Abstract
The idea of using nucleotide sequences as barcodes for species identification has stirred up debates in the community of taxonomists and systematists. We argue that barcodes are potentially extremely useful tools for taxonomy for several reasons. Barcodes may, for example, help to identify cryptic and polymorphic species and give means to associate life history stages of unknown identity. Barcode systems would thus be particularly helpful in cases when morphology is ambiguous or uninformative and would provide tools for higher taxonomic resolution of disparate life forms. Comparative analysis of short DNA sequences may also represent heuristic access cards to a deeper understanding of evolutionary relationships between organisms. However, barcodes are the “essence” of species identities no more than taxonomic holotypes are “the species”. It makes no sense to think that morphology and other biological information about organisms can be made obsolete by barcode systems. The biological significance of matching or diverging nucleotide sequences will still have to be the subject of taxonomic decisions that must be open for scrutiny. It is imperative, therefore, that barcodes are associated with specimen vouchers.
Decline of diversity and identifiers?
We are currently facing a biodiversity crisis. Each day habitats are changed due to human activity, and each day organisms are disappearing forever. Marine habitats are no exception to this. At the same time, the number of taxonomists who are to study the remaining biodiversity is dwindling (Iseley Citation1972; Gaston & May Citation1992; Daly Citation1995; Buyck Citation1999; Lammers Citation1999; McAllister Citation2000; Hopkins & Freckleton Citation2002). Recruitment of a new generation of taxonomists has been poor. There are several reasons for this situation, many of which can probably be boiled down to some degree to indifferent attitudes, both in society and the educational systems, and to organisms that are “invisible” from the perspective of immediate economic and medical human interest. Even within the scientific community there seems to be a tendency to regard taxonomy as merely a sort of stock-keeping of stamp collections with limited intellectual contribution. Poor funding and an uncertain future are sufficient in themselves to discourage students from pursuing a career in taxonomy, and if the intellectual realms of taxonomy are additionally perceived as a metaphorical equivalent of dusty museum collections (Brooke Citation2000), it certainly does not compensate for the lack of incitements.
With a greater need for biological inventories and studies of biodiversity than ever and with fewer researchers to do the job, something obviously has to be done, but what? One suggestion is to facilitate organisms by using parts of the genome as a marker in a sort of molecular typification. By this molecular approach, a small fraction of an organism's total genome is used as an identifying tag, a barcode, for the organism (Blaxter Citation2003, Citation2004; Hebert et al. Citation2003a; Stoeckle Citation2003). To let molecular typification complement traditional descriptions was also suggested by one of us (CS) at the beginning of the Swedish Taxonomy Initiative (http://www.artdata.slu.se/Svenska_artprojektet.htm), but was not considered possible at that time.
Barcoding animals
Cytochrome c oxidase subunit 1 (COI) has been proposed as the main barcoding gene for metazoans. Based on initial screening and theoretical considerations, Hebert et al. (Citation2003a,Citationb) have suggested that an approximately 650 bp stretch of this gene may be sufficient to obtain resolution on all levels between species and phylum for the majority of groups. Barcoding is already employed for complex groups such as nematodes and mosquitoes (Floyd et al. Citation2002; Floyd & Abebe Citation2002; Besansky et al. Citation2003), and recommendations for a standard protocol are published and updated on-line by the Census of Marine Life (CoML) project (http://phe.rockefeller.edu/PDF_FILES/DNAbarcode.pdf).
The success of a barcode database as a general identification tool is highly dependent on a dense representation of diverse taxa and this sampling would be the primary advantage of massive sequencing efforts on one gene. Unfortunately, exclusive promotion of COI as an all-purpose species diagnostic gene would be potentially deceptive. The Cnidaria, for instance, seem to have a unique DNA repair system that results in low levels of variability and few distinctive features in COI (Hebert et al. Citation2003a,Citationb). The levels of intra- and interspecific variability are also unknown for the great majority of organisms. Additionally, the phenomena of lineage sorting and genetic introgression may represent cases where a particular genetic marker is inadequate to identify recognized species. Despite such difficulties, COI is an attractive candidate gene for organism diversity screening. The target fragment is relatively easy to amplify with standard primers (Folmer et al. Citation1994), and accumulated data from many research groups have already made starting points for the comparison of new sequences. COI is frequently being used for phylogenetic and evolutionary inference and is considered by some researchers (Miya & Nishida Citation2000) as a “very good” gene for that purpose. Molecular barcoding may not be the sharpest tool for forming robust phylogenetic hypotheses (even if the sequences occasionally may be used so), but phylogenetic signal would potentially help to identify sequences that are not already in a barcode database. A range of other genes may also be used as barcodes, depending on the purpose and the need for taxonomic resolution. Many practical applications would need diagnostic tools that are less expensive than sequencing, and DNA fragment-based techniques may be developed to be used, for instance, in fisheries food industries and stock management (e.g. Sotelo et al. Citation2001)
“Black holes” in fauna inventories
In marine biological inventories, samples are sieved and sorted and the flora and fauna are carefully determined. For some regions with a long history of taxonomy, determining species is an easier task than for areas with a shorter biological tradition. Some groups are better investigated than others, resulting in readily available keys and species lists. But there are still surprisingly many organisms in most faunal investigations that are only determined to genus, family or some higher taxon. We made a compilation of 138 reports and inventories published from the North Atlantic, the Baltic Sea and the Mediterranean Sea between 1960 and 2004 (). Only studies including two or more higher taxa (i.e. phyla) or the total fauna were included. The compilation shows that the percentage of specimens determined to species varies greatly between different organism groups, but in total about one third of the specimens are not determined to species. These numbers must not be taken as absolute, and the true part of undetermined species is probably higher. For example, Botnen et al. (Citation1991) reported more than 3000 specimens of nematodes in a single sample but provided only one diagrammatic entry. All of these species being conspecific is unlikely and therefore should not be represented as a single entry.
Figure 1. Comparison between the percentage of determined (white) and undetermined (black) species in 138 published papers and reports from the North Atlantic between 1960 and 2004 (references are available on request from the first author). n indicates the cumulative number of taxa included in the studies.
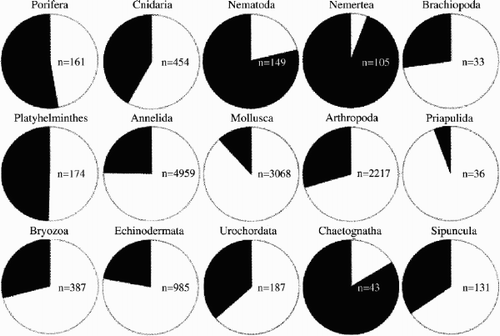
It is obvious that different organism groups are not equally well determined. Molluscs, annelids, echinoderms and crustaceans are examples of groups that are common in the samples and are identified to a large part because they are well studied and documented with descriptions and identification keys. Nematodes, flatworms and nemerteans are examples of organism groups that are mostly undetermined, and they are also good examples of groups with more difficult anatomy from the identification viewpoint. Nematodes and nemerteans are known to be groups where biodiversity is often underestimated due to the great similarity between different species (e.g. Envall & Sundberg Citation1998; Dorris et al. Citation1999) and barcoding is particularly endorsed by students of animal groups with poor diagnostic features in morphology (Blaxter Citation2004). Even within the groups with the largest part of determined species there is great variation between different taxa. Among the molluscs, for example, the aplacophorans are seldom, if ever, determined to species in more general inventories (Scheltema Citation1997). Spionids are in a similar situation when it comes to the annelids. Such “black holes” in fauna inventories may indicate a lack of expertise or that the taxa are generally problematic and difficult to identify. (There is also a financial side and a judgement as to whether work-intensive identification is worth the efforts.)
A common reason for not being able to determine species fully is also that samples get damaged during collection, so that the diagnostic characters are lost. Individuals may also happen to be in a life history stage or of a sex that does not have the diagnostic morphological characters to separate it from other species. We feel that molecular taxonomy has a lot to offer when it comes to the identification of organisms that are beyond the reach of morphological demarcation and diagnostics. The association of sex and life history stages is a prerequisite to understanding the biology of species. Barcode approaches may also help to reveal cryptic species and to understand the role of cryptics in ecological and evolutionary processes (Whiteman et al. Citation2004).
In broad-scaled fauna inventories, the identification of well-known taxa by traditional means will prevail because using keys and descriptions will still be the fastest, cheapest and most efficient way. However, molecular identification will certainly be a useful tool for difficult groups, and an expected outcome of barcoding is the means for non-specialists to discriminate taxa that are otherwise difficult to identify.
Barcoding does not mean the end of morphological studies
The philosophy of barcoding has been criticized on various grounds, and obvious problems have been pointed out (e.g. Seberg et al. Citation2003; Lipscomb et al. Citation2003; Lee Citation2004; Will & Rubinoff Citation2004). Opponents are particularly provoked by rhetoric that may seem to express a promise that barcodes would make morphology-based approaches to taxonomy obsolete. However, morphological and molecular characterizations of taxa do not have to be exclusive activities. Taxonomic groups are recognized from a broad range of characteristics, including morphology, ecology, distributions and so on. In the process of producing barcode diagnostics, the taxonomic significance of nucleotide characters will still have to be evaluated in the context of a pre-existing taxonomy. Because taxonomies are dynamic and change as a result of taxonomic decisions based on evidence from various sources, barcoding is not just a question of assigning appropriate molecular tags to already given named and unnamed entities. The traditional knowledge and skills of specialists in various groups are thus indispensable. There are many examples of molecular studies that have changed the concepts of species diversity in the study group and required the re-analysis of morphological and distributional data (Colborn et al. Citation2001). The mutualism between molecular systematics and traditional taxonomy is clearly an obligate relationship.
Identifications, whether made by producers or users of taxonomies, represent taxonomic decisions that must be open to scrutiny. Linking DNA sequences to specimens in museum collections is therefore critical for the success of barcoding. The deposition of voucher specimens will ensure that all results entered into GenBank or a similar database can be checked and corrected. Voucher specimens are not a requirement by GenBank today, which is a known problem as errors are frequently discovered in the submissions without any possibility of checking the original material (e.g. Harris Citation2003). A barcoding programme linked to natural history collections would mean a new role for many museums, where not only the traditional material (e.g. formalin-fixed, alcohol-preserved) would have to be housed, but also frozen and alcohol-preserved tissue and possibly also extractions of DNA.
A molecular barcoding programme leaves us with many challenges. For smaller animals in particular, new techniques for non-destructive DNA sampling must be developed to preserve the link between specimen morphology and DNA sequences (e.g. Ekrem & Willassen Citation2004, for insects). Other technical problems also need to be addressed. For a long time formaldehyde has been the number one fixative for natural history collections, and this material is difficult to work with even if new protocols are being developed (e.g. Schander & Halanych Citation2003). It is also common practice to bulk-fix in formalin material collected in the field. Alternatives have to be developed. Storage is another problem. How do we best store the material in the long term? Alcohol sometimes seems to work well, and one of us (CS, unpublished), has successfully extracted useful DNA from more than 150-year-old polyplacophorans from the collections of the Zoological Museum in Copenhagen using standard extraction kits, but are there alternatives? Another question is how extracted DNA is best stored for long periods of time.
Barcodes and the future of taxonomy
DNA characterization has given us new means to understand the morphological disparity (and lack thereof) among organisms. It has also provided new methods for grasping the evolutionary context and phylogenetic history of diversity. These new technologies have already made a large impact in most fields of biology, not least in systematics, and many recent taxonomic revisions are based on insights from DNA studies. Hence, a plea for DNA studies in taxonomy (Tautz et al. Citation2002, Citation2003) does not at all seem to be an undue recommendation without implying that DNA sequences are the only valid currency in taxonomy. The implementation of a barcoding programme will certainly provide new discoveries that must be pursued by research that requires a multitude of data about organisms and also wise taxonomic judgement. Discoveries may, for example, include species complexes and unexpected genetic variability. As such, barcodes may be conceived of as access cards to new insights in organism diversity and the patterns and processes of evolution. Such discoveries serve well to raise attention to the importance of taxonomy studies and help us to focus on what additional research is needed. A well-functioning programme for barcoding marine organisms would have far-reaching scientific advantages and provide an excellent opportunity to contribute to the training of a new generation of taxonomists. We see many possibilities with a marine barcoding programme.
Acknowledgments
We thank Amélie H. Scheltema at the Woods Hole Oceanographic Institution, Kjersti Sjøtun at the Department of Biology, University of Bergen, and three anonymous reviewers for valuable suggestions on the manuscript. The work was in part supported by grants from the Meltzer Foundation and from the Swedish Research Council (to CS).
Notes
Published in collaboration with the University of Bergen and the Institute of Marine Research, Norway, and the Marine Biological Laboratory, University of Copenhagen, Denmark
Editorial responsibility: Kjersti Sjøtun
References
References
- Besansky , NJ , Severson , DW and Ferdig , MT . 2003 . DNA barcoding of parasites and invertebrate disease vectors: what you don't know can hurt you . Trends in Parasitology , 19 : 545 – 6 .
- Blaxter , ML . 2003 . Counting angels with DNA . Nature , 421 : 122 – 4 .
- Blaxter , ML . 2004 . The promise of a DNA taxonomy . Philosophical Transactions of the Royal Society of London, B , 359 : 669 – 79 .
- Botnen , HB , Tvedten , Ø and Johannessen , PJ . 1991 . Resipientundersøkelse i Øklandsosen, Bømlo kommune . IFM rapport , 21 : 1 – 18 .
- Brooke , MD . 2000 . Why museums matter . Trends in Ecology and Evolution , 15 : 136 – 7 .
- Buyck , B . 1999 . Taxonomists are an endangered species in Europe . Nature , 401 : 321
- Colborn , J , Crabtree , RE , Shaklee , JB , Pfeiler , E and Bowen , BW . 2001 . The evolutionary enigma of bonefishes (Albula spp.): cryptic species and ancient separations in a globally distributed shorefish . Evolution , 55 : 805 – 20 .
- Daly , HV . 1995 . Endangered species: doctoral students in systematic entomology . American Entomologist , 1995 : 55 – 9 .
- Dorris , M , De Ley , P and Blaxter , ML . 1999 . Molecular analysis of nematode diversity and the evolution of parasitism . Parasitology Today , 15 : 188 – 93 .
- Ekrem , T and Willassen , E . 2004 . Exploring Tanytarsini relationships (Diptera: Chironomidae) using mitochondrial COII gene sequences . Insect Systematics and Evolution , 35 : 263 – 76 .
- Envall , M and Sundberg , P . 1998 . Phylogenetic relationships and genetic distances between some monostiliferous interstitial nemerteans (Ototyphlonemertes, Hoplonemertea, Nemertea) indicated from the 16S rRNA gene . Zoological Journal of the Linnean Society , 123 : 105 – 15 .
- Folmer , O , Black , M , Hoeh , W , Lutz , R and Vrigenhoek , R . 1994 . DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates . Molecular Marine Biology and Biotechnology , 3 : 294 – 9 .
- Floyd , R and Abebe , E . 2002 . Nematode molecular barcodes . Soil Biodiversity , 8 : 3
- Floyd , RM , Abebe , E , Papert , A and Blaxter , ML . 2002 . Molecular barcodes for soil nematode identification . Molecular Ecology , 11 : 839 – 50 .
- Gaston , KJ and May , RM . 1992 . Taxonomy of taxonomists . Nature , 356 : 281 – 2 .
- Harris , JD . 2003 . Can you bank in GenBank? . Tends in Ecology and Evolution , 18 : 317 – 9 .
- Hebert , PDN , Cywinska , A , Ball , SL and deWaard , JR . 2003a . Biological identification through DNA barcodes. . Proceedings of the Royal Society, London B , 270 : 313 – 21 .
- Hebert , PDN , Ratnasingham , S and deWaard , JR . 2003b . Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. . Proceedings of the Royal Society, London B (Suppl.) , 270 : S96 – 9 .
- Hopkins , GW and Freckleton , RP . 2002 . Declines in the numbers of amateur and professional taxonomists: implications for conservation . Animal Conservation , 5 : 245 – 9 .
- Iseley , D . 1972 . The disappearance . Taxon , 21 : 3 – 12 .
- Lammers , TG . 1999 . Plant systematics today: all our eggs in one basket? . Systematic Botany , 24 : 494 – 6 .
- Lee , MSY . 2004 . The molecularisation of taxonomy . Invertebrate Systematics , 18 : 1 – 6 .
- Lipscomb , D , Platnik , N and Wheeler , Q . 2003 . The intellectual content of taxonomy: a comment on DNA taxonomy . Trends in Ecology and Evolution , 18 : 65 – 6 .
- McAllister , D . 2000 . Biodiversity awareness: people, museums and the web . Biodiversity , 1 : 38 – 9 .
- Miya , M and Nishida , M . 2000 . Use of mitogenomic information in teleostean molecular phylogenetics: a tree-based exploration under the maximum-parsimony optimality criterion . Molecular Phylogenetics and Evolution , 17 : 437 – 55 .
- Schander , C and Halanych , KM . 2003 . DNA, PCR and formalinized animal tissue – a short review and protocols . Organisms, Diversity and Evolution , 3 : 195 – 205 .
- Scheltema , AH . 1997 . Aplacophoran molluscs: deep-sea analogs to polychaetes . Bulletin of Marine Science , 60 : 575 – 83 .
- Seberg , O , Humphries , CJ , Knapp , S , Stevenson , DW , Petersen , G , Scharff , N and Andersen , NM . 2003 . Shortcuts in systematics? A commentary on DNA-based taxonomy . Trends in Ecology and Evolution , 18 : 63 – 5 .
- Sotelo , CG , Calo-Mata , P , Chapela , MJ , Perez-Martin , RI , Rehbein , H , Hold , GL , Russell , VJ , Pryde , S , Quinteiro , J , Izquierdo , M , Rey-Mendez , M , Rosa , C and Santos , AT . 2001 . Identification of flatfish (Pleuronectiformes) species using DNA-based techniques . Journal of Agricultural and Food Chemistry , 49 : 4562 – 9 .
- Stoeckle , M . 2003 . Taxonomy, DNA, and the bar code of life . BioScience , 53 : 796 – 7 .
- Tautz , D , Arctander , P , Minelli , A , Thomas , RH and Vogler , AP . 2002 . DNA points the way ahead in taxonomy . Nature , 418 : 479
- Tautz , D , Arctander , P , Minelli , A , Thomas , RH and Vogler , AP . 2003 . A plea for DNA taxonomy . Trends in Ecology and Evolution , 18 : 70 – 4 .
- Whiteman , NK , Santiago-Alarcon , D , Johnson , KP and Parker , PG . 2004 . Differences in straggling rates between two genera of dove lice (Insecta: Phthiraptera) reinforce population genetic and cophylogenetic patterns . International Journal of Parasitology , 34 : 1113 – 9 .
- Will , KW and Rubinoff , D . 2004 . Myth of the molecule: DNA barcodes for species cannot replace morphology for identification and classification . Cladistics , 20 : 47 – 55 .