7:1–8.
- Ilves KL, Huang W, Wares JP, Hickerson MJ. 2010. Colonization and/or mitochondrial selective sweeps across the North Atlantic intertidal assemblage revealed by multi-taxa approximate Bayesian computation. Molecular Ecology 19:4505–19. doi:10.1111/j.1365-294X.2010.04790.x
- Imamura H, Satapoomin U, Kimura S. 2009. Mullidae. Family account in: Kimura S, Satapoomin U, Matsuura K, editors. Fishes of Andaman Sea: West Coast of Southern Thailand. Tokyo: National Museum of Nature and Science, p 173–76.
- Jordan DS, Seale A. 1907. Fishes of the islands of Luzon and Panay. Bulletin of the Bureau of Fisheries 26:1–48.
- Kimura M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. Journal of Molecular Evolution 16:111–20. doi:10.1007/BF01731581
- Lachner EA. 1954. A revision of the goatfish genus Upeneus with descriptions of two new species. Proceedings of the United States National Museum 103:497–532. doi:10.5479/si.00963801.103-3330.497
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–48. doi:10.1093/bioinformatics/btm404
- Mabragaña E, Díaz de Astarloa JM, Hanner R, Zhang J, González Castro M. 2011. DNA barcoding identifies Argentine fishes from marine and brackish waters. PLoS One 6:e28655. 11 pages.
- Meyer CP. 2003. Molecular systematics of cowries (Gastropoda: Cypraeidae) and diversification patterns in the tropics. Biological Journal of the Linnean Society 79:401–59. doi:10.1046/j.1095-8312.2003.00197.x
- Motomura H, Yamashita M, Itou M, Haraguchi Y, Iwatsuki Y. 2012. First records of the two-tone goatfish, Upeneus guttatus, from Japan, and comparisons with U. japonicus (Perciformes: Mullidae). Species Diversity 17:7–14. doi:10.12782/sd.17.1.007
- Okamura O, Amaoka K. 1997. Sea Fishes of Japan. Tokyo: Yama-Kei. 784 pages.
- Posada D, Crandall KA. 1998. Modeltest: Testing the model of DNA substitution. Bioinformatics 14:817–18. doi:10.1093/bioinformatics/14.9.817
- Randall JE, Heemstra E. 2009. Three new goatfishes of the genus Parupeneus from the Western Indian Ocean, with resurrection of P. seychellensis. Smithiana Bulletin 10:37–50.
- Randall JE, King DR. 2009. Parupeneus fraserorum, a new species of goatfish (Perciformes: Mullidae) from South Africa and Madagascar. Smithiana Bulletin 10:31–35.
- Randall JE, Kulbicki M. 2006. A review of the goatfishes of the genus Upeneus (Perciformes: Mullidae) from New Caledonia and the Chesterfield Bank, with a new species and four new records. Zoological Studies 45:298–307.
- Randall JE, Allen GR, Steene RC. 1997. Fishes of the Great Barrier Reef and Coral Sea. Bathurst, Australia: Crawford House Publishing. 557 pages.
- Randall JE, Williams JT, Rocha LA. 2008. The Indo-Pacific tetraodontid fish Canthigaster coronata, a complex of three species. Smithiana Bulletin 9:3–13.
- Richardson J. 1846. Report on the ichthyology of the seas of China and Japan. In: Report of the Fifteenth Meeting of the British Association for the Advancement of Science; held at Cambridge in June 1845. London: John Murray, p 187–320.
- Sabaj Pérez MH. 2012. Standard symbolic codes for institutional resource collections in herpetology and ichthyology: An online reference. Version 3.0 (23 February 2012). Washington DC: American Society of Ichthyologists and Herpetologists. http://www.asih.org/node/204 (accessed 25 February 2013).
- Sainsbury KJ, Kailola PJ, Leyland GG. 1985. Continental Shelf Fishes of Northern and Northwestern Australia. An Illustrated Guide. Canberra: CSIRO Division of Fisheries Research. 375 pages.
- Saitou N, Nei M. 1987. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Molecular Biology and Evolution 4:406–25.
- Swofford DL. 2002. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4. Sunderland, MA: Sinauer Associates. Computer Program.
- Thomas PA. 1969. Goatfishes (Mullidae) of the Indian seas. Marine Biological Association of India Memoir 3:1–174.
- Uiblein F. 2007. Goatfishes (Mullidae) as indicators in tropical and temperate coastal habitat monitoring and management. Marine Biology Research 3:265–88. doi:10.1080/17451000701687129
- Uiblein F. 2011. Taxonomic review of Western Indian Ocean goatfishes of the genus Mulloidichthys (Family Mullidae), with description of a new species and remarks on colour and body form variation in Indo-West Pacific species. Smithiana Bulletin 13:51–73.
- Uiblein F, Causse R. 2013. A new deep-water goatfish of the genus Upeneus (Mullidae) from Vanuatu, South Pacific. Zootaxa 3666:337–44. doi:10.11646/zootaxa.3666.3.4
- Uiblein F, Heemstra PC. 2010. A taxonomic review of the Western Indian Ocean goatfishes of the genus Upeneus (Family Mullidae), with descriptions of four new species. Smithiana Bulletin 11:35–71.
- Uiblein F, Heemstra PC. 2011a. A new goatfish, Upeneus seychellensis sp. nov. (Mullidae), from the Seychelles Bank, with remarks on Upeneus guttatus and a key to Western Indian Ocean Upeneus species. Marine Biology Research 7:637–50. doi:10.1080/17451000.2010.547202
- Uiblein F, Heemstra PC. 2011b. Description of a new goatfish species, Upeneus randalli sp. nov. (Mullidae), from the Persian Gulf, with remarks on and keys for Western Indian Ocean Upeneus species. Scientia Marina 75:585–94. doi:10.3989/scimar.2011.75n3585
- Uiblein F, Lisher M. 2013. A new goatfish of the genus Upeneus (Mullidae) from Angoche, northern Mozambique. Zootaxa 3717:85–95. doi:10.11646/zootaxa.3717.1.7
- Uiblein F, McGrouther M. 2012. A new deep-water goatfish of the genus Upeneus (Mullidae) from northern Australia and the Philippines, with a taxonomic account of U. subvittatus and remarks on U. mascareinsis. Zootaxa 3550:61–70.
- Uiblein F, Winkler H. 1994. Morphological variability among Vimba in Austrian waters: Quantitative examination of a taxonomic and a functional hypothesis (Pisces: Cyprinidae). Senckenbergiana Biologica 73:57–65.
- Uiblein F, Köhler C, Tian MC. 1998. Quantitative examination of morphological variability among goatfishes of the genus Upeneus from the Malayan Province (Pisces: Perciformes: Mullidae). Senckenbergiana Maritima 28:123–32. doi:10.1007/BF03043143
- Ward RD, Zemlak TS, Innes BH, Last PR, Hebert PDN. 2005. DNA barcoding Australia's fish species. Philosophical Transactions of the Royal Society B 360:1847–57. doi:10.1098/rstb.2005.1716
- Weigt LA, Baldwin CC, Driskell A, Smith DG, Ormos A, Reyier EA. 2012. Using DNA barcoding to assess Caribbean reef fish biodiversity: Expanding taxonomic and geographic coverage. PLoS One 7:e41059. 7 pages. doi:10.1371/journal.pone.0041059
- Yamashita Y, Golani D, Motomura H. 2011. A new species of Upeneus (Perciformes: Mullidae) from southern Japan. Zootaxa 3107:47–58.
- Zhang J. 2011. Species identification of marine fishes in China with DNA barcoding. Evidence-Based Complementary and Alternative Medicine 978253:1–10.
- Zhang J, Hanner R. 2012. Molecular approach to the identification of fish in the South China Sea. PLoS One 7:e30621. 9 pages.
