Abstract
Background and purpose — Recent advances in artificial intelligence (deep learning) have shown remarkable performance in classifying non-medical images, and the technology is believed to be the next technological revolution. So far it has never been applied in an orthopedic setting, and in this study we sought to determine the feasibility of using deep learning for skeletal radiographs.
Methods — We extracted 256,000 wrist, hand, and ankle radiographs from Danderyd’s Hospital and identified 4 classes: fracture, laterality, body part, and exam view. We then selected 5 openly available deep learning networks that were adapted for these images. The most accurate network was benchmarked against a gold standard for fractures. We furthermore compared the network’s performance with 2 senior orthopedic surgeons who reviewed images at the same resolution as the network.
Results — All networks exhibited an accuracy of at least 90% when identifying laterality, body part, and exam view. The final accuracy for fractures was estimated at 83% for the best performing network. The network performed similarly to senior orthopedic surgeons when presented with images at the same resolution as the network. The 2 reviewer Cohen’s kappa under these conditions was 0.76.
Interpretation — This study supports the use for orthopedic radiographs of artificial intelligence, which can perform at a human level. While current implementation lacks important features that surgeons require, e.g. risk of dislocation, classifications, measurements, and combining multiple exam views, these problems have technical solutions that are waiting to be implemented for orthopedics.
Despite CT and MRI being commonplace, conventional radiographs remain central in orthopedics due to their availability, speed, price, and low radiation. Extracting all available information from these requires years of training and there is always the question of inter-observer reliability (Andersen et al. Citation1996, Audigé et al. Citation2005, Sayed-Noor et al. Citation2011, Shehovych et al. Citation2016). Aiding image interpretation using computers is therefore highly interesting from a clinical perspective.
Recent advances in artificial intelligence (AI) have shown remarkable results, even reaching superhuman performance at certain image interpretation tasks; for example He et al. (Citation2015) were able to surpass human test subjects (Russakovsky et al. Citation2015) when selecting the 5 most likely out of 1,000 categories where the most common error by the test subjects was failure to consider relevant classes. Deep learning is the primary method most often used and is a new take on traditional neural networks where the main difference is the number of layers. Neural networks are computational versions of biological nervous systems. They use mathematics to weigh the input and output from individual neurons and compute the best way to process information to reach a desired outcome, i.e. least errors. This could for example be to scan and recognize features of a radiograph (Erickson et al. Citation2017). Originally the networks consisted of a single layer of neurons but the rapid increase in computational power allows newer networks to have many layers of neurons communicating with each other. Modern networks can have anything ranging from 8 to 1,000 layers, hence the deep in “deep learning”, enabling complex interpretation and decision-making. While deep learning and AI has been a hot topic in mainstream media (DeAngelis Citation2014, Hardy Citation2016, Hern Citation2016, Rhodes Citation2016) and believed by many to be the next technological revolution, it has so far never been applied in an orthopedic setting for radiographs.
The aim of this study was to see if standard deep learning networks can be trained to identify fractures in orthopedic radiographs. Secondarily we also examined whether deep learning could be used to determine additional features such as body part, exam view, and laterality.
Methods
Setting
We extracted 256,458 hand (including scaphoid projections), wrist, and ankle radiographs, with associated radiologist reports, taken between the years 2002 and 2015 from Danderyd Hospital’s Picture Archiving and Communication System (PACS) identified through the Radiology Information System (RIS).
Exposure
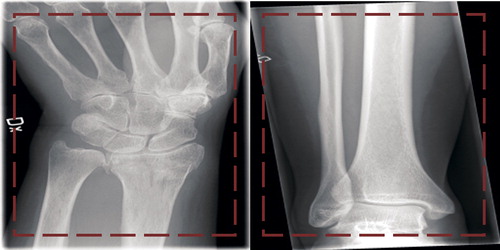
The deep learning networks were exposed to a single image at a time. Each image was cropped and rescaled to 256 x 256 pixels, about 10–20% of original size (see for example images). The rescaling was performed to match the predefined image size of each network (see Supplementary data for explanation) and the images were stretched to retain maximum information. While the images are distorted after this pre-processing, it is important to keep in mind that the tasks the network needed to perform did not require a non-distorted image, e.g. measuring angles.
Figure 1. 2 images from the dataset. The area within the red box is the section presented to the network in order to classify the image. The left image is of a wrist fracture while the right image is without any apparent fracture.
Outcome labels
The images, image metadata, and radiologist reports were merged and anonymized (see Supplementary data for details) before deducing the labels. We identified 4 basic outcome labels that were used for training the networks:
Fracture: the presence of a fracture was deduced from a combination of multiple visits together with identification of keywords and expressions identified through an automated language extraction software applied to the radiologist’s report.
Laterality: whether it was left or right. This information was generally present in the image meta-data (the DICOM header, see Supplementary data for details), otherwise it was assumed that if the examination before and after was of the same side, the intermediate would most likely also belong to that side.
Exam view: The type of view (anteroposterior/frontal, lateral, oblique (2 different types), and 4 scaphoid specific views (proximal, distal, ulnar, and radial) was also identified via the image meta-data.
Body part: Body part refers to the general body area: ankle, wrist, and hand. The latter had also subgroups of scaphoid, thumb, or finger. These were also identified via the image meta-data.
Fracture was the primary outcome while the latter 3 were chosen for comparing the noisy fracture label with high-quality labels specific to orthopedic radiographs. These secondary outcomes are also important to any practical implementation where they can serve as quality control and validation, to minimize errors and for more advanced fracture classification.
Deep learning framework
We selected 5 common, freely available, deep networks from a popular online library (the Caffe library (Jia et al. Citation2014)):
BVLC Reference CaffeNet network (8 layers) (Krizhevsky et al. Citation2012);
VGG CNN S network (8 layers) (Chatfield et al. Citation2014);
VGG CNN (16 and 19 layers networks) (Shi Zhong et al. Citation2014);
Network-in-network (14 layers) (Lin et al. Citation2013).
The networks were adapted to the outcomes above and retrained for 13 epochs (1 epoch =1 run through all images). The training was performed using stochastic gradient descent on 70% of the original images, where neurons are corrected by a tiny portion after each image (see Supplementary data for details).
Manual review of radiographs
To assess a network’s performance and understand where it fails, we manually investigated the errors for the best performing network. We benchmarked the network against a gold standard for the primary outcome for 400 images from the test dataset by reviewing each image in full resolution together with its alternative views and the radiologist’s report.
We compared the network with human performance by allowing 2 senior orthopedic consultants (AS, OS) to identify fractures in the same 400 images at the same resolution as the network. The reviewers were blinded to the network’s and the other reviewer’s labels. We calculated the accuracy for comparison with the network and Cohen’s kappa as an interrater reliability estimate.
For secondary outcomes, unlike fractures, the label outcome was extracted from the DICOM image header set for each individual image during the examination. Apart from random human error they were assumed to be correct and therefore only investigated for the network’s errors in order to understand the underlying cause of the errors. We randomly selected 200 misclassified exam views and 200 misclassified lateralities for review. The exam views were categorized as either correctly classified or misclassified, for all statistical and computational purposes. To understand network errors for the image review the misclassified images were divided into two subcategories, unrelated view and closely related view. Images in the closely related view were further divided into subcategories describing the most common noticed misclassification errors. We furthermore reviewed all 86 misclassified body part images. The review was performed by AS, MG, and JO.
Statistics
Confidence intervals were computed using bootstrapping with 10,000 bootstraps using the 2.5 and 97.5 percentile. Inter-observer reliability for fractures was computed using Cohen’s exact Kappa between the observers and the best performing network.
Ethics, funding and potential conflicts of interest
The Stockholm Regional Ethical Review Board approved the study (2014/453-31/3). Financial support for the study was from funding from the Swedish Association of Local Authorities and Regions. We also wish to gratefully acknowledge the support of the NVIDIA Corporation with the donation to this research of 2 Tesla K40 GPUs. MG, AS, OS, and AJ are shareholders in DeepMed AB.
Results
We included 256,458 images, of which 56% of the images contained fractures. Ankles were the most common body part (38%), with right extremity (52%) slightly more common than left. The anteroposterior was the most common view ().
Table 1. Raw image and label data for a total of 256,458 images. 70% were reserved for training, 20% for validation, and 10% for testing
General network performance
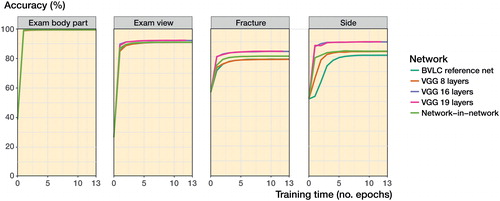
All networks performed with an accuracy of above 90% when identifying body part and exam view (). For fractures there was a clear improvement in the networks with more layers and newer design principles (Network-in-Network). The best raw performance was exhibited by the VGG 16 layered network with an accuracy of 83%.
As VGG 16 layers had the best performance in the fracture class, our primary outcome variable, we selected it for manual review. When comparing the network with 2 senior orthopedic surgeons we found that the network performed on par with the humans (). The interrater reliability was adequate (); the 2 human observers generally agreed with each other (kappa =0.8).
Table 2. Observer fracture outcome compared with gold standard
Table 3. Outcomes compared between observers. Accuracy is the percentage of outcomes where both observers agree, presented with Cohen’s kappa
Table 4. Manual review of classifications where the network failed
Outcome-specific details ()
The most common causes for misclassification were (1) lack of information within the tested image and (2) image ambiguity. In images labeled with fracture, we could frequently not see the fracture when reviewing the images. This was often because the fracture was only visible in another exam view within the same series.
For our secondary outcomes, we reached accuracies of over 99% on body part (6 outcomes), 95% on exam view (7 outcomes), and 90% on laterality.
For the “wrong, but similar views” category, projections were often ambiguous due to incorrectly positioned limbs, i.e. somewhere in between two standard projections. Many of the scaphoid images were noted to be non-standard projections. For ankle projections, the network often caught the intention of the image, i.e. the label, but poor angles ultimately resulted in a different projection. This was especially true for oblique and lateral views.
Body part, with 6 categories, had close to perfect accuracy with only 86 images not matching the original label. Studying the errors, we found that the errors were often not errors at all, but rather that the network found “hidden” or “equivalent” features of the image. Common errors were a mix-up of hand and wrist in projections where both were clearly visible. Other errors were finger/thumb or fingers/hand.
For laterality, the dominant error—two-thirds of the cases—was that the marker with the “Sin”/”Dx” text was not visible due to the image cropping. For the remaining cases, in 5 times out of 6 the network was correct and the metadata incorrect, i.e. human error.
Discussion
We found that standard deep learning networks could adequately identify key image properties in orthopedic radiographs despite the limited image quality. The best network had a similar accuracy to the performance of 2 senior orthopedic consultants when compared under the same conditions. The most common causes for errors were lack of data and ambiguity in the image.
The ability to classify an unlimited amount of radiograph images will most likely have a major impact on orthopedics. We can now review images on an unprecedented scale in our digital picture archives and link them to outcomes. Apart from identifying traditional orthopedic measures such as wrist angles we can also let the algorithms search for new patterns, for example we can go beyond simple angles into complex patterns that combine angles, comminution, and bone quality. As many of our fracture classifications lack prognostic value (Shehovych et al. Citation2016), often with questionable inter-observer reliability (Audigé et al. Citation2005), the option of aiding the classification using a computer algorithm is of great interest.
Furthermore, since machine-learning algorithms do not have preconceived notions of what is interesting within an image, it is possible that we will find new, previously unknown predictors. For instance, when predicting breast cancer prognosis a machine-learning algorithm found that in addition to the already known histological aspects of tumor cells the surrounding stroma was also of value for prognosis (Beck et al. Citation2011). It is therefore likely that orthopedics will be substantially influenced in the coming decade as this technology evolves.
Our study shows that networks originally developed for other tasks than skeletal imaging can be applied to skeletal radiographs with minimal intervention. While to our knowledge our study is the first to show that deep learning works for orthopedic radiographs, it has previously been investigated for other medical domains, including spine MRIs (Jamaludin et al. Citation2017). Others have identified pathologies in 2-dimensional slices of CT -images (Shin et al. Citation2015, Citation2016, Tajbakhsh et al. Citation2016) as well as in chest radiographs (Bar et al. Citation2015).
While the deeper layered models outperform the older and shallower models in every category, suggesting that the extra computational work of modern networks is worthwhile, for some groups the difference was small. In our networks () we see that the initial progress in training is rapid, with most progress taking place within the first 5 passes (epochs) over the training data. Overfitting means that the network learns to identify each image with its particular outcome instead of the pattern that makes up the outcome. When new images are introduced the network fails to interpret them correctly and accuracy falls. We saw no tendencies towards overfitting during training or testing. This was likely due to the large data set and the fact that the training and validation sets were resampled at each epoch.
We were surprised that the top networks also performed better that the others on trivial tasks such as “laterality” where the main task is to identify the “Sin” and “Dx” markers. Since the training images were randomly mirrored, effects such as a right hand mostly appearing with the thumb to the left should have been eliminated. Our interpretation is that there must be additional laterality indicators apart from the markers that we did not expect to find. This could possibly be due to the dominant side being used more and having a different bone structure.
Limitations
The neural networks use labelled images as input, and the quality of image labels is therefore a fundamental limitation. The current data are labeled by trained specialists with years of experience but come in the form of raw text from which labels need to be extracted. This is due to the complex language being inherently difficult, and in this study we have manually identified key phrases. Improved use of the reports and information is therefore of great interest. Using Natural Language Processing (NLP), another field of machine learning, more information could be extracted.
The current networks used only a single image for classification. Often an examination will contain a series of images in different projections, since pathologies may not be visible in 1 image but very well seen in another. This limitation was also supported by our manual review. Expanding the networks to manage multiple images is therefore a natural next step. Possible network designs could be reusing the same network multiple times and then gathering the data by a separate network that classifies the image, also known as Siamese networks (Zbontar and LeCun Citation2016), or using a network with a memory that remembers the previous images, also known as recurrent neural network (LeCun et al. Citation2015).
The number of labels/classes is limited and needs to be increased to be clinically useful. It would be possible to include more advanced screening of the reports for other diagnostic criteria to label images. It could also be possible to use automated language interpretation technologies to extract information for image labels (Shin et al. Citation2015). This does, however, raise a second more fundamental issue. Radiologists’ reports are usually written as answers to questions (usually a series of diagnoses) posed by the physician in the exam referral. Thus some information contained in the image might be purposely omitted in the report as it has no bearing on the current question or might already be clearly stated in the referral. As these pathologies are not mentioned in the report, they will be impossible to extract for image labelling and assumed to be non-present. This can inhibit network training for labels rarely asked for in trauma referrals but that may still be common, e.g. osteoarthritis in wrists. Including other journal data such as referral and other patient records would be an excellent way to improve labels but may raise privacy concerns.
Strengths
We used a large data set with 256,000 images where the networks’ potential was no’t limited by overfitting but by the network design and quality of the training labels.
We believe that deep learning will have a great impact on orthopedics in the coming years. We propose that our current results could be useful in the emergency room as a method for fast screening when radiologists or orthopedists are not readily available, e.g. night-time at smaller hospitals. Future results will most likely allow us to improve diagnostic accuracy, classify fractures on a large scale, and identify new prognostic features.
Supplementary data
Supplementary data are available in the online version of this article, http://dx.doi.org/10.1080/17453674.2017.1344459
JO preprocessed the radiological reports and created labeling in collaboration with MG and NF; he also helped create the gold standard and supervised the image reviews performed by the senior authors. JO synthesized the manuscript together with MG and conducted the data analysis. AM helped with planning of the study, necessary computational resources, and supported with the technical aspects. ASR aided with network designs and the technical details behind transfer learning. AJ helped with setting up the image-extraction procedure from the PACS. AS, OS, MG, and AJ reviewed images. OS also aided with the study design and ethical application for the study. He also wrote the introduction to the manuscript and prepared the manuscript for submission. MG initiated the study, wrote the ethical application and coordinated the project. He collected and created the data set for the study, and was responsible for the image and neural network code base and functionality, including network training.
Acta thanks Hans Berg and other anonymous reviewers for help with peer review of this study.
IORT_A_1344459_Supp.pdf
Download PDF (51.7 KB)- Andersen D J, Blair W F, Stevers C M, Adams B D, El-Khouri G Y, Brandser E A. Classification of distal radius fractures: An analysis of interobserver reliability and intraobserver reproducibility. J Hand Surg. 1996; 21 (4): 574–82.
- Audigé L, Bhandari M, Hanson B, Kellam J. A concept for the validation of fracture classifications. J Orthop Trauma. 2005; 19 (6): 404–9.
- Automation and anxiety. The Economist [Internet]. 2016 Jun 25 [cited 2016 Nov 22]; Available from: http://www.economist.com/news/special-report/21700758-will-smarter-machines-cause-mass-unemployment-automation-and-anxiety
- Bar Y, Diamant I, Wolf L, Greenspan H. Deep learning with non-medical training used for chest pathology identification. 2015 [cited 2016 Jun 2]. p. 94140V–94140V–7. Available from: http://dx.doi.org/10.1117/12.2083124
- Beck A H, Sangoi A R, Leung S, Marinelli R J, Nielsen T O, van de Vijver M J, West R B, van de Rijn M, Koller D. Systematic analysis of breast cancer morphology uncovers stromal features associated with survival. Sci Transl Med 2011; 3 (108): 108ra113–108ra113.
- Chatfield K, Simonyan K, Vedaldi A, Zisserman A. Return of the devil in the details: Delving deep into convolutional nets. British Machine Vision Association; 2014 [cited 2016 Aug 19]. p. 6.1-6.12. Available from: http://www.bmva.org/bmvc/2014/papers/paper054/index.html
- DeAngelis S F. Machine learning: Bane or blessing for mankind? [Internet]. WIRED. 2014 [cited 2016 Nov 22]. Available from: https://www.wired.com/insights/2014/06/machine-learning-bane-blessing-mankind/
- Erickson B J, Korfiatis P, Akkus Z, Kline T L. Machine learning for medical imaging. Radiogr Rev Publ Radiol Soc N Am Inc. 2017; 37(2): 505–15.
- Hardy Q. Artificial intelligence software is booming: But why now? N Y Times [Internet]. 2016 Sep 18 [cited 2016 Nov 22]; Available from: http://www.nytimes.com/2016/09/19/technology/artificial-intelligence-software-is-booming-but-why-now.html
- He K, Zhang X, Ren S, Sun J. Delving deep into rectifiers: Surpassing human-level performance on imagenet classification. 2015 [cited 2016 Aug 19]. p. 1026-34. Available from: http://www.cv-foundation.org/openaccess/content_iccv_2015/html/He_Delving_Deep_into_ICCV_2015_paper.html
- Hern A. Why data is the new coal. Guardian [Internet]. 2016 Sep 27 [cited 2016 Nov 22]; Available from: https://www.theguardian.com/technology/2016/sep/27/data-efficiency-deep-learning
- Jamaludin A, Lootus M, Kadir T, Zisserman A, Urban J, Battié M C, Fairbank J, McCall I; Genodisc Consortium. ISSLS prize in bioengineering science 2017: Automation of reading of radiological features from magnetic resonance images (MRIs) of the lumbar spine without human intervention is comparable with an expert radiologist. Eur Spine J 2017; 26(5): 1374–83.
- Jia Y, Shelhamer E, Donahue J, Karayev S, Long J, Girshick R, Guadarrama S, Darrell T. Caffe: Convolutional architecture for fast feature embedding. New York: ACM Press; 2014 [cited 2016 Aug 19]. p. 675-8. Available from: http://dl.acm.org/citation.cfm?doid=2647868.2654889
- Krizhevsky A, Sutskever I, Hinton G E. ImageNet classification with deep convolutional neural networks. In: Pereira F, Burges CJC, Bottou L, Weinberger KQ, editors. Adv Neural Inf Process Syst 25 [Internet]. Curran Associates, Inc.; 2012 [cited 2014 Sep 11]. p. 1097–1105. Available from: http://papers.nips.cc/paper/4824-imagenet-classification-with-deep-convolutional-neural-networks.pdf
- LeCun Y, Bengio Y, Hinton G. Deep learning. Nature 2015; 521 (7553): 436–44.
- Lin M, Chen Q, Yan S. Network in network. ArXiv13124400 Cs [Internet]. 2013 Dec 16 [cited 2016 Aug 19]; Available from: http://arxiv.org/abs/1312.4400
- Rhodes M. Whoa, Google’s AI is really good at Pictionary [Internet]. WIRED. 2016 [cited 2016 Nov 22]. Available from: https://www.wired.com/2016/11/woah-googles-ai-really- good-pictionary/
- Russakovsky O, Deng J, Su H, Krause J, Satheesh S, Ma S, Huang Z, Karpathy A, Khosla A, Bernstein M, Berg A C, Fei-Fei L. ImageNet large scale visual recognition challenge. Int J Comput Vis 2015; 115 (3): 211–52.
- Sayed-Noor A S, Ågren P-H, Wretenberg P. Interobserver reliability and intraobserver reproducibility of three radiological classification systems for intra-articular calcaneal fractures. Foot Ankle Int 2011; 32 (9): 861–6.
- Shehovych A, Salar O, Meyer C, Ford D. Adult distal radius fractures classification systems: Essential clinical knowledge or abstract memory testing? Ann R Coll Surg Engl 2016; 98 (8): 525–31.
- Shi Zhong, Li K, Rui Feng. Deep convolutional hamming ranking network for large scale image retrieval. IEEE; 2014 [cited 2016 Aug 19]. p. 1018–23. Available from: http://ieeexplore.ieee.org/lpdocs/epic03/wrapper.htm?arnumber=7052856
- Shin H-C, Lu L, Kim L, Seff A, Yao J, Summers R M. Interleaved text/image deep mining on a very large-scale radiology database. 2015 [cited 2016 Aug 19]. p. 1090-9. Available from: http://www.cv-foundation.org/openaccess/content_cvpr_2015/html/Shin_Interleaved_TextImage_Deep_2015_CVPR_paper.html
- Shin H-C, Roth H R, Gao M, Lu L, Xu Z, Nogues I, Yao J, Mollura D, Summers R M. Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. ArXiv160203409 Cs [Internet]. 2016 Feb 10 [cited 2016 Aug 19]; Available from: http://arxiv.org/abs/1602.03409
- Tajbakhsh N, Shin J Y, Gurudu S R, Hurst R T, Kendall C B, Gotway M B, Liang J. Convolutional neural networks for medical image analysis: Full training or fine tuning? IEEE Trans Med Imaging 2016; 35 (5): 1299–312.
- Zbontar J, LeCun Y. Stereo matching by training a convolutional neural network to compare image patches. J Mach Learn Res 2016; 17 (1-32): 2.