Abstract
Fast DNA detection remains of great interest in human genetics, medicine, and drug discovery. The detection of DNA hybridisation makes the screening of point mutations in potential cancer genes or DNA fingerprinting for phylogenesis purposes possible (S.W. Yeung, T.M.H. Lee, H. Cai, and I.M. Hsing, A DNA biochip for on-the-spot multiplexed pathogen identification, Nucleic Acids Research 34 (2006), p. e118; P. Liepold, H. Wieder, H. Hillebrandt, A. Friebel, G. Hartwich, DNA-arrays with electrical detection: a label-free low cost technology for routine use in life sciences and diagnostics, Bioelectrochemistry, 67 (2005), pp. 143–150). The speed, cost and reliability of the hybridisation detection is of high importance. Electronic detection of hybridisation events using standard CMOS-fabricated devices such as Field Effect Transistors (FETs) promises fast, label-free and multiplexed read-out systems. Moreover, they hold the advantage of high-throughput and minimalisation, which makes them ideal for implementation in fast diagnostic tools such as lab-on-chip systems. Field-effect devices, however, imply the necessity of low-ionic strength buffer solutions for signal maximisation because of the occurrence of charge screening effects near the electrolyte-oxide interface layer. In this article, we present a surface chemistry-based methodology that allows FET-based recordings of hybridisation events in low-ionic strength solutions. Quartz Crystal Microbalance results show that positively-charged surfaces promote DNA hybridisation, even when performed in lower salt concentrations then commonly used. Fluorescence measurements were performed on the different surfaces to reveal the optimal DNA adsorption conditions on the surface. For proof-of-principle, the surface chemistry was applied on the surface of a floating-gate field-effect transistor, and online recordings of DNA hybridisation events were performed in low-ionic strength solutions.
1. Introduction
Fast, in situ DNA detection remains of great interest in biomedical applications nowadays Citation1–4. DNA detection is typically based on monitoring hybridisation events. DNA hybridisation occurs when a single stranded DNA (ssDNA) target is identified by its complementary ssDNA probe to form double stranded DNA (dsDNA). The hybridisation between two ssDNA strands is known as a very efficient binding with a high affinity and specificity, and is therefore often used as a model system for DNA detection.
Although, multi-analyte optical detection systems based upon fluorescent or radioactive markers are widely used in research applications Citation5, Citation6, a growing interest in electronic detection systems is observed. Electronic detection techniques have the advantage of direct and label-free detection of DNA hybridisation events Citation7–9. Hence, the elimination of labelling has the advantage of simplifying the readout, making the technique much faster and less expensive. This is of increasing importance in nucleic acid assays in DNA genotyping and polymorphism detection Citation10. Microfabricated field-effect devices are often used for biological sensing purposes such as enzymatic sensors, immunosensors, and DNA sensors Citation11. Essentially, the ISFET operation principle is the same as the MOSFET, except for the fact that the standard metal (polysilicon) gate is replaced by an ‘ionic’ gate formed by the electrolyte in direct contact with the gate dielectric. An external reference electrode (RE) is used in order to fix the potential of the solution. The proton concentration in the electrolyte influences the potential drop at the electrolyte/dielectric interface, which in turn modifies the transistor threshold voltage. In this way, the proton concentration in the electrolyte exercises an electrostatic control on the drain-source current. The use of standard silicon (CMOS) technology has the advantage of miniaturisation, and therefore mass production and cost effectiveness can be realised. Furthermore, the implementation of these devices in small and portable read-out systems could make the development of lab-on-a-chip applications for field DNA diagnostics feasible. Moreover, the effect of DNA adsorption on the gate of the ISFET can be evaluated by a simple parameter such as the variation in the transistor threshold voltage. As an alternative for silicon-based ISFETs, organic thin film transistors (OTFTs) could be used in the future for the construction of disposable DNA sensors, provided that the current stability problems of organic semiconductors are overcome and appropriate packaging methodologies are developed Citation12.
Besides having appropriate physico-chemical transducers, reliable DNA hybridisation can only be assured when using a surface chemistry methodology optimised for binding a sufficient amount of probe DNA on the surface. The attachment of the probe DNA can be done either by covalent attachment or by adsorption on the surface. Covalent attachment has the advantage that the DNA probe can be positioned in a way that the complementary strand can easily bind. This method, using self-assembled monolayers, polymers, or crosslinkers is often used in DNA hybridisation assays Citation13. Although, the adsorption of the DNA strands may not ensure the strongest binding to the surface, the advantage of adsorption lies in the fact that the DNA strands lay down on the surface films instead of standing up, and in that way more negative charges of the DNA strand are positioned closer to the channel of the transistor. The importance of the distance of the hybridisation events on top of the surface of field-effect devices has been described Citation14. The sensitivity of the Ion Sensitive FET (ISFET) depends on the possibility of sensing charged molecules within the length of the double layer formed at the electrolyte-insulator interface, which depends on the salt concentration of the solution. Due to screening effects of the buffer solution, charges that are found outside the double layer can not be detected by the ISFET Citation14. On the contrary, the presence of a sufficient concentration of counter ions is necessary for DNA hybridisation. Indeed, a DNA molecule consists of a large effective negative charge, caused by the sugar-phosphate backbone on the exterior of the molecule, that is surrounded in buffered solutions by positive ions to enhance the probability of binding.
In this article, we report an approach to reduce the necessity of high-strength solutions needed for DNA hybridisation based on surface chemistry, and therefore make DNA hybridisation detection with ISFET devices feasible. By immobilising an excess positive charge onto the gate of the transistor, the electrostatic repulsion between target and probe DNA can be reduced Citation15. We have investigated this principle by means of QCM measurements and fluorescent characterisation of the surface and the DNA immobilisation on that particular surface. The developed surface chemistry was then transferred to the surface of a floating gate (FG) FET transducer in order to show the feasibility of DNA hybridisation in low ionic-strength solutions.
2. Materials and methodology
2.1. Materials
All cleaning chemicals were obtained from Sigma (MO, USA). Poly-L-Lysine (PLL) (MW = 70,000), sodium cyanoborohydride (CNBH) and ethanolamine were also obtained from Sigma (MO, USA). The ssDNA (probe, target and noncomplementary) was obtained from Eurogentec (Seraing, Belgium). Glutaraldehyde (25% in water) was obtained from Fluka (Buchs SG, Switzerland), and was further diluted to a concentration of 2.5%. 3-amino-propyl triethoxy silane (APTES) and Triethoxtsilylundecanal (TESU) were obtained from Geleste Inc. (PA, USA). The Alexa Fluor 555 amine-reactive and thiol-reactive dye came from Invitrogen (CA, USA). Purification of the labelled DNA from the excess dye was done using a NAP-10 filtration column from GE Healthcare. Tris[2-carboxyethyl]phosphine hydrochloride (TCEP) was obtained from Acros (NJ, USA).
2.2. Surface chemistry
The QCM crystals were cleaned before each experiment with Piranha solution (H2SO4:H2O2, 3 : 1) for 15 min and subjected to an UV/O3-treatment for 15 min. The crystals were then immersed in an ethanol-based solution (5% water, 93% ethanol) containing 2% APTES for 30 min, or likewise in a 2%TESU solution for 5 min. After silanisation, all crystals were rinsed plentifully with ethanol and baked at 110°C on a hotplate for 30 min. To carry out the PLL-coating, samples with the APTES coating were treated further by immersing them into a solution containing the amine-reactive homobifunctional crosslinker glutaraldehyde and CNBH (2,5% glutaraldehyde:CNBH, 1 : 1) for 30 min. After rinsing, the samples were immersed in a 1 : 1 solution containing Poly-L-Lysine (2 mg/ml in 10 mM borate buffer, pH8) and CNBH. TESU-treated samples were used as a control by immersing them into a 1M ethanolamine solution for 30 min, creating hydroxyl end groups.
The DNA model system used for all experiments was a 50 bp SH-C6-DNA strand with the following content: 12 × A, 16 × T, 11 × C, 11 × G. The 25 bp target DNA consisted of 10 × A, 5 × T, 7 × C, 3 × G and the 25 bp bis target DNA contained 6 × A, 7 × T, 4 × C, 8 × G. The sequence of the 25 bp noncomplementary strand contained 6 × A, 7 × A, 4 × C, 8 × G.
2.3. Fluorescent labelling and imaging of surface chemistry and probe DNA
1 × 1 cm pieces of SiO2 covered with 50 nm of Ta2O5 were used as test samples. Labelling of the amine groups was investigated by placing a drop of amine-reactive Alexa Fluor dye (Invitrogen, A20187) on the different surfaces. Analysis of the relative fluorescence intensity at 555 nm was performed with a confocal microscope (Axioscope, Zeiss). Labelling of the absorbed DNA was carried out using a thiol-reactive dye Alexa Fluor (Invitrogen, A20346), which was coupled to the thiol end of the 50 bp probe DNA. Purification of the labelled DNA was performed with a NAP10 column filtration unit. TCEP was used to make sure that possible disulfide bonds between single DNA probes were eliminated. Fluorescent analysis was done at 555 nm; the drops were monitored using the same system described above. Relative fluorescence units were divided by the background intensity of each substrate.
2.4. QCM mass adsorption
The quartz crystal microbalance (QCM) biosensor and 5 MHz crystal sensors (qsx301, Au, 50 nm) were obtained from Qsense AB (Västra Frölunda, Sweden). The QCM technique has been used extensively to study the molecular adsorption to different surfaces in order to investigate the properties of biomaterials and functionalised surfaces. The QCM technique determines the mass of very thin surface bound layers and simultaneously gives information about their viscoelastic properties.
QCM Crystals were covered in house with a 50 nm Ta2O5 layer by means of reactive sputtering. The liquid flow rate in the QCM flow cell was maximum 2 ml/min with external temperature stabilisation, the measurement volume was ∼80 µl, and the optimal exchange volume was around 2 ml. After deposition of each new layer, the sensor crystal was rinsed with the corresponding buffer. If the film on the surface is thin and rigid (which was the case), the decrease in frequency is proportional to the mass of the film. The mass of the adhering (PLL and enzyme) layer was calculated using the Sauerbrey relation:
2.5. FG FET transducer
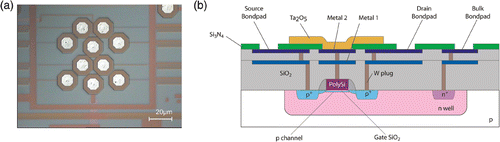
A chip containing eight individually addressable floating-gate PMOS transistors with an extended gate configuration was fabricated in house in 0.25 µm CMOS technology. After the standard CMOS processing (two metal layers interconnects), a Ta2O5-layer with a thickness of 100 nm is deposited by means of reactive sputtering, in order to increase the gate capacitance and provide a good anchoring layer for the surface chemistry. Having a large buffer capacity, Ta2O5 has been proven to be one of the best charge sensitive oxides in ISFET devices Citation16. The purpose of the two level metal interconnect scheme is to provide sufficient isolation of the transistor from the potentially corrosive electrolyte environment. The device structure is illustrated schematically in . After dicing, the chips have been wire-bonded on standard dual-in-line (DIL) packages and the electrical connections have been protected against liquid exposure by a two-components epoxy (H54, Epotek, MA, USA), which was baked for 2 h at 95°C.
2.6. FG-FET measurement set-up
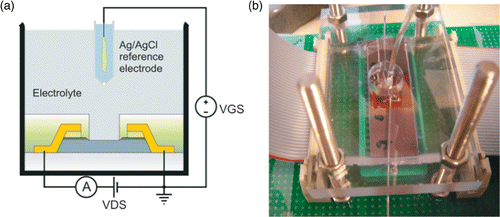
The FG FETs were first evaluated for pH sensitivity using buffers with different pH values. The device was brought in contact with the pH buffers together with an Ag/AgCl RE in an open flow-cell configuration. Solutions were pumped over the sensor surface using peristaltic pumps. The device, immersed together with the RE in buffer, was biased as a typical FET in common source configuration. The RE is used to ensure a stable electrical contact with the solution phase when it is biased with respect to the source electrode. This electrode plays the role of the control gate for the FG transistor. Probe, target and noncomplementary DNA were injected with a micropipette in the open flow cell; rinsing was done by pumping. shows the measurement principle and a photograph of the flow-cell set-up. All FET characteristics and time lapse experiments were recorded with an Agilent/HP 4156A Semiconductor Parameter Analyser using a software programmable protocol (Matlab, The Mathworks).
3. Results and discussion
3.1. Fluorescent imaging of surface chemistry and immobilised DNA
Because of the net negative charge of DNA, a positively charged surface layer is desired on the sensor surface to ensure a reliable adsorption. Self-assembled monolayers are frequently being used in biosensor technology to ensure reproducible and robust surface layers Citation12. Here, silane chemistry (APTES) was used to create a monolayer of amine endgroups on the oxide surface. To further increase the number of amine groups for DNA adsorption, Poly-L-Lysine was coupled via the homobifunctional crosslinker glutaraldehyde. CNBH was added as a reducing agent to stabilise the amide binding between glutaraldehyde and APTES, and between glutaraldehyde and PLL. As a control condition, samples were coated with TESU silane, creating aldehyde endgroups on the surface, which were then reformed to hydroxyl groups using ethanolamine to ensure a nonpositively charged surface.
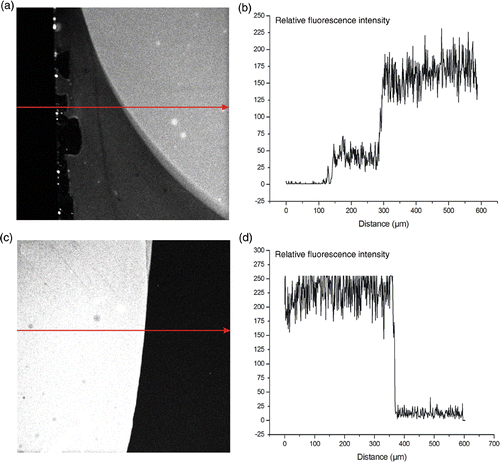
shows the relative fluorescent intensity of the APTES surface compared to the PLL surface after addition of the amine-reactive dye. The PLL surface contained four times more amine endgroups then the APTES surface, taking the background in consideration. This result shows that the PLL surface consists of much more positive charges then the APTES surface, providing more binding sites for DNA adsorption. This can be explained by the 3D structure of the PLL polymer.
Figure 3. Fluorescent imaging of the presence of amine groups on the surface of different substrates. The arrows indicate the direction in which the relative fluorescence was monitored. (a) and (b): relative fluorescence of the amine groups on the APTES surface. (c) and (d): relative fluorescence image of the amine groups on the PLL surface.
shows the adsorption of the labelled DNA for three tested surfaces: PLL, APTES, and hydroxyl. The relative fluorescence intensity of the absorbed DNA on the PLL surface was slightly higher then on the APTES surface. The ratio of fluorescence between the drop of DNA and the background for the APTES surface is approximately 4, while this ratio is circa 6 for the PLL surface. In other words, the DNA adsorption is 50% higher on the PLL layer.
Figure 4. Fluorescent imaging of the adsorption of Alexa555 labelled DNA on different surfaces. The arrows indicate the direction in which the relative fluorescence was monitored. (a) and (b): relative fluorescence of DNA on the control surface with hydroxyl groups. (c) and (d): relative fluorescence of DNA on the APTES surface. (e) and (f): relative fluorescence of DNA on the PLL surface.
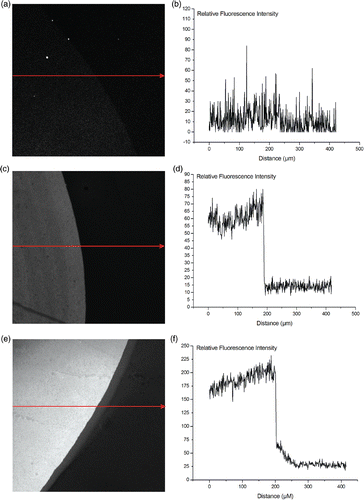
The control surface, displaying hydroxyl groups on the surface, only showed a very limited increase in fluorescence. The change in intensity of the PLL layer compared to the APTES layer is not as drastical as the difference in amine groups. This can be explained by the saturation level of DNA adsorption on the PLL layer. The already absorbed DNA strands will eventually prevent more DNA strands from binding due to steric hinder.
3.2. QCM measurements
The QCM technique is a commonly used method to characterise various surface chemistries based upon the mass adsorption on the surface.
The probe DNA (2 µM in phosphate buffer, pH7) was first absorbed onto the PLL layer. An excess amount of noncomplementary DNA of 50 bp was then added to visualise the nonspecific binding on the surface. The nonspecific binding on the PLL layers increases with the decreasing salt concentration in the solution. This can be explained by the fact that the DNA will be surrounded by a smaller amount of counterions, which allows them to bind to the large amount of amine groups on the PLL surface. At the concentration NaCl that was used in the FET experiments (50 mM, experiment shown below), the nonspecific adsorption seen in QCM was 7 Hz for 1 µM noncomplementary DNA. As can be seen in , the response for 250 nM complementary DNA in this condition was 11 Hz. Because the concentration of the noncomplementary DNA used in the experiment was four times higher than the highest concentration of complementary DNA that was applied in the experiments, we can expect a higher response on the sensor when applying the complementary DNA, compared to the injection of noncomplementary DNA.
Figure 5. QCM monitoring of DNA hybridisation in buffers with different salt concentrations for the 3 above presented surfaces and for different target DNA concentrations: PLL layers are represented by the full line, APTES by the striped line, and the hydroxyl surface with the dotted line. All buffers were 10 mM phoshate buffers, pH7. The DNA hybridisation was allowed for 30 min. Regeneration of the hybridised DNA was performed with 2.5 mM HCl for 5 min.
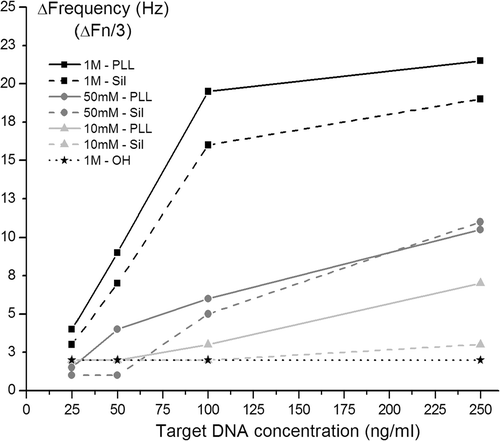
Different concentrations (25-50-100-250 nM) of the target DNA (mixture of 25 bp and 25 bp bis DNA) were used to optimise the DNA hybridisation. Using a mixture of the complementary DNA, we attempted to optimise the hybridisation. After all, we may assume that most of the DNA probe lays flat on the surface, and therefore, not one complementary 50 bp strand was chosen that binds the 3′ or the 5′ side, but two strands of 25 bp that bind on the 3′ and the 5′ strand of the probe DNA. Our assumption was that the hybridisation signal should be enhanced in this way. The hybridisation time was limited to 30 min for all conditions. Regeneration of the hybridised DNA was done by flowing a HCl solution (2.5 mM in water) over the sensor.
Different concentrations of salt were tested to determine the minimal salt concentration needed for DNA hybridisation to occur on this specific surface chemistry. shows the PLL layer to be the best layer for DNA hybridisation. Obviously, hybridisation is most effective at higher salt concentrations, but these experiments show that it is still possible to perform hybridisation in lower ionic strength soulutions. The control surface, with hydroxyl groups exposed to the DNA, did not show a significant DNA hybridisation.
3.3. Hybridisation detection with FG FETs
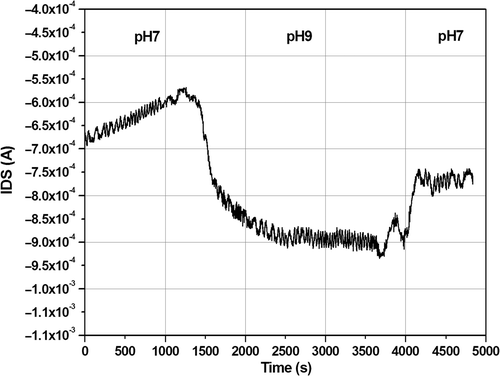
The developed surface chemistry has been transferred to the surface of the FG FET, which consists of the same Ta2O5 layer. At first, the transistor transfer characteristics have been recorded in buffer solutions in order to select the optimal operation conditions required for the DNA hybridisation experiments. Second, pH sensitivity experiments have been carried out to evaluate the pH sensitivity of the top Ta2O5 layer. The pH response of the device is shown in (the pH sensitivity of the PLL-functionalised device is 100 µA/pH unit).
Figure 6. pH response of the Ta2O5 FG FET device in flow conditions. The bias applied ont the transistors were V DS = V GS = −1 V.
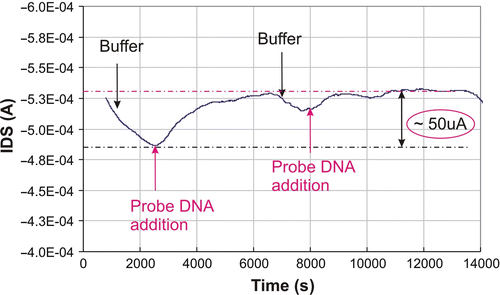
After stabilisation of the device in buffer solution, 2.5 µM of the 50 bp SH-DNA strand was allowed to absorb on the surface of the transducer by injecting the DNA into the flow cell from the top. As shown in , this adsorption caused an increase in the drain current (I DS) flowing the channel of the transducer. The shift in I ds could be explained by the strong electrostatic attraction of the negatively charged DNA strands by the positively charged PLL on the active area of the sensor. The DNA adsorption on the sensor surface generates a charge variation at the electrolyte/PLL interface, which, by field-effect, will induce the charge reorganisation in the semiconductor layer, in the close vicinity of the oxide/semiconductor interface. A further rinsing step (buffer is pumped through the flow cell) removes small amounts of the adsorbed DNA.
Figure 7. Adsorption of probe DNA (2.5 µM) in 10 mM phosphate buffer, pH7 on the surface of the FG FET. The biases are V DS = −2 V and V GS = −1.5 V. DNA was injected with a micropipette, rinsing was done by peristaltic pumping. The I DS shift caused by DNA addition is relatively large (∼50 µA).
In the hybridisation experiments, 2.5 µM from a mixture of 25 bp and 25 bp bis complementary strands was injected in the flow cell and allowed to absorb on the probe DNA already present on the surface of the sensor. The hybridisation buffer was a 10 mM phosphate buffer (pH = 7), containing 50 mM NaCl. The electrical response of the sensor is shown in . While rinsing with the same buffer for several minutes, some of the excess DNA (nonspecifically adsorbed) was removed from the sensor. After that, the same concentration was added again on the sensor surface to check whether more hybridisation can occur. The small current variation demonstrates that the majority of the probes present on the sensor surfaces were already hybridised. Again, afterwards, some of the DNA seems to be removed by the pumping during the following rinsing step.
Figure 8. Hybridisation response after injection of a mixture of 2.5 µM complementary target DNA on the surface. Injection of noncomplementary DNA did not show a significant response in the channel of the transistor. The hybridisation buffer was a 10 mM phosphate buffer, containing 50 mM NaCl. The bias was V DS = −2 V and V GS = −1.5 V.
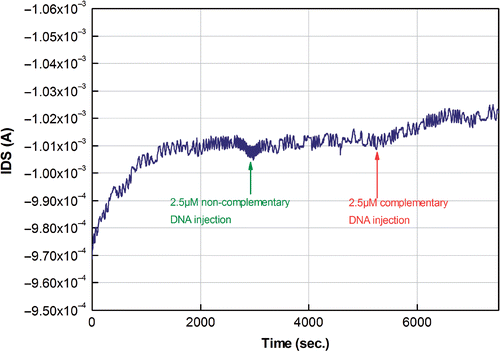
As a control, 2.5 µM of a noncomplementary DNA strand of 50 bp was injected in the flow cell and allowed to absorb on the sensor. This caused only a small response due to a limited amount nonspecifically adsorbed onto the PLL layer.
The time interval required to reach the signal saturation was ∼10 min, which is reasonable for hybridisation experiments. The reason for the efficient hybridisation occurring in low-ionic strength solutions might be the large positive charge brought by the PLL layer on the gate of the transducer. In this way, the negatively charged DNA strands are rapidly attracted at the surface despite the electrostatic repulsion between them.
4. Conclusions and outlook
Electronic DNA hybridisation detection is believed to be a very promising approach towards integrated lab-on-chip applications.
To increase the reliability of the DNA hybridisation experiments, we performed all experiments online. This means that the sensor was never removed from the flow cell set-up. Removing the sensor, for instance to do the hybridisation at high salt concentration, makes the measurement results more untrustworthy. When the hybridisation is done offline in different conditions (high salt) than the measurement condition (low salt), there is a chance that a considerable amount of the hybridised DNA will detach again because of the change in ionic strength. Moreover, variations in the electrical characterisation of the FET (caused by charge redistribution, memory effects, etc.) mean that the measurement outcome cannot be trusted.
Future experiments should be performed in order to investigate the detection limit of the sensor system, and the feasibility of DNA hybridisation detection in different ionic strength solutions. We used in-house fabricated FG transistors for availability reasons. This sort of gate architecture is responsible for instability effects in the online measurements, caused by the charging of this floating gate. For efficient online recordings, other FET architectures should be more promising, such as open-gate configurations. Also, the use of a reference FET on the same chip would improve the hybridisation set-up.
References
- Yeung , SW , Lee , TMH , Cai , H and Hsing , IM . 2006 . A DNA biochip for on-the-spot multiplexed pathogen identification . Nucleic Acids Res , 34 : e118
- Sun , H , Zhang , YY and Fung , YS . 2006 . Flow analysis coupled with PQC/DNA biosensor for assay of E-coli based on detecting DNA products from PCR amplification . Biosens. & Bioelect. , 22 : 506 – 512 .
- Liepold , P , Wieder , H , Hillebrandt , H , Friebel , A and Hartwich , G . 2005 . DNA-arrays with electrical detection: a label-free low cost technology for routine use in life sciences and diagnostics . Bioelectrochem. , 67 : 143 – 150 .
- Rodriguez-Mozaz , S , de Alda , MJL and Barcelo , D . 2006 . Biosensors as useful tools for environmental analysis and monitoring . Analyt. Bioanalyt. Chem. , 386 : 1025 – 104 .
- Heller , DA , Jeng , ES , Yeung , TK , Martinez , BM , Moll , AE , Gastala , JB and Strano , MS . 2006 . Optical detection of DNA conformational polymorphism on single-walled carbon nanotubes . Science , 311 : 508 – 511 .
- Wu , ZS , Jiang , JH , Fu , L , Shen , GL and Yu , RQ . 2006 . Optical detection of DNA hybridization based on fluorescence quenching of tagged oligonucleotide probes by gold nanoparticles . Analyt. Biochem. , 353 : 22 – 29 .
- Patolsky , F , Lichtenstein , A and Willner , I . 2001 . Detection of single-base DNA mutations by enzyme-amplified electronic transduction . Nature Biotechnol. , 19 : 253 – 257 .
- Shiigi , H , Tokonami , S , Yakabe , M and Nagaoka , T . 2005 . Label-free electronic detection of DNA-hybridization on nanogapped gold particle film, . J. Amer. Chem. Soc. , 127 : 3280 – 3281 .
- Kayyem , JF . 1999 . DNA diagnostics using electronic detection . Clin. Chem. , 45 : S12 – S12 .
- Berger , MF , Philippakis , AA , Quereshi , AM , He , FXS , Estep , PW and Bulyk , ML . 2006 . Compact, universal DNA microarrays to comprehensively determine transcription-factor binding site specificities . Nature Biotechnol. , 24 : 1429 – 1435 .
- Izquierdo , A and Luque de Castron , MD . 1995 . Ion-sensitive field-effect transistors and ion-selective electrodes as sensors in dynamic systems . Electroanal. , 6 : 505 – 519 .
- Bartic , C and Borghs , G . 2006 . Organic thin-film transistors as transducers for (bio) analytical applications . Analytical and Bioanalytical Chemistry , 384 : 354 – 365 .
- Oh , SJ , Hong , BJ , Choi , KY and Park , JW . 2006 . Surface modification for DNA and protein microarrays . OMICS , 10 : 327 – 343 .
- Poghossian , A , Cherstvy , A , Ingebrandt , S , Offenhausser , A and Schoning , MJ . 2005 . Possibilities and limitations of label-free detection of DNA hybridization with field-effect-based devices . Sensors and Actuat. B-Chem. , 111 : 470 – 480 .
- Fritz , J , Cooper , EB , Gaudet , S , Sorger , PK and Manalis , JR . 2002 . Electronic detection of DNA by its intrinsic molecular charge . Proc. National Acad. Sci. United States of Amer. , 99 : 14142 – 14146 .
- Vanhal , REG , Eijkel , JCT and Bergveld , P . 1995 . A novel description of ISFET sensitivity with the buffer capacity and double-layer capacitance as key parameters . Sensors and Actuat. B-Chem. , 24 : 201 – 205 .