Abstract
Ophyotrocha is easy to keep in the laboratory and has therefore been used in several studies of evolution and speciation. The phylogenetic relationships within the group are, however, still not clear and morphological and molecular data are contradictory. Here we attempt to shed light on the phylogeny by adding an additional gene (cytochrome c oxidase subunit I) to the previous analyses of the group. However, the results are still incongruent with the results from the morphological data. We also include a species of the genus Iphitime, and conclude that this species falls within the Ophryotrocha clade. The implications are discussed.
Published in collaboration with the University of Bergen and the Institute of Marine Research, Norway, and the Marine Biological Laboratory, University of Copenhagen, Denmark
Introduction
The genus Ophryotrocha was first described by Clarparède & Mencznikow in 1869 from material from a seawater aquarium in Naples. The first species, named Ophryotrocha puerilis (Clarparède & Mencznikow, Citation1869) was later split into two subspecies, O. puerilis puerilis from the Mediterranean and O. puerilis siberti from the Atlantic (Bacci & La Greca Citation1953). Since the description of the genus, more than 40 additional species of Ophryotrocha from various habitats have been described.
Many Ophryotrocha species are found in shallow warm-water habitats and often in polluted areas. Accordingly, the effects of pollution (Åkesson & Ehrenström Citation1984; Brooks et al. Citation2003; Conlan et al. Citation2004) and changes in temperature and salinity (Åkesson & Costlow Citation1978) have been investigated. Ophryotrocha have also been found in temperate regions (Oug Citation1978, Citation1994), Arctic and Antarctic regions (Fournier & Conlan Citation1994; Varella Petti et al. Citation2006), deep-sea regions (Hilbig & Blake Citation1991) and in association with hydrothermal vents (Blake Citation1985; Dreyer et al. Citation2005). The speciation has also been studied (e.g. Åkesson & Paxton Citation2005).
Many Ophryotrocha species are easy to maintain in laboratory culture, which has greatly facilitated the possibility of conducting scientific investigations. As the species are morphologically similar, jaw morphology, reproductive mode and egg morphology are widely accepted as some of the most important key characters for distinguishing between species. These structures have undergone numerous investigations (jaw morphology: Bonnier Citation1893, Paxton Citation2004, Citation2005; reproductive mode and egg morphology: Åkesson Citation1973, Ockelmann & Åkesson Citation1990, Sella et al. Citation1997, Cassai & Prevedelli Citation1999, Sella & Lorenzi Citation2000, Citation2003, Schleicherová et al. Citation2005, Prevedelli et al. Citation2006, Lorenzi et al. Citation2006). Based on reproductive mode and jaw morphology, Ophryotrocha have been divided into three informal groups:
-
The labronica group is gonochoristic. When reaching adulthood, the p-type (primitive) jaws are replaced by k-type (kompliziert) jaws.
-
The hartmanni group is hermaphroditic and has a soft, irregularly shaped egg mass. In addition their p-type jaws become replaced by k-type jaws when the individuals become adults.
-
Species belonging to the gracilis group are also hermaphrodites. The gracilis group is recognized by a fusiform egg mass surrounded by a harder cover. The p-type jaws are never replaced.
The division of Ophryotrocha into three species groups has not been supported by genetic investigations. A division between gonochoristic and hermaphroditic species was found by Dahlgren et al. (Citation2001), who utilized the mitochondrial 16S gene to study the relationship between 18 Ophryotrocha species. Their results confirmed the monophyly of the labronica group, but the hartmanni and gracilis groups were shown to be paraphyletic.
The placement of Ophryotrocha within Annelida is debated. Ophryotrocha was originally placed within the Dorvilleidae. This placement has been challenged by Orensanz (Citation1990), who proposed the establishment of the family Iphitimidae, including Ophryotrocha and Iphitime. This view was not supported by Eibye-Jacobsen & Kristensen (Citation1994), who found Iphitimidae not to be monophyletic and a subtaxon of Dorvilleidae sensu Orensanz (Citation1990), but still favoured a close relationship of Ophryotrocha and Iphitime. Caryological (Robotti et al. Citation1991; Sella et al. Citation1995) and genetic (Dahlgren et al. Citation2001) methods have been used to investigate the relationships among ophryotrochan species, as well as their relationships to other genera. These studies have indicated close relationships among species within Ophryotrocha and between Ophryotrocha and other genera placed in Dorvilleidae.
In order to shed light on the incongruent results from the molecular and morphological phylogenetic analyses (Pleijel & Eide 1996; Dahlgren et al. Citation2001), the phylogenetic relationships among some species of Ophryotrocha were addressed using the mitochondrial gene cytochrome c oxidase subunit I (COI).
Two different data sets were used, one set using only COI data and one set using combined COI and 16S data. In the first analysis, a sequence from Iphitime paguri Fage & Legendre, Citation1934 was also included in order to be able to address the question of the relationship between Iphitime Marenzeller, Citation1902 and Ophryotrocha. Several outgroup taxa were chosen for this investigation. Dorvillea albomaculata was used as this was one of the outgroup species used by Dahlgren et al. (Citation2001). Alvinella pompejana Desbruyères & Laubier, Citation1980 was used to aid the aligning, and was therefore also included in the analysis.
Material and methods
In total, 20 specimens belonging to 14 different Ophryotrocha species were included in the phylogenetic analyses. Additionally, one specimen of Iphitime paguri and one specimen of Dorvillea albomaculata fixed in 98% alcohol were included. A list of included species is provided in . All samples were extracted by use of the Qiagen DNeasy Tissue kit following the animal tissue protocol. All species are deposited in the Bergen Museum, Natural History Collections.
Table I. Summary of samples included in the survey. The catalogue numbers are vouchers deposited in the Bergen Museum, Natural History Collections of Bergen.
All amplification reactions were carried out in a final volume of 50 µl. The primer pairs LCO 1490 (forward: 5′-GGT CAA CAA ATC ATA AAG ATA TTG G-′3) and HCO 2198 (reverse: 5′-TAA ACT TCA GGG TGA CCA AAA AAT CA-′3) (Folmer et al. Citation1994) were used. The protocol followed included 1 µl of each primer, 5 µl 10×buffer, 4 µl dNTPs, 0.25 µl Takara (polymerase) and 35.75 µl ddH2O. In addition, 4 µl template was used. Polymerase chain reactions (PCR) were carried out in 0.2 ml PCR tubes (Eppendorf®) in a thermal cycler (Applied Biosystems GeneAmp® PCR System 2700 or MJRESEARCH PTC-200 Pelitier Thermal Cycler) both under split temperature cycle conditions: denaturation temperature 94°C for 1.5 min; five cycles of 94°C for 30 s, 45°C for 1.5 min, 72°C for 1 min; 34 cycles of 94°C for 30 s, 51°C for 1.5 min, 72°C for 1 min; finally, 72°C for 7 min and 4°C indefinitely. The PCR products were loaded on to a 1% agarose gel stained with ethidium bromide. For visualization, an ultraviolet transilluminator and Grab-IT v2.59 computer software were used. The QIAquick PCR Purification Kit was used to clean the PCR products, according to the manufacturer's recommendations, using 30 µl of the elution buffer. Purified PCR products were loaded on to a 1% agarose gel for verification of the PCR product.
Sequencing of the PCR product was performed using the following protocol: Big-dye, 1.0 µl; sequencing buffer, 1.5 µl; primer 1, 0.5 µl; primer 2, 0.5 µl; ddH2O, 6.0 µl. A volume of 9 µl mix and 1 µl template per sample was used. The temperature cycle was initially set to 95°C for 5 min, followed by 25 cycles of 96°C for 10 s, 50°C for 5 min and 60°C for 4 s. Finally, the temperature was set to 4°C indefinitely. Ten microlitres ddH2O was added before the sequences were delivered to the Sars Centre at the University of Bergen where the sequencing was performed using an ABI 3700 DNA Sequencer (Applied Biosystems). Cleaning the sequenced product was also performed by the Sars Centre, using Millipore Montage seq cleanup and a BIOMEK 2000 robot (Beckmann). Sequences were assembled and aligned using the Seqman package in Lasergene 6.1 (DNASTAR, Inc.). Gap penalties were set to 45 and gap extension penalties were set to 30 to ease the aligning. Alvinella pompejana (GeneBank accession no. AY645988) was downloaded from the NCBI GeneBank and used to ease the aligning. ClustalX (Thompson et al. Citation1997) and BioEdit (Hall Citation1999) were used for further editing and trimming the alignments.
Phylogenetic analysis was applied to two different data matrices. Dataset one (COI analysis) comprised only sequences obtained in this study. The second dataset (combined analysis) comprised 16S sequences from Dahlgren et al. (Citation2001) and some of the COI sequences from this study. GeneBank accession numbers are provided in . Alignments of COI and 16S were carried out separately and were joined afterwards for the phylogenetic analysis.
Table II. Taxon and authors for the species included in the analysis. GeneBank accession numbers for cytochrome c oxidase subunit I (COI) and the 16S sequences included in the combined analysis are also given.
Phylogenetic analyses were conducted in the program PAUP 4.0 beta 10 win (Swofford Citation2002). The following analyses were applied to both datasets. A simple parsimony analysis was conducted by a heuristic search. Modeltest 3.06 (Posada & Crandall Citation1998) was applied to find the best model for maximum likelihood. Furthermore, MrModeltest 2.2 (Nylander Citation2004) was applied to find the best model for Bayesian analysis. The results from this analysis were used in MrBayes 3 (Huelsenbeck & Ronquist Citation2003) for Bayesian inference.
Each COI sequence contained 628 base pairs. Both a Modeltest search and an iterative procedure for model optimization were conducted. The iterative procedure was based on a most-parsimonious tree as the starting tree. A GTR + I+G model was obtained as the best fitting model. The best fitting model obtained from the Modeltest search (hierarchical likelihood ratio tests) was used as a model when iteratively searching for optimized parameter values. The optimized log likelihood value was 6783.70707. Base frequencies (A = 0.2957, C = 0.1840, G = 0.1680 and T = 0.3523) and substitution rates ([A–C] = 1.1518, [A–G] = 7.5098, [A–T] = 2.4235, [C–G] = 3.4645 and [C–T] = 9.6478) were estimated. The proportion of invariable sites (I) was 0.2015 and the gamma distribution shape parameter was 0.4966.
A bootstrap analysis was conducted for COI with a neighbour-joining tree as the starting tree and by branch swapping with neighbour interchanges. Computing time allowed for only 100 bootstrap replicate searches.
A GTR + G+I model was also suggested by MrModeltest 2.2. MrBayes 3 was employed for Bayesian inference. A MCMC search was performed with 1,000,000 generations, and the sampling frequency set to 100. Burnin was set to 250 based on the equilibrium of log likelihood values.
For the combined analysis, the same procedure as used in the COI analysis was applied. Modeltest 3.06 and an iterative maximum likelihood search were used to find the model that best fit the sequence matrix. The best fitted model resulting from hierarchical likelihood ratio tests was the GTR + G+I model. The optimized log likelihood value was estimated to be 8734.7275 in the iterative searches for the best fitting model. The base frequencies were estimated to be A = 0.2964, C = 0.1744, G = 0.1834 and T = 0.3457 and the resulting rate matrix values were [A–C] = 1.1817, [A–G] = 5.3021, [A–T] = 2.3637, [C–G] = 1.5677 and [C–T] = 4.8543. The gamma distribution shape parameter was estimated to be 1.0508 and the proportion of invariable sites was estimated to be 0.2520.
MrModeltest 2.2 was used to find the model that best fitted the data for inferring phylogeny with the program MrBayes. The result was a GTR + G+I model. Phylogenetic analyses were conducted in MrBayes 3.0 with MCMC searches and with 1,000,000 generations, and the sample frequency set to 100. Burnin was set to 250 based on the equilibrium of log likelihood values.
Alignments have been submitted to treebasee, submission number SN3407-15085.
Results
COI parsimony
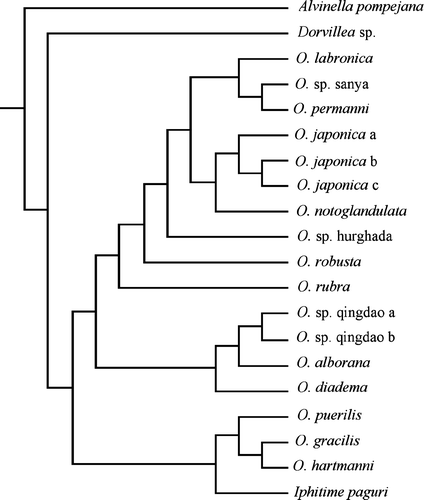
The sequence matrix was made up of a total of 20 different specimens from 17 different species (). The parsimony search resulted in four equally parsimonious trees, 1760 steps long, in which 202 characters were constant. Of the resulting variable characters, 59 were parsimony uninformative, whereas 334 were parsimony informative. The consistency index was 0.4630 and the retention index was 0.5539. The consensus () of these trees was used as the starting tree in the maximum likelihood analyses.
COI maximum likelihood
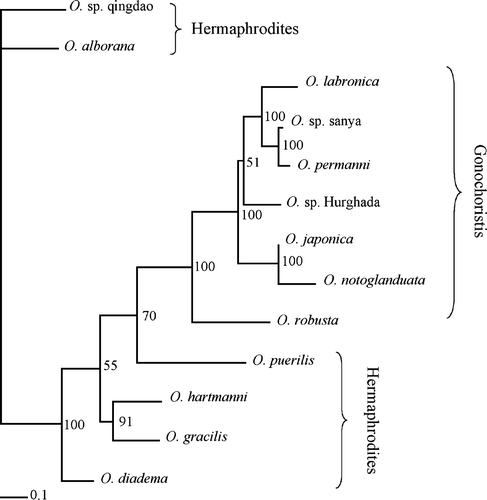
The maximum likelihood analysis was conducted through a heuristic search. The first maximum likelihood search was based on the consensus tree from the parsimony analysis. One tree was obtained by stepwise addition and branch-swapping algorithm set to tree-bisection-reconnection. The analysis resulted in three distinct clades (). Clade A consists of O. robusta, O. rubra, O. sp. Hurghada, O. labronica, O. sp. Sanya, O. notoglandulata and O. japonica, which all are all consistent with the so-called labronia group. Clade B consists of O. puerilis, Iphitime paguri, O. hartmanni and O. gracilis. Finally, clade C is made up of O. alborana, the two O. sp. Qingdao, and O. diadema.
Figure 2. The single best tree from the maximum likelihood analysis of the cytochrome c oxidase subunit I data. Branch support values given in italics indicate bootstrap values, whereas those in bold are Bayesian values.
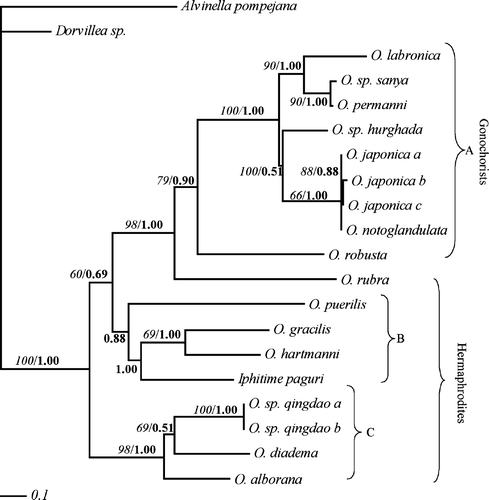
The bootstrap consensus tree included several collapsed branches, but most of the branch support values were fairly high (). An exception was the support value of the branch leading to clade A and B. Also interesting is the branch support value of the clade O. gracilis and O. hartmanni, which shows low bootstrap support. Because the branch of O. puerilis and I. paguri is collapsed, the branches leading to clade B and to I. paguri, O. gracilis and O. hartmanni are lacking bootstrap support values.
COI Bayesian analysis
The Bayesian analysis resulted in a phylogenetic consensus tree identical to the maximum likelihood tree (not shown). The support values resulting from the Bayesian analysis are given in bold in .
Combined analysis – parsimony
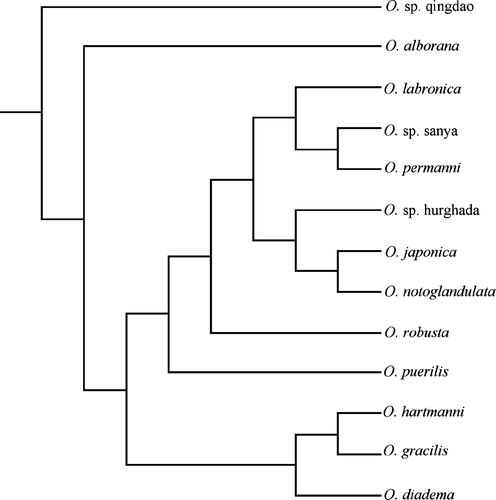
The data matrix for the combined analysis was constructed from 13 taxa and 1061 characters. In the combined analysis, the sequence of O. japonica A was chosen to represent the O. japonica species, whereas O. sp. Qingdao A was chosen to represent the O. sp. Qingdao species. A single most-parsimonious tree with length 1907 resulted when stepwise addition and branch-swapping algorithm set to tree-bisection-reconnection were applied (). Of the characters estimated, 408 were constant. The number of variable characters which were parsimony uninformative was 138, whereas 515 of the variable characters were parsimony informative. The resulting tree had a consistency index of 0.5868 and a retention index of 0.5247. This tree was used as the starting tree for the iterative search of a maximum likelihood tree.
Combined analysis – maximum likelihood and bootstrap support
Through a stepwise addition and branch-swapping algorithm set to tree-bisection-reconnection, one maximum likelihood tree was obtained (). The resulting phylogenetic tree showed no division of species into the clades indicated by morphology. Not all of the distinct clades that were obtained in the COI analysis were found in the combined analysis. The same species that were placed closely in the phylogenetic tree resulting from the COI analysis were also grouped closely in the combined analysis.
Combined analysis – Bayesian analysis
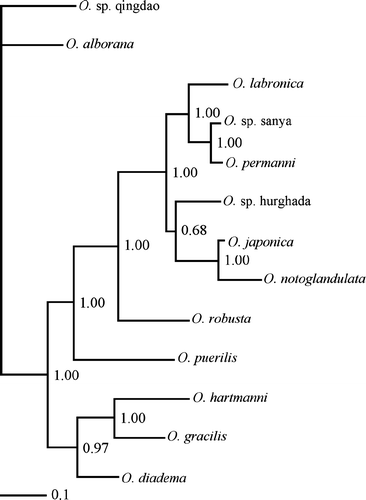
The Bayesian analysis resulted in a tree similar to the maximum likelihood tree. The resulting tree showed high posterior probability on branches. An exception was the branch leading to O. sp. Hurghada, O. japonica and O. notoglandulata, with a value of 0.68. The result of the analysis is shown in .
Discussion
Morphological congruence
The COI analysis and the combined analysis resulted in similar trees. The species were placed into two or three groups. The tree with two groups indicated one gonochoristic group: O. rubra, O. robusta, O. sp. Hurghada, O. labronica, O. sp. Sanya, O. notoglandulata, O. japonica A–C; and one hermaphroditic group: O. puerilis, O. hartmanni, O. gracilis, O. alborana, O. diadema, and O. sp. Qingdao A and B. The tree that resulted in three groups had an additional subdivision of the hermaphroditic species. One of the hermaphroditic groups contained O. puerilis, O. harmanni, O. gracilis and Iphitime paguri, whereas the other group incorporated O. sp. Qingdao A and B, O. diadema and O. alborana. By excluding O. sp. Qingdao A and B and O. alborana, which comprise most of the species in one of the two hermaphroditic groups, O. diadema was forced to regroup. As a consequence, one of the hermaphroditic clades was excluded.
Bootstrap and Bayesian analyses indicated good support for the branch leading to O. hartmanni and O. gracilis. Former analyses based on morphological data are incongruent with the grouping of O. hartmanni and O. gracilis resulting from this analysis. Ophryotrocha hartmanni and species similar to it posses k-type jaws and have an irregularly shaped egg mass. Species similar to O. gracilis possess p-type jaws and a fusiform-shaped egg mass.
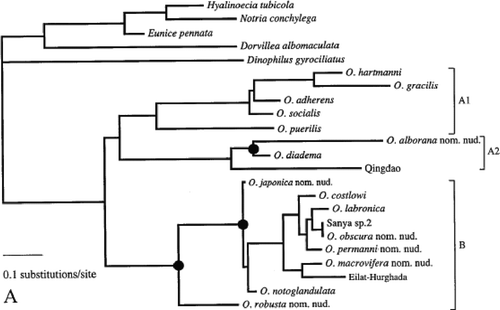
Molecular investigations conducted earlier have also resulted in the placement of O. hartmanni and O. gracilis as sister species (Dahlgren et al. Citation2001) (). Dahlgren et al. did not have any suggestions to explain this pattern. The constrained analysis also resulted in a significant difference between the tree obtained by Dahlgren et al. and the results obtained here. Clearly, more studies are needed to finally clarify the phylogenetic relationships within Ophryotrocha.
Figure 6. Phylogenetic tree resulting from the maximum likelihood analysis conducted by Dahlgren et al. (Citation2001). The filled circles indicate nodes congruent with the investigation of Pleijel & Eide (Citation1996).
The placement of Iphitime paguri
The phylogenetic analysis placed I. paguri within the Ophryotrocha clade. The relationship between these groups has previously been argued for by Orensanz (Citation1990). Not all agree on treating the genera as closely related. Eibye-Jacobsen & Kristensen (Citation1994) argued that ophryotrochans and iphitimids should be placed into different families, Dorvilleidae and Iphitimidae, respectively. Høisæter & Samuelsen (Citation2006) have investigated iphitimids and believe that the conclusions of Orensanz (Citation1990) may deserve more credit. Investigations conducted by Paxton (Citation2004) based on jaw morphology have also justified the conclusions of Orensanz.
The placement of I. paguri in this study indicates that this species could be placed within Ophryotrocha. Because the relationship between I. paguri and the type species I. doederleinii Marenzeller, Citation1902, that is the type species by monotypy, has not been investigated, we cannot at this stage conclude anything about the relative positions of Ophryotrocha and Iphitime. New investigations are necessary to resolve the iphitimid–ophryotroch relationship.
Editorial responsibility:Kenneth Halanych
Acknowledgements
Jon Kongsrud, Bergen Museum, Fredrik Pleijel, Göteborg University, and Eyvind Oug NIVA are thanked for help with obtaining references. Kenneth Meland and Endre Willassen, University of Bergen are thanked for critically reading and commenting on the manuscript. Critical comments from the subject editor and two anonymous referees greatly improved the manuscript. The study was funded by grants from the University of Bergen and from the Meltzer Foundation.
Notes
Published in collaboration with the University of Bergen and the Institute of Marine Research, Norway, and the Marine Biological Laboratory, University of Copenhagen, Denmark
References
- Åkesson , B . 1973 . Reproduction and larval morphology of five Ophtyotrocha species . Zoologica Scripta , 2 : 145 – 55 .
- Åkesson , B . 1976a . Temperature and life cycle in Ophryotrocha labronica (Polychaeta, Dorvilleidae) . Ophelia , 15 : 37 – 47 .
- Åkesson , B . 1976b . Morphology and life cycle of Ophryotrocha diadema, a new polychaete species from California . Ophelia , 15 : 23 – 35 .
- Åkesson , B . 1982 . A life table study of three genetic strains of Ophryotrocha diadema (Polychaeta, Dorvilleidae) . International Journal of Invertebrate Reproduction , 5 : 59 – 69 .
- Åkesson , B and Costlow , JD . 1978 . Effects of temperature and salinity on the lifecycle of Ophryotrocha diadema (Polychaeta, Dorvilleidae) . Ophelia , 17 : 215 – 29 .
- Åkesson B , Ehrenström F . 1984 . Avoidance reactions of dorvilleid polychaetes when exposed to chemical-contaminated sediments . In : Persoone G , Jaspers E , Claus C Ecotoxiological Testing for the Marine Environment 2 . Bredene : State University of Gent and Institute for Marine Scientific Research . p 3 – 12 .
- Åkesson , B and Paxton , H . 2005 . Biogeography and incipient speciation in Ophryotrocha labronica (Polychaeta, Dorvilleidae) . Marine Biology Research , 1 : 127 – 39 .
- Åkesson , B and Rice , SA . 1992 . Two new Dorvillea species (Polychaeta, Dorvilleidae) with obligate asexual reproduction . Zoologica Scripta , 21 : 351 – 62 .
- Bacci , G and La Greca , M . 1953 . Genetic and morphological evidence for subspecific differences between Naples and Plymouth populations of Ophryotrocha puerilis . Nature , 237 : 1208 – 9 .
- Blake , JA . 1985 . Polychaeta from the vicinity of deep-sea geothermal vents in the Eastern Pacific I: Euphrosinidae, Phyllodocidae, Hesionidae, Nereididae, Glyceridae, Dorvilleidae, Orbiniidae, and Maldanidae . Bulletin of the Biological Society of Washington , 6 : 67 – 101 .
- Bonnier , J . 1893 . Notes sur les annélids du Boulonnais. 1.-L’Ophyotrocha puerilis (Claparède et Metschnikoff) et son appareil maxillaire . Bulletin scientifique de la France et de la Belgique , 25 ( 4 ) : 198 – 226 .
- Brooks , KM , Stierns , AR , Mahnken , CVW and Blackburn , DB . 2003 . Chemical and biological remediation of the benthos near Atlantic salmon farms . Aquaculture , 219 : 355 – 77 .
- Cassai , C and Prevedelli , D . 1999 . Fecundity and reproductive effort in Ophryotrocha labronica . Marine Biology , 133 : 489 – 94 .
- Claparède , É and Metschnikow , E . 1869 . Beitraege zur Kenntnis der Entwicklungsgeschichte der Chaetopoden . Zeitschrift für wissenschaftliche Zoologie , 19 : 163 – 205 .
- Conlan , KE , Kim , SL , Lenihan , HS and Oliver , JS . 2004 . Benthic changes during 10 years of organic enrichment by McMurdo Station, Antarctica . Marine Pollution Bulletin , 49 : 43 – 60 .
- Dahlgren , TG , Åkesson , B , Schander , C , Halanych , KM and Sundberg , P . 2001 . Molecular phylogeny of the model annelid Ophryotrocha . Biological Bulletin , 201 : 193 – 203 .
- Desbruyeres , D and Laubier , L . 1980 . Alvinella pompejana gen. sp. nov., aberrant Ampharetidae from East Pacific Rise hydrothermal vents . Oceanologica Acta , 3 : 267 – 74 .
- Dreyer , JC , Knick , KE , Flickinger , WB and Van Dover , CL . 2005 . Development of macrofaunal community structure in mussel beds on the northern East Pacific Rise . Marine Ecology Progress Series , 302 : 121 – 34 .
- Eibye-Jacobsen , D and Kristensen , RM . 1994 . A new genus and species of Dorvilleidae (Annelida, Polychaeta) from Bermuda, with a phylogenetic analysis of Dorvilleidae, Iphitimidae and Dinophilidae . Zoologica Scripta , 23 : 107 – 31 .
- Fage , L and Legendre , R . 1934 . Les annélides polychètes du genre Iphitime. A propos d'une espece nouvelle commensale des pagures, Iphitime paguri n.sp . Bulletin de la Société Zoologique de France , 58 : 299 – 305 .
- Folmer , O , Black , M , Hoeh , W , Lutz , R and Vrijenhoek , R . 1994 . DNA primers for amplification of mitochondrial cytochrome c oxidace subunit I from diverse metazoan invertebrates . Molecular Marine Biology and Biotechnology , 3 : 294 – 9 .
- Fournier , JA and Conlan , KE . 1994 . A new species of Ophryotrocha (Polychaeta, Dorvilleidae) associated with ice scours in Canadian Arctic Archipelago. Actes de la 4ème Conférence internationale des Polychètes. Mémoires du Muséum National d . Histoire Naturelle , 162 : 185 – 90 .
- Hall TA. 1999 . Bioedit: a user friendly biological sequence alignment editor and analysis . Department of Microbiology, North Carolina State University ( http://www.mbio.ncsu.edu/BioEdit/page2.html ).
- Hilbig , B and Blake , JA. 1991 . Dorvilleidae (Annelida: Polychaeta) from the U. S. Atlantic Slope and Rise. Description of two new genera and 14 new species, with a generic revision of Ophryotrocha . Zoologica Scripta , 20 : 147 – 83 .
- Huelsenbeck , RF and Ronquist , F . 2003 . MrBayes 3: Bayesian phylogenetic inference under mixed models . Bioinformatics , 19 : 1572 – 4 .
- Huth , W . 1933 . Ophryotrocha-studien. Zur Cytologie der Ophryotrochen . Zeitschrift für Zellforschung und Mikroskopische Anatomie , 20 : 309 – 81 .
- Høisæter , T and Samuelsen , TJ . 2006 . Taxonomic and biological notes on a species of Iphitime (Polychaeta, Eunicida) associated with Pagurus prideaux from Western Norway . Marine Biology Research , 2 : 333 – 54 .
- La Greca , M and Bacci , G . 1962 . Una nuova specie di Ophryotrocha delle coste tirreniche (Annelida, Polychaeta) . Bollettino di Zoologia , 29 : 7 – 18 .
- Lorenzi , NC , Schleicherová , D and Sella , G . 2006 . Life history and sex allocation in the simultaneously hermaphroditic polychaete worm Ophryotrocha diadema: the role of sperm competition . Integrative and Comparative Biology , 46 : 381 – 9 .
- Marenzeller E. 1902 . Südjapaische Anneliden . III. Aphroditea, Eunicea. Denkschriften der Kaiserlichen Akademie der Wissenschaften (Wien) : 563 – 82 .
- Nylander JAA. 2004 . MrModeltest v2. Program distributed by the author . Evolutionary Biology Centre, Uppsala University ( http://www.ebc.uu.se/systzoo/staff/nylander.html ).
- Ockelmann , KW and Åkesson , B . 1990 . Ophryotrocha sosialis n. sp, a link between two groups of simultaneous hermaphrodites within the genus . Ophelia , 31 : 145 – 62 .
- Orensanz , JM . 1990 . The eunicemorph polychaete annelids from Antarctic and Subantarctic seas. Biology of the Antarctic Seas XXI . Antarctic Research Series , 52 : 1 – 183 .
- Oug , E . 1978 . New and lesser known Dorvilleidae (Annelida, Polychaeta) from Scandinavian and Northeast American waters . Sarsia , 63 : 285 – 303 .
- Oug , E . 1994 . The genus Ophryotrocha sensu lato (Polychaeta, Dorvilleidae) in the Tromsø area, northern Norway. Actes de la 4ème Conférence internationalse des Polychètes. Mémoires du muséum national d . histoire naturelle , 162 : 251 – 7 .
- Paavo , B , Bailey-Brock , JH and Åkesson , B . 2000 . Morphology and life history of Ophryotrocha adherens sp. nov. (Polychaeta, Dorvilleidae) . Sarsia , 85 : 251 – 64 .
- Paxton , H . 2004 . Jaw growth and replacement in Ophryotrocha labronica (Polychaeta, Dorvilleidae) . Zoomorphology , 123 : 147 – 54 .
- Paxton , H . 2005 . Moulting polychaete jaws – ecdysozoans are not the only moulting animals . Evolution and Development , 7 ( 4 ) : 337 – 40 .
- Pfannenstiel , H-D . 1972 . Eine neue Ophryotrocha Art (Polychaeta, Eunicidae) aus Japan . Helgoländer wissenschaftliche Meeresuntersuchungen , 23 : 117 – 24 .
- Pleijel , F and Eide , R . 1996 . The phylogeny of Ophryotrocha (Dorvilleidae: Eunicida: Polychaeta) . Journal of Natural History , 30 : 647 – 59 .
- Posada , D and Crandall , KA . 1998 . MODELTEST: testing the model of DNA substitution . Bioinformatics , 14 : 817 – 8 .
- Prevedelli , D , N′siala , GM and Simonini , R . 2006 . Gonochorism vs. hermaphroditism: relationship between life history and fitness in three species of Ophryotrocha (Polychaeta: Dorvilleidae) with different forms of sexuality . Journal of Animal Ecology , 75 : 203 – 12 .
- Prevedelli , D and Simonini , R . 2003 . Life cycles in brackish habitats: adaptive strategies of some polychaetes from the Venice Lagoon . Oceanologica Acta , 26 : 77 – 84 .
- Robotti , CA , Ramella , L , Cervella , P and Sella , G . 1991 . Chromosome analysis of nine species of Ophryotrocha . Ophelia (Suppl.) , 5 : 625 – 32 .
- Scheicherova , D , Lorenzi , MC and Sella , G . 2005 . How outcrossing hermaphrodites sense the presence of conspecifics and suppress female allocation . Behavioural Ecology , 17 : 1 – 5 .
- Sella , G and Lorenzi , MC . 2000 . Partner fidelity and egg reciprocation in the simultaneously hermaphroditic polychaete worm Ophryotrocha diadema . Behavioural Ecology , 11 : 260 – 4 .
- Sella , G and Lorenzi , MC . 2003 . Increased sperm allocation delays body growth in a protandrous simultaneous hermaphrodite . Biological Journal of the Linnean Society , 78 : 149 – 54 .
- Sella , G , Premoli , M and Turri , F . 1997 . Egg trading in the simultaneously hermaphroditic polychaete worm Ophryotrocha gracilis (Huth) . Behavioural Ecology , 8 : 83 – 6 .
- Sella , G , Vitturi , R , Ramella , L and Colomba , MS . 1995 . Chromosomal nucleolar organizer region (NOR) phenotypes in nine species of the genus Ophryotrocha . Marine Biology , 124 : 425 – 33 .
- Swofford DL. 2002 . PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods) . Version 4 . Sunderland : Sinauer Associates .
- Thompson , JD , Gibson , TJ , Plewniak , F , Jeanmougin , F and Higgins , DG . 1997 . The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools . Nucleic Acids Research , 24 : 4876 – 82 .
- Varella Petti , MA , Ferraz Nonato , E , Skowronski , RSP and Navajas Corbisier , T . 2006 . Bathymetric distribution of the meiofaunal polychaetes in the nearshore zone of Martel Inlet, King George Island, Antarctica . Antarctic Science , 18 : 163 – 70 .