1. Introduction
Graphics processing units (GPUs) have been used in the computer game and movie industries for several decades. Though initial applications of GPUs were exclusively for creation of computer graphics, GPUs have matured to a state where they can be applied for non-graphical purposes. The newer general purpose graphics processing units (GPGPUs) have received considerable attention in computational chemistry, notably as molecular dynamics simulations have exploited the vector and matrix processing capabilities of GPGPUs to improve the time to compute the per-frame trajectories of individual atoms in a system by orders of magnitude [Citation1,Citation2]. Another pharmaceutically relevant application of GPU technology has been to construct systems for analysis of high-content imaging systems, notably phenotypic cellular response to chemical perturbation. As images are matrices of values, leveraging matrix capabilities of GPUs has aided automatic image segmentation, annotation, and analysis.
Initial generations of GPUs required dedicated personnel to understand their architectures and accessibility. Recent developments in programming libraries have succeeded in abstracting away the technical details of individual GPU hardware. For drug discovery and design rooted in computational chemistry or machine learning, the advances in programming abstractions have now matured to the point where the interface to GPGPU hardware can be a launch pad to novel computational drug discovery efforts, rather than the bottleneck.
In parallel with GPGPU advancements, theoretical developments in the concept of auto-encoding have gained attention. In contrast to chemical descriptors engineered to describe specific aspects of compounds (e.g., indicating the presence or absence of certain functional groups or physicochemical features), auto-encoding seeks to automatically and dynamically generate a representation of molecular structure such that a minimalistic description of the compounds is achieved that yet still retains differentiability among individual compounds in a collection. This approach provides an alternative to strictly application-specific perspectives of molecular representation. Chemoinformatics can benefit from investigating an expanded breadth of different auto-encodings of molecules, accelerated by GPU technology.
The concept of using artificial neural networks (ANNs) for prediction of compound properties and drug discovery is well explored. Essentially, ANNs contain a collection of layers, where the outputs of one layer serve as the inputs to another layer, until an output layer which predicts a property is reached. Pairs of connected layers are represented by a matrix reflecting the weights of connections between the layers, making ANNs amenable to matrix operations. Since their inception, layer depth, connectivity, and computability have been constrained by the processing capabilities of traditional central processing units (CPUs).
GPGPU computing has recently alleviated these constraints, opening up the possibility of computing networks of considerably greater complexity and depth. The abstraction of patterns in data by use of increased-depth ANNs has rapidly gained attention in recent years under the banner name ‘deep learning’, and such ANNs are referred to as DNNs. The potential of DNNs to achieve abstractions in chemistry, coupled with the concept of auto-encoding, has delivered us to a stimulating point in computer-assisted drug discovery and design. What technical underpinnings of GPGPUs are enabling new computational research, and why should the field of drug discovery be interested in these advancements?
2. Accessing GPUs with applications to neural networks
Access to GPGPUs is provided via application programming interfaces (APIs), such as NVIDIA’s Compute Unified Device Architecture (CUDA), Apple/Khronos group’s OpenCL, and Microsoft’s Direct Compute. These interfaces enable the parallelization of specified functions (kernels) from existing programs in widely used programming languages including C++ and Fortran, and allow software to dispatch computations across GPU cores. An early adoption of GPGPU programming in the life sciences was the Folding@home project developed by researchers at Stanford University, initially created for distributed CPU computing but has since evolved such that GPUs contribute the majority of its compute power today [Citation3].
ANN/DNN calculations are parallelizable and often are formulated as successive matrix operations [Citation4]. To facilitate development of ANN methodologies and their applications, open-access software libraries, including Theano and Tensorflow, have been developed [Citation5,Citation6]. In combination with the ability to directly control a GPGPU via API, extra library functions efficiently allocate and manage the various types of memory on GPUs, whilst providing support for programming languages such as Python. The abstractions provided by high-level language support help teams focus on scientific concepts rather than implementation specifics. In practice, the application will dictate the direction of implementation, as there are different strengths between CPUs and GPUs ().
Table 1. A comparison of technical aspects of two recent GPU and CPU implementations.
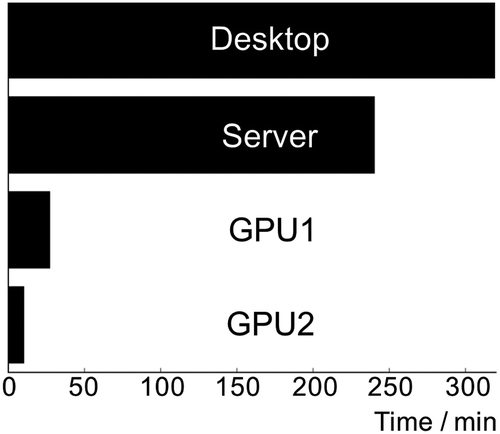
To consider how GPGPUs and their APIs could impact the field of drug discovery, we execute the task of training a fixed-topology neural network classifier on a set of 11,798 small molecules annotated as active (inhibitors) or inactive against coagulation factor Xa. Four configurations of CPU and CPU+GPU were tested for speed in determining the weights for the network layers divided into two convolution layers, a feedforward layer, and an output layer. Significant gain in speed was achieved with the two machines extending conventional CPU computing to also employ GPUs ().
Figure 1. Comparison of ANN compute times using CPU and CPU+GPU hardware. Hardware used: (Desktop) Dual 2.66 GHz 6-core Intel Xeon X5650 processors; (Server) Four 2.3 GHz 16-core AMD Opteron 6376 processors; (GPU1) Dual 2.4 GHz Intel Xeon E5-2620 processors and an ASUS GeForce GTX1080 STRIX graphics card; (GPU2) Four 3.6 GHz Intel Core i7 6850K processors and an ASUS GeForce GTX1080Ti STRIX graphics card.
3. GPUs and DNN
Having demonstrated that speedups can be achieved by investing in GPU technology, the next key aspect we consider is the set of ANN-based computational drug discovery workflows that become accessible (). The first approach considers the replacement or extension of well-established ‘shallow’ machine learning methods [Citation7]. Such an example is the DeepTox network for compound toxicity prediction by utilizing a DNN trained on molecular fingerprints. DeepTox won the Tox21 toxicity prediction challenge [Citation8].
Table 2. Directions and possibilities in DNN-based drug discovery.
A second approach, which we have alluded to above, employs auto-encoding with a sufficient DNN topology to fuel automated representation and abstraction. First applications include representation of molecular graphs and virtual screening, representation generation of compound SMILES strings, and estimation of quantum chemical potentials [Citation9–Citation11].
A third approach concerns training DNNs on input data encoding molecules with respect to different experimental and theoretical metrics (e.g. measured binding affinity and molecular weight), or data from different input sources (e.g. simultaneous molecular descriptor and microscopy image representations). This approach is reminiscent of ensemble methods and has generated expectations as demonstrated by Merck’s ligand classification machine learning challenge [Citation12].
Orthogonal to compound and related input data representation, the topology of the network and its resultant capabilities should be kept in mind. Specific topologies including long-short term memory networks (LSTMs) or generative adversarial networks (GANs) have the capability to generate new data according to a learned representation, and can therefore be employed for automated de novo molecule design [Citation9,Citation13,Citation14]. In a pioneering prospective application, a generative DNN autonomously generated novel potent RXR and PPAR receptor agonists [Citation15].
4. Expert opinion
While GPU computing is not new, it has been inaccessible to the public at large, and even major molecular dynamics projects have invested heavily in dedicated development efforts to tap GPU power. As generalization to GPU interfaces has advanced, ANNs, and DNNs in particular, have re-entered center stage in recent years. While some successes with ANNs and DNNs for drug discovery are being reported, a fundamental question exists about the topologies of the networks that lead to success or failure [Citation16]. Not only must basic parameters of layer depth and layer size be considered, but additionally how upstream layers are connected and potentially recombined in downstream layers, and how those topologies then lead to drug discovery and knowledge discovery must be addressed. With an intractable number of topologies possible, the GPGPU can help accelerate evaluation.
Then, the question of how compound representation and sufficiently descriptive topology are interrelated requires our attention. Is it possible that auto-encoding of chemical space or subspaces will generate representations that change or augment our view of pharmacologically active compounds? GPUs can accelerate generation of multiple and possibly complementary representations of drugs, which should ideally be human-accessible and interpretable via suitable man-machine interfaces. Further, given the fact that pairs of compounds when viewed as molecular graphs may have a difference of 10- or 100-fold activity when measured in a given biological context, it is a challenge to drive the representation generator in a way that not only incorporates this reality, but critically enables the exploitation of the learned representation for molecule design. The requirement that multiple distinct representations of compounds converge on the identification of an activity cliff has already been proposed [Citation17]. At this point, deep chemical knowledge is required to address the fundamental question of how to represent molecular structure for DNN. Molecular graphs only capture parts of reality, and it will be wise to consider multiple alternatives.
Irrespective of GPUs and APIs for a drug discovery project, it is essential to keep in mind that any ANN-based project will fail without suitable training data. Meticulous data curation is as important as ever. Therein, likely lies the greatest challenge for machine learning in drug discovery, which is to adapt these algorithms to heterogeneous, pharmaceutically relevant data. Novel, auto-generated molecular representations hold potential to augment our view of bioactive molecules by one-to-one mapping between representations and resulting properties. Multi-criteria optimization for compound generation and selection reflects the reality of pharmaceutical design, where methods such as Pareto curve analysis can identify the set of compounds such that no compound is more superior to another compound in all design objective criteria [Citation18,Citation19]. Deep network models are able to inherently capture multiple design criteria (e.g. by means of multitask and transfer learning) [Citation20]. We would expect answers as to their broad practical applicability from prospective molecular design campaigns.
Novel representations to aid in classification or regression property prediction have already been manifested in the idea of kernel method-based algorithms such as the support-vector machine (SVM), and GPU implementations of SVMs have been written. However, a big difference between kernel methods and the recent expectation on auto-encoded neural networks is the nature of manual piloting versus auto-piloting. Advanced GPU technology may in fact be game-changing in this regard, not only by leading to the necessary massive computational speedup, but by enabling us to perform studies that have previously been formulated merely as Gedankenexperiments. For the current applications of GPU-based DNNs in drug discovery we agree with Ekins [Citation21] in that more prospective testing is needed to evaluate the actual usefulness of this technology for the field. If DNNs can lead to encapsulation of molecular representations which correlate with heterogeneous, multi-criteria pharmaceutical data and can be subsequently applied for multi-objective design (whether machine-driven or manual), then DNN-driven virtual screening may become a formidable tool for accelerating drug discovery efforts. The next 5–10 years will bring answers and inspirations [Citation22].
Declaration of interest
G Schneider declares a potential financial conflict of interest in his role as life–science industry consultant and cofounder of inSili.com GmbH, Zurich. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed. Peer reviewers of this manuscript have no relevant financial or other relationships to disclose.
Acknowledgments
The authors thank Petra Schneider for technical support.
Additional information
Funding
References
- Korb O, Finn PW, Jones G. The cloud and other new computational methods to improve molecular modelling. Expert Opin Drug Discov. 2014;9:1121–1131.
- Martínez-Rosell G, Giorgino T, Harvey MJ, et al. Drug discovery and molecular dynamics: methods, applications and perspective beyond the second timescale. Curr Top Med Chem. 2017;17:2617–2625.
- Beberg AL, Ensign DL, Jayachandran G, et al. Folding@home: lessons from eight years of volunteer distributed computing. IEEE International Symposium on Parallel & Distributed Processing. 2009;1–8.
- Goodfellow I, Bengio Y, Courville A. Deep learning. Cambridge, MA: MIT Press; 2016.
- Theano Development Team. Theano: a Python framework for fast computation of mathematical expressions. arXiv e-prints 2016.
- Abadi M, Barham P, Chen J, et al. TensorFlow: a system for large-scale machine learning. 12th USENIX Symp Oper Syst Des Implement. 2016. DOI:10.1038/nn.3331
- Korotcov A, Tkachenko V, Russo DP, et al. Comparison of deep learning with multiple machine learning methods and metrics using diverse drug discovery data sets. Mol Pharm. 2017;14:4462–4475.
- Mayr A, Klambauer G, Unterthiner T, et al. DeepTox: toxicity prediction using deep learning. Front Environ Sci. 2016;3:1–15.
- Gupta A, Müller AT, Huisman BJH, et al. Generative recurrent networks for de novo drug design. Mol Inf. 2018;37:1700111.
- Schütt KT, Arbabzadah F, Chmiela S, et al. Quantum-chemical insights from deep tensor neural networks. Nat Commun. 2017;8:13890.
- Goh GB, Hodas NO, Vishnu A. Deep learning for computational chemistry. J Comput Chem. 2017;38:1291–1307.
- Ma J, Sheridan RP, Liaw A, et al. Deep neural nets as a method for quantitative structure–activity relationships. J Chem Inf Model. 2015;55:263–274.
- Olivecrona M, Blaschke T, Engkvist O, et al. Molecular de-novo design through deep reinforcement learning. J Cheminform. 2017;9:48.
- Blaschke T, Olivecrona M, Engkvist O, et al. Application of generative autoencoder in de novo molecular design. Mol Inf. 2018;37:1700123.
- Merk D, Friedrich L, Grisoni F, et al. De novo design of bioactive small molecules by artificial intelligence. Mol Inf. 2018;37:1700153.
- Gawehn E, Hiss JA, Schneider G. Deep learning in drug discovery. Mol Inf. 2016;35:3–14.
- Peltason L, Bajorath J. SAR index: quantifying the nature of structure-activity relationships. J Med Chem. 2007;50:5571–5578.
- Nicolaou CA, Kannas C, Loizidou E. Multi-objective optimization methods in de novo drug design. Mini Rev Med Chem. 2012;12:979–987.
- Schneider G, Fechner U. Computer-based de novo design of drug-like molecules. Nat Rev Drug Discov. 2005;4:649–663.
- Simões RS, Maltarollo VG, Oliveira PR, et al. Transfer and multi-task learning in QSAR modeling: advances and challenges. Front Pharmacol. 2018;9:74.
- Ekins S. The next era: deep learning in pharmaceutical research. Pharm Res. 2016;33:2594–2603.
- Schneider G. Automating drug discovery. Nat Rev Drug Discov. 2018;17:97–113.
