ABSTRACT
Introduction: The use of semantic web technologies to aid drug discovery has gained momentum over recent years. Researchers in this domain have realized that semantic web technologies are key to dealing with the high levels of data for drug discovery. These technologies enable us to represent the data in a formal, structured, interoperable and comparable way, and to tease out undiscovered links between drug data (be it identifying new drug-targets or relevant compounds, or links between specific drugs and diseases).
Areas covered: This review focuses on explaining how semantic web technologies are being used to aid advances in drug discovery. The main types of semantic web technologies are explained, outlining how they work and how they can be used in the drug discovery process, with a consideration of how the use of these technologies has progressed from their initial usage.
Expert opinion: The increased availability of shared semantic resources (tools, data and importantly the communities) have enabled the application of semantic web technologies to facilitate semantic (context dependent) search across multiple data sources, which can be used by machine learning to produce better predictions by exploiting the semantic links in knowledge graphs and linked datasets.
1. Introduction
Drug discovery is a complex and long-term scientific investigation involving interdisciplinary research methods coupled with large heterogeneous datasets [Citation1,Citation2]. Recognition of drug targets [Citation3] and the identification of new compounds that can be used in drugs against specific diseases are major aspects of drug discovery [Citation4]. All of these processes are complex within their own right and involve analyzing large quantities of data [Citation5]. With such a substantial volume of data relating to different areas of drug discovery [Citation6,Citation7] it is unsurprising that frequently scientists have research questions that cannot be addressed using a single dataset and require the integration of multiple sources to obtain the answers they need. The drug discovery process has become increasingly more reliant on creating and using computational methods to find innovative ways of managing, curating and integrating these datasets to ensure that researchers have access to the relevant knowledge they need [Citation4].
There has been a wave of computation-based innovation around creating tools to aid drug discovery [Citation8,Citation9,Citation10] and many of these tools utilize semantic web technologies. This is both due to data management capabilities of the semantic tools coupled with the researcher’s recognition that purely integrating sources together is not enough. Semantic web technologies are needed to facilitate interoperability, to interpret the information in the correct context [Citation11], to make use of inferencing and the richer data structures to provide a superior level of knowledge management [Citation12,Citation13] and the ability to both search and mine large integrated datasets [Citation14], and effectively utilize ‘Big Data’ and Artificial Intelligence and Machine Learning [Citation15–Citation17].
2. What is the semantic web? the current state of the technologies
The conceptualization of the web broke down some of the barriers to entry into data driven fields (such as drug discovery), as it provided a key information channel to reaching public data [Citation18], and this combined with movements towards making scientific datasets open provided access to vital resources to conduct research in this area. However, these resources could only be of limited use due to a lack of data standards and interoperability across different data resources and domains.
The Semantic Web was conceptualized by Tim Berners-Lee [Citation19] with the main goal of creating the underlying technology to support a `Web of Data’ that enables machines to better interpret information and provide a set of common interoperable data formats to be used across different platforms [Citation20]. In order to achieve this aim, several core formats were created to facilitate a richer way of modelling, characterizing and querying data.
2.1. Knowledge representation
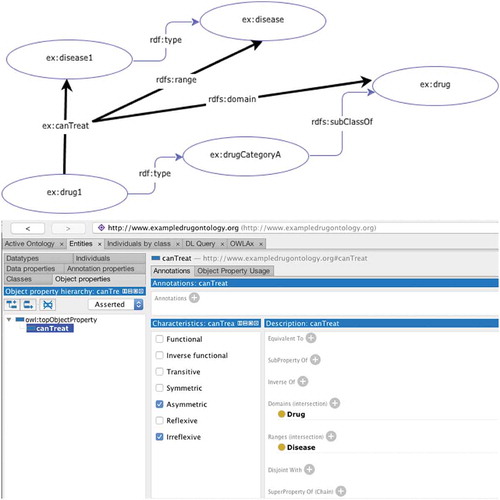
RDF (Resource Description Framework) is the semantic web linked data format [Citation21]. A graph model is used to store data in triples (subject → predicate → object) whereby the predicate defines the relationship between the subject and object, thus permitting almost any dataset to be broken down into these triples (as demonstrated in ) [Citation20].
Using this linked data format, data relating to drug discovery can be formalised and linked in a standard interoperable format within a knowledge base that provides a much richer description. Below is a textual and graph example of an RDF triple illustrating the relationship between a drug that can treat a disease.
Subject: http://www.exampledrugontology.org/drugs/drug1 (the URI for the drug in question)
Predicate: http://www.exampledrugontology.org/terms/#cantreat (The URI that provides the term ‘can treat’ which links together drugs and diseases
Object: (the URI for the disease in question)
It is possible to produce code to export data from a relational database model into RDF using the Relational Database Model (RDB) to RDF Mapping Language (R2RML) [Citation22], which facilitates defining customizable mappings between relational datasets to RDF. The creation of this language enabled a substantially faster method of converting large datasets into RDF as opposed to being required to manually make the conversion.
RDF supports storing data in a linked graph format, but is not sufficient by itself to represent the required domain knowledge that provides the context and meaning behind the data. In order to achieve this, ontologies need to be created. Ontologies are vocabularies or dictionaries for the semantic web, which provide a formal definition of the common terms used within a specific domain; essentially describing the hierarchy of classes used to define the relationships and restrictions of different concepts within the ontology. This technology allows reusable terms to be built up for use within other systems employing the same terminology [Citation23]. Ontologies are typically written in the Web Ontology Language (OWL) [Citation24] due to its expressive capabilities but can also be written in the simple ontology language RDF(S) [Citation25]. These two vocabularies are similar, but OWL has a much larger vocabulary and stronger syntax than RDF. With RDF(S) one can define classes, properties and restrict the type of the subject (domain) and object (range) linked together by a property (as demonstrated in ).
With OWL however, it is possible to define the semantic relationships. As can be shown in , the domain and range of a property can be set, which restricts which types of classes this property can be used to link. However, with RDF(S) that’s as far as the description capabilities go, whereas OWL has description logic capabilities, which have been enhanced since the latest version of OWL (OWL2) was formalized [Citation26]. Classes and object properties (that define relationships between classes) can be given qualified cardinality restrictions; data range restrictions, and a wider range of defined properties. In addition to using description logic, rules can also be made in ontologies using other languages such as SWRL (Semantic Web Rule Language) [Citation27]. Ontologies are useful in drug discovery because they facilitate a formal description of concept relations in a set domain [Citation28]. There have been many drug discovery ontologies developed over the last decade and these are starting to be utilized in further applications; as researchers are starting to extend these ontologies, their usefulness is advancing. For example, work has been undertaken to extend the Gene Ontology to facilitate describing subcellular structure concepts in the extracelluar RNA (exRNA) domain [Citation29] which enabled a simplified process to annotate and query exRNA data as there was a standardized shared set of terms and relationships for this domain.
2.2. Semantic annotation
RDFa (Resource Description Framework in Attributes), is a W3C recommendation that adds a new set of extensions at attribute level to HTML, XHTML and certain XML document types to allow rich metadata to be embedded within web pages. In addition to RDFa, two other lightweight formats have emerged for semantic metadata, and these are the JavaScript Object Notation for Linked Data (JSON-LD) [Citation30], and more recently Microdata [Citation31], a HTML-extension specification that defines HTML attributes to embed machine-readable data in HTML documents (although this is even more lightweight and less expressive than RDFa or JSON-LD). This means that HTML documents containing drug discovery information can have semantic metadata embedded within them, this enabling search engines like Google to return more accurate, comprehensive results for users seeking answers to drug discovery questions. Using the same terms as , below are examples of using RDFa and JSON-LD for semantic annotations in this domain ( and ):
Box 1. Simple example of using rdfa for semantic annotations in drug discovery.
Box 2. Simple example of using JSON-LD for semantic annotations in drug discovery.
2.3. Semantic queries for search
SPARQL (Simple Protocol and RDF Query Language) is the query language for RDF and enables linked data to be retrieved using significantly more complex queries than using single datasets with an unlinked structure. Once drug discovery data has been put into a linked data format using domain specific ontologies, SPARQL can be used to make complex queries over these combined datasets to pull out new, previously unexplored, connections. An increasing amount of work has been done to create new semantic knowledgebases which can be searched across using SPARQL, and to combine the use of SPARQL with other technologies such as text mining and similarity scoring to enable smarter semantic searches [Citation32]
2.4. Rules, reasoning & inferencing
Another facet of the semantic web toolkit is the rule creation, and reasoning and inferencing capabilities. As touched on above, the rules regarding the relationships defined in ontologies can be made more complex using either the inbuilt description logic capabilities of OWL (which were extended in OWL2) or by using one of the rule-based languages developed for the Semantic Web. The data that exists within the drug discovery domain is multi-faceted and in addition to having many different concepts and relationships to represent, many different rulesets and modelling techniques are used on this data when conducting research. There are several rule languages that have been created as part of the semantic web toolkit to facilitate the inclusion of mathematical expressions and more complex rule-based constraints. The Semantic Web Rule Language (SWRL) [Citation27] was created to extend OWL axioms to include Horn-like rules [Citation33] by combining the OWL DL and OWL Lite versions of OWL with the Unary/Binary Rule ML version of the Rule Markup Language. SPIN [Citation34] (SPARQL Inferencing Notation) and its successor SHACL [Citation35] (Shapes Constraint Language) were created as an RDF syntax to represent SPARQL complex constraints by specifying inferencing rules. Thus far the available research only demonstrates the use of SWRL in drug discovery, however SHACL is a relatively new language (proposed in 2017) and given the increase in semantic search applications over the last few years, it is feasible that future applications could begin to include the use of these languages.
2.5. Increasing use of semantic web technologies
These core technologies were developed nearly 20 years ago, and still make up the backbone of the semantic web. However, in the last decade, improvements have been made to advance these core technologies [Citation26,Citation36,Citation37], and as semantic web technologies are being further recognized as necessary enablers to support broader technologies [Citation38] further innovations that utilize these technologies are being made across multiple domains, including drug discovery.
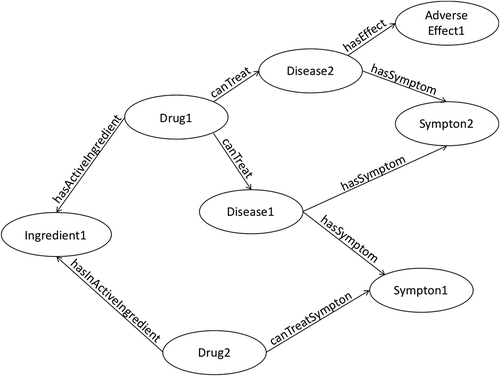
As the use of semantic technologies has become more commonplace, other novel tools, which are based on semantics, have started to emerge. A classic example of this is the knowledge graph. This in itself is not an overtly ‘new’ advancement, as Google introduced the concept in 2012 [Citation39]. However, similarly to the core technologies themselves, this concept has taken a while to gain traction, and it is only in the last three years or so that drug discovery applications that utilize knowledge graphs have started to emerge. Knowledge graphs are essentially graph network structures to describe real world entities and their relationships through the combination of linked data and ontologies. These graphs of linked entities are powerful structures and enhancing these has led to the creation of new applications that exploit the networks of links to apply advanced learning techniques on the data. provides a simplified example of how drug discovery data could be represented in a knowledge graph.
Using semantic web technologies has the potential to be of great value, however there is a certain time cost involved in formalizing the ontologies and putting datasets into a linked data form such that they can be linked, searched and used in conjunction with other technologies such as machine learning. Semantic web technologies have been around since 2001, and slow and steady work began to use these technologies to aid and advance drug discovery shortly after. However, in recent years this work has gained momentum and a new wave of innovation in semantic web-based tools for drug discovery have come to the forefront. This paper will consider the recent advancements that have been made in this area, both with regards to creating and advancing ontologies and in creating new semantic web-based tools for drug discovery. An overview of the progress that has been made in recent years will be presented, and an expert opinion on where the advances have been made and how they are aiding drug discovery further will be given.
3. Advances in semantic web tools for drug discovery
In 2012 the latest version of OWL (OWL2) was created, and since then advancements have also been made in the latest versions of RDF, OWL and SPARQL, to enhance their capabilities and offer additional features. Moreover, there has been a steady uptake of these technologies in the area of drug discovery. Initial research in this domain when the Semantic Web was first conceptualized proposed that semantic web technologies would support ‘flexible, extensible and evolvable knowledge transfer and reuse of scientific data’ [Citation12]. As semantic web technologies have developed, researchers have periodically evaluated their use within the drug discovery domain.
Borkum and Frey in 2014 [Citation40] expressed the value of linked datasets and ontologies to aid chemical research, giving examples of general vocabularies such as SKOS [Citation41] and Dublin Core [Citation42], in addition to describing their own work to create some controlled vocabularies for specific chemical terminology. This line of thinking has gained traction in recent years as many more linked repositories, datasets and ontologies are being curated and created in the area of drug discovery, and in general the uptake of semantic web technologies to aid managing and leveraging higher value from drug discovery data has increased significantly. An example of one of these semantic repositories is Chem2BioRDF [Citation43], a cross domain chemical biology resource which contains aggregated data from multiple chemogeonomics repositories that have been cross linked into BioRDF (a platform for querying biological data in RDF). This repository was created to enable users to make specific chemical/biological queries, which as the authors demonstrate, could have strong uses in areas such as multiple pathway inhibition and adverse drug reaction pathway mapping.
In 2010 Chen and Xie [Citation44] cited creating linked networks of open data, ad hoc collaboration to improve pipeline productivity, and efficient semantic data mining as the main benefits of using semantic web technology at the time of their article. These remain valid points, although since these articles the amount of available semantic resources has increased, meaning that there have been notable advancements in semantic web tools for drug discovery. There have been endeavors to create new ontologies and make extensions to those that already exist to improve their coverage in certain areas [Citation45–Citation47]. Since these ontologies have gained traction and both increased and improved, new platforms have been created to make enhanced predictive systems that utilize the semantic links afforded by ontologies and network graphs in conjunction with other computational techniques such as network analysis and supervised learning approaches. Since Google introduced knowledge graphs in 2012, their use in drug discovery has increased and advanced the previous data mining capabilities.
In 2015 Machado et al [Citation18] reviewed 11 different applications from the last decade that make use of semantic web technologies in drug discovery, illustrating that the main areas that these technologies can be utilized is through data and knowledge representation, and resource mapping and integration services. Companies have also started to realize the potential of using these technologies in their drug discovery work. OntoText (a semantic web company) agreed a partnership with DrugBank (a bioinformatics and cheminformatics drug data resource) in 2018 [Citation48] to provide DrugBank’s database in an RDF format, thus providing a new semantic resource for researchers.
All three reviews of semantic web technologies within the scientific domain had a strong focus on the linking of data through the use of RDF and Ontologies [Citation18,Citation40,Citation44]. Machado et al illustrated that out of the 11 semantic drug discovery applications that they surveyed, seven of these used controlled vocabularies in their systems; demonstrating that this was a common method of using these technologies. However, one of the key benefits of producing this type of linked data is the enhanced semantic search capability that it affords. As new semantic knowledge bases, ontologies, and knowledge graphs have started to emerge, as has new and improved semantic search. Semantic applications in drug discovery have evolved past purely using knowledgebases to become systems that use intelligent semantic search techniques, and use other technologies such as machine learning to exploit the semantic links in the data.
The following sections detail the work done to make more drug discovery data available in a linked format, to create and extend ontologies and to create community-driven knowledge bases for semantically annotating data with drug discovery information. Following this, the more notable advances of new semantic search and machine learning based applications will be described, including providing exemplars of applications from both industry and academia that utilize these technologies.
3.1. Enhanced knowledge representation for drug discovery
Much of today’s scientific knowledge and data has progressed from being kept in the heads of scientists, through being written down in books, to being put into a digital form that (depending on its openness and format) can be searched and consumed much faster than on paper. However, just because something is in a digital form, does not mean it is necessarily going to be useful. Humans process data to turn it into meaningful information, which enables them to build knowledge; the semantic web provides these capabilities by offering the mechanisms to markup data in a linked format, using ontologies to add context and meaning. These techniques have been used in drug discovery to improve the knowledge representation of drug discovery data, thus providing enhanced knowledge bases that can be used to explore undiscovered links and be used in conjunction with other technologies.
In order to create well-formed, consistent, semantic drug discovery data, there is a need to create and maintain ontologies that define the many different concepts and relationships within this space [Citation49]. Much fundamental groundwork has been done to create these ontologies over the last 15 years [Citation50,Citation51], creating both specific ontologies for a certain area (e.g. The Drug Ontology [Citation52], or the Vaccine Ontology [Citation53]) and also the underlying ontologies that define core scientific terms such as the Basic Formal Ontology [Citation54]). This reuse and extension of ontologies is vitally important, as it promotes a standard for certain terms and means that similar terminology is not needlessly replicated. Some of the ontologies detailed above have been created with these ideals in mind, however there is still an issue of interoperability between some ontologies in the biomedical domain, as noted by Shen and Lee [Citation55].
With regards to using existing ontologies, one of the most well-used set of ontologies in the drug discovery space – and the one that has spawned the most advancements as a result of its creation – is the Gene Ontology Group (GO) [Citation28,Citation56]. The Gene Ontology consortium is a collaborative project that aims to provide consistent descriptions of gene products across different databases, and includes controlled vocabulary terms for cellular components, biological processes and molecular functions. There have been a number of projects either to extend the functionality of the Gene Ontology, or to create new semantic platforms that utilize it. Foulger et al [Citation57] undertook some work to extend the Gene Ontology with new classes to model processes relating to virus-host interactions by describing microbial and viral gene products. The creation of the ontology in itself was not explicitly for drug discovery, but nonetheless it has many potential future applications. Drug discovery is a complex process that involves the consideration and combination of many different types of data including viral receptors. A potential future application of this ontology could be to construct complex search queries across datasets annotated using these ontology terms to locate all of the viral receptors for a specific cell type. Therefore these new ontologies are vastly important to the overall advancement of drug discovery.
Another area of vital importance to drug discovery is drug-target identification. Research has suggested that only a very small amount of potential druggable targets have been extensively studied within this space, leaving a great deal of unexplored territory to be investigated [Citation46]. With such a vast amount of data and potential untapped knowledge, it is unsurprising that there have been many endeavors to improve this process and facilitate the prediction and identification of relevant drug-target pairs using semantic web technologies. Lin et al [Citation46] recently created the Drug Target Ontology (DTO) which provides a formal semantic model for druggable targets. This was created to facilitate integrating, navigating and analyzing drug discovery data using consistent standards and classifications, thus facilitating a more efficient well-defined method of utilizing this data to its greatest potential.
These new ontologies have been created in part due to the lack of standards to formally and consistently represent aspects of drug discovery. The BioAssay Ontology (BAO) [Citation58] was created by Visser et al [Citation59] to provide a common vocabulary for describing drug and probe screening assays and results. Subsequent work has been conducted over the last five years to enhance and extend this ontology [Citation45]. This was to both integrate with further external ontologies, and to modularize the ontology such that the computing costs of the description logic capabilities are spread across different aspects of the ontology, thus reducing these costs where possible depending on which module of the ontology is used. Additionally, after the ontology was improved it was utilized in a new application in 2016 by Clark et al [Citation60] to create a platform to allow domain experts who do not necessarily possess ontology expertise to semantically annotate assay protocols. This is an example of how building these underlying data models facilitates advances in the technology for users within the drug discovery domain.
These new and extended ontologies facilitate advancement in drug discovery, enabling new computational methods to be created that require the rich, interoperable, clearly defined data terms made available by using a combination of RDF and OWL [Citation50].
3.2. Community knowledge bases and semantic annotation for drug discovery
In addition to small groups of researchers creating ontologies, there have been a number of community efforts to create shared schemas for a wide range of disciplines to improve the amount of structured data on the web. These vocabularies are then used to markup data on websites or in emails to create structured data with semantic annotations to define their context and meaning. Some of these community knowledge bases are domain-specific, such as WikiPathways [Citation61] whereas others are more general purpose such as WikiData [Citation62] which has over 52 Million data items, and schema.org which is used by over 10 million websites [Citation63]. These large vocabularies have grown to contain data items relevant to drug discovery (for example schema.org has a specific health-lifesci extension to its core vocabulary, and WikiData contains some data items related to drug discovery) and now that this initial work has been put in, use cases for these vocabularies are starting to emerge.
Earlier in 2018 Xin et al [Citation44] published their work on using JSON-LD with their API, BioThings [Citation45], which has three APIs to provide annotation for Genes, Variants and Chemicals/Drugs. This was to provide interoperability and cross linking between the different APIs and provide more complex query capabilities; thus allowing links and answers to be extracted from multiple datasets. These authors postulate that there is still more to be done to standardize URIs for biological concepts, and recognizes schema.org as one of the entities that provides some of the vocabularies for the standard concepts. Ekins et al [Citation64]identified WikiData among others as a data source for information on the Zika virus, illustrating that multiple data sources would need to be linked together in order to achieve the proposed research on this virus.
Crowdsourcing these vocabularies is a way to bring domain experts (both technical and scientific) together and divide the workload. Furthermore this to an extent decouples the process of adding semantic metadata from creating the vocabularies themselves, and defining how they can be used to markup data. Once these processes are put in place this removes some of the expertise barrier that comes with the use of semantic web technologies. Typically, creating ontologies and semantic data requires the use of both domain and technical experts [Citation65] to capture the appropriate domain knowledge, and also to use it correctly to create the necessary semantic resources. However, once these have been put in place applications can be made that mean domain experts that do not necessarily have the required level of technical experience can still partake in these activities. Tang et al [Citation66] have worked on a community effort to create a knowledge base of drug-target interactions whereby users can help each other annotate their data, working with data approvers who will ensure that this process is performed correctly.
3.3. Semantic search engines for drug discovery
As detailed in Section 2, one of the outcomes of the increase in the availability of linked drug discovery data, and the creation and use of ontologies to markup this data, is being able to design semantic search engines to exploit this data [Citation67]. Semantic search is an advanced technique that facilitates more complex queries, and enables searching via concepts and concept relationships rather than purely by text matching [Citation68]. Sometimes just having the data is not enough, it needs to be combined in ways that are useful to scientists; undiscovered links need to be found. Furthermore, there are many scientific questions that cannot be addressed with a single data source, but instead require multiple sources. In this vein, Elkseth et al [Citation69], took 37 datasets and marked them up semantically into a linked data format, then normalized them such that they used the same concepts across the different datasets; thus enabling the researchers to build KnittingTools, a semantic search engine for these datasets. Similarly, Djokic-Petrovic et al [Citation69] created a web based application called PIBAS FedSPARQL which uses semantic technologies to enable researchers to search across multiple chemical, biological and pharmacological datasets.
Along similar lines, Open PHACTS (Open Pharmacological Concept Triple Store) [Citation70], was created as part of the Open PHACTS project to answer complex questions in drug discovery [Citation71]. It essentially provides an application layer on top of the Open PHACTS API, such that users can search for drug discovery data without needing to understand the RDF data behind it, or even understand SPARQL [Citation72]. This platform combines a number of the well-used datasets in drug discovery (including the Gene Ontology and ChEBI – Chemical Entities of Biological Interest Ontology [Citation73]), such that it is possible to see the relationships between compounds, targets, pathways etc. This work has subsequently been extended by Miller et al [Citation74] to use WikiPathways data [Citation61] (which is a database of biological pathways). López-Massaguer [Citation75] used data extracted from Open PHACTS in their work to obtain QSAR-ready compounds using semantic web technologies.
These are clear examples of how the previous work undertaken to create semantic web foundations for drug discovery has led to new innovations. The initial ontologies needed to be created to put the data into a linked format using the correct concepts, and the linked data needed to be available before it could be linked with other data. Creating these resources enabled these datasets and ontologies that each had their own individual uses to be combined to provide a new layer of knowledge.
3.4. Rules, reasoning, inferencing & learning for drug discovery
Building new ontologies and vocabularies has paved the way for new applications to be created that can make use of not only the semantic links facilitated by the combination of OWL and RDF, but also the rules and reasoning aspects of ontologies. Rule-based reasoning and Inferencing is a powerful aspect of the Semantic Web toolkit. By defining hierarchies, relationships, and the nature of these relationships between concepts, reasoning can be performed to infer where relationships could exist, and what classes different data concepts belong to. Furthermore, researchers have started to use machine learning techniques in conjunction with semantic web technologies, as their training algorithms and neural networks can exploit the semantic links and relationships between the data concepts to find new patterns and make better predictions [Citation76].
The Gene Ontology has been used to facilitate several drug-target identification and classification systems. For example, Chen et al [Citation77] used GO in conjunction with KEGG (Kyoto Encyclopedia of Genes and Genomes, a drug-target classification system) to score and identify drug-target-based classes and determine which GO terms are the most important for this process.
As with any data-driven domain, time is a key element to conducting investigations and making important discoveries; thus making data and literature mining a vital part of this process [Citation78–Citation80]. Semantic web technologies can be used to enhance and improve the data mining process [Citation81], as data can be mined based on concepts and relationships rather than just text [Citation82]. In this vein, the knowledge graph has been found to be a very useful aspect of semantic data mining [Citation83]. Knowledge graphs can be constructed to provide the relevant graph pathways between related concepts to yield new information and links. Malas et al [Citation84] highlighted how semantic knowledge graphs can be useful in identifying drug candidates, by pointing out that they can connect different databases and reflect relationships between different concepts, in this instance, gene pathways and diseases. In a similar vein, Sang et al [Citation85] developed a knowledge graph based on literature mining, in order to discovery new drug therapies from literature. A knowledge graph was created with extractions from PubMed abstracts, which was then used by SemTyP (Semantic Path Type – an intelligent drug discovery method) to exploit the semantic paths to discovery new drug therapies.
These projects illustrate the power of linking and reasoning in ontologies that have helped to further drug discovery capabilities. They also demonstrate how different learning techniques can be used in conjunction with semantic web technologies to exploit those much-needed links between data concepts to make new discoveries.
3.5. Semantic applications in drug discovery
In addition to the work conducted to extend these resources and technologies, a number of new semantic web applications for drug discovery have been created, both in industry and academia. New systems are being built that demonstrate creating and using ontologies for drug discovery and using semantic search and machine learning techniques for drug discovery. Furthermore, scientific companies are also making partnership agreements with semantic web companies to enable them to use semantic capabilities with their data and products. As noted in the Laboratory Informatics Guide 2019: Semantic search and metadata alongside other software approaches are being increasingly used to bridge the gap between traditional methods and the new data-driven research methods that are being used today.
Passi et al [Citation86] created the drug repurposing platform RepTB, which uses molecular function correlations among known drug-target pairs to enable predictions about new novel drug-target pairings that could be repurposed for tuberculosis (TB). To facilitate this platform, a new Gene Ontology Network was created (based on the Molecular Function Ontology – part of the GO set) and Network Based Inference (NBI) [Citation87] was used to predict which existing drugs could potentially be repurposed to combat TB by identifying new drug-target interactions and computing their association scores. Whilst NBI in itself is not a semantic technology, research has shown that the semantic links between the data can significantly improve the prediction capabilities of supervised learning models, as demonstrated by [Citation88]. In a similar vein, Olier et al [Citation89] made use of ontologies and semantic datasets in their QSAR (Quantitative Structure Activity Relationships) learning algorithm, illustrating that their techniques could improve performance in these areas by up to 13%.
Han et al [Citation9], harnessed the power of ontology inferencing combined with network analysis to identify new immune-relevant drug-target genes for Alzheimer’s Disease (AD). This was working on the theory that due to the strong relation of AD to the immune system, there were potentially undiscovered immune-relevant AD drug-target genes that could help combat the disease. Han et al created an ontology of AD drug, gene, SNOP, disease, and haplotype data, and reasoned over it to identify which target genes to further analyze. Enrichment analysis was then performed on these targets, making use of the Gene, PANTHER and Reactome ontologies, in addition to eight other databases. In addition to using description logic-based rules in OWL, work has also been undertaken to use SWRL (Semantic Web Rule Language). Herreo-Zao et al [Citation90] created DINTO (Drug-Drug Interactions Technology) to formally represent different types of drug-drug interactions (DDIs) using SWRL rules to infer different types of DDIs. By using this rule-based inferencing technique, the researchers were able to demonstrate that DDIs and their mechanisms could be inferred on a much larger scale than with previous knowledge bases.
Vitrana [Citation91] are now using semantic and ontology enabled search in their HiLIT Platform (which delivers specialized clinical and healthcare information management services) [Citation92]. BioMax – a semantic search company, partnered with Royal DSM (a company working in health, nutrition and materials) to provide them with a sophisticated AI-Semantic search platform which uses AI augmented search algorithms over their semantic network. Zheng et al [Citation93] recently reported successfully using ontologies to help identify factors in Schizophrenia and NuMedi [Citation94] announced their adoption of semantic web technologies to accelerate drug discovery in complex and rare diseases [Citation95].
These endeavors and applications illustrate the new wave of innovation in semantic web tools for drug discovery. Industry and academia alike have demonstrated an increased uptake in the use of semantic web technologies in this domain, both through the efforts that have been made to create new drug discovery semantic resources (knowledgebases, platforms and ontologies), and also through the ways they have progressed to use these technologies.
4. Expert opinion
There is a circular issue in using semantic web technologies to aid in data-driven spaces such as drug discovery. Very powerful applications can be made that harness semantic capabilities and use them in conjunction with other technologies to make predictions and identify new links in drug discovery. However, in order to do this, the underlying standards and formal descriptions of the data are required, which involves first creating and then curating the linked datasets and ontologies. Therefore, whilst these technologies have been in existence for nearly two decades, it is only recently that real advancements in terms of utilizing novel computational methods within drug discovery have emerged.
Much of the initial work in the drug discovery space was to produce ontologies and create and integrate linked datasets related to this domain. Furthermore, some of these ontologies (such as the Basic Formal Ontology (BFO) [Citation35]) were not intended directly for drug discovery but were created for general purpose representation of scientific data; and many of the specifically developed drug discovery ontologies have built upon this early work. These underlying ontologies may not have seemed like overt advancements at the time of creation, but the overall set of ontologies and integrated linked datasets that now exist for drug discovery have paved the way for some real advancement in what semantic tools can do to aid in this space. Furthermore, researchers have been taking steps to enhance and extend existing ontologies with necessary information, and reuse ontologies where possible, rather than creating new ones that replicate the same information. For example the Ontology of Adverse Vaccine Events [Citation96] was created to extend the original Ontology of Adverse Events [Citation97] and the Vaccine Ontology [Citation53]; demonstrating that the initial work done to create these ontologies helped enable further development of a formal vocabulary to describe these terms. There have also been many community endeavors to provide open schemas and vocabularies with which to embed semantic metadata within HTML documents, thus improving the general representation of data on existing websites. Due to these efforts, much of the recent work undertaken in this area has been to create novel applications that make use of semantic links, logical capabilities and inferencing and reasoning capabilities of these technologies for drug discovery (as illustrated in ).
Figure 4. A variant on the Semantic layer cake [Citation98] illustrating the added value of using Semantic web technologies for drug discovery.
![Figure 4. A variant on the Semantic layer cake [Citation98] illustrating the added value of using Semantic web technologies for drug discovery.](/cms/asset/20e06d32-071f-495f-b59f-674a686dcffb/iedc_a_1586880_f0004_oc.jpg)
The capabilities of the core semantic web technologies, and indeed researchers’ understanding of how best to get value out of their use, have been advanced in the past six years. Newer, improved versions of RDF, OWL and SPARQL have all been developed with more advanced capabilities: more clearly defined data types, better ways of representing and structuring the rules around the data, and improved methods for querying linked data. Furthermore, new formats have become available to embed metadata within existing websites, and of course the knowledge graph has been conceptualized, providing new ways of creating and exploiting semantic links in and between datasets. However, whilst these advancements have been useful, the authors assert that the main ‘advancement’ has been human-driven rather than technology-driven. The core concepts of the Semantic Web have been around for nearly two decades, and it has taken that time for its value to be appreciated on a larger scale, both as a whole, and for specific endeavors such as drug discovery. The drug discovery community has now reached a state where a lot of useful groundwork has been put in, both to make specific ontologies and drug discovery knowledge bases, and also for the overall semantic web community to crowdsource large-scale knowledge bases.
This paper has shown a shift towards using more powerful semantic searching capabilities for drug discovery (based on the plethora of new semantic resources available) and the work conducted by industry and academia to produce superior semantic search tools. It has also become clear that researchers and companies are starting to realize the power of harnessing machine learning capabilities alongside semantic web technologies to exploit linked datasets of drug discovery information.
Further work should be undertaken, not only to extend these endeavors now that there is a solid starting point, but also to exploit the nature of semantic web technologies in conjunction with other technologies, such as machine learning, to create intelligent semantic systems to aid and advance drug discovery. This can only be achieved if researchers continue to work together to ensure that the necessary data and ontologies are made available in the areas of drug discovery that need to be addressed. When considering problems that need to be solved in drug discovery, researchers should consider what semantic data sources are available to them, and exploit their links in their applications. If the data sources that they require are not available, efforts should be made to create them. It is important to note that, where possible, reuse of ontologies should be encouraged. When creating new vocabularies, researchers should consider extending existing resources rather than duplicating information. To this end, the ontology communities that facilitate collaborative ontology and vocabulary development have great potential to help in this area.
There are a number of community endeavours such as schema.org that allow the public to request new items for vocabularies and query what vocabularies to use, and how best to use them. These resources should be utilized, ensuring a strong interdisciplinary collaboration between technical and domain experts. The work to date that has been considered in this article illustrates the importance of the human factor in using semantic web technologies for drug discovery. The core technologies of linked data, annotations and ontologies are key enablers for facilitating semantic search and enhancing the capabilities of other intelligent technologies. As demonstrated in this article, these technologies will be of considerable value to the drug discovery process. If the momentum of semantic web development in this domain continues along its current trajectory there is the potential to make a real difference to drug discovery.
Article highlights
The uptake of using semantic web technologies to aid and advance drug discovery has increased in both academia and industry over recent years.
Semantic web applications for drug discovery began as predominantly knowledge bases, and they have evolved to include powerful semantic search capabilities, and have made use of other technologies such as machine learning to exploit the links in semantic datasets to make better predictions.
A significant driver in creating new semantic web applications for drug discovery has been the increased availability of shared semantic resources (tools, data and the community). Creating (and extending existing) semantic drug discovery resources enabled datasets and ontologies that each had their own individual uses to be combined to provide a new layer of knowledge.
The drug discovery community has now reached a state where a lot of useful groundwork has been put in, both to make specific ontologies and drug discovery knowledge bases, and also for the overall semantic web community to crowdsource large-scale knowledge bases. This means that there is now a potential for exploitation; as evidenced by the uptake of commercial semantic drug discovery applications.
Further work should be undertaken, not only to extend these endeavors now that there is a solid starting point, but also to exploit the nature of semantic web technologies in conjunction with other technologies, such as machine learning, to create intelligent semantic systems to aid and advance drug discovery.
Declaration of interest
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
Reviewer disclosures
Peer reviewers on this manuscript have no relevant financial or other relationships to disclose.
Acknowledgements
The authors would like to thank Dr Colin Bird for his invaluable help with editing and reviewing the manuscript.
Additional information
Funding
References
- Taglang G, Jackson DB. Use of “big data” in drug discovery and clinical trials. Gynecol Oncol. 2016;141:17–23.
- Lu P, Bevan DR, Leber A, et al. Computer-aided drug discovery. In: Bassaganya-Riera J, editor. Accelerated path to cures [Internet]. Cham: Springer International Publishing; 2018 [cited 2018 Dec 17]. p. 7–24. Available from: http://link.springer.com/10.1007/978-3-319-73238-1_2
- Ramsay RR, Popovic-Nikolic MR, Nikolic K, et al. A perspective on multi-target drug discovery and design for complex diseases. Clin Transl Med [Internet]. 2018 [cited 2018 Dec 17];7. Available from: https://clintransmed.springeropen.com/articles/10.1186/s40169-017-0181-2
- Katsila T, Spyroulias GA, Patrinos GP, et al. Computational approaches in target identification and drug discovery. Comput Struct Biotechnol J. 2016;14:177–184.
- Jing Y, Bian Y, Hu Z, et al. Deep learning for drug design: an artificial intelligence paradigm for drug discovery in the big data era. Aaps J [Internet]. 2018 [cited 2018 Dec 17];20. Available from: http://link.springer.com/10.1208/s12248-018-0210-0
- Brown N, Cambruzzi J, Cox PJ, et al. Chapter five - big data in drug discovery. In: Witty DR, Cox B, editors. Progress in Medicinal Chemistry [Internet]: Elsevier; 2018 [cited 2018 Nov 7]. p. 277–356. Available from: http://www.sciencedirect.com/science/article/pii/S0079646817300243
- Chen B, Butte A. Leveraging big data to transform target selection and drug discovery. Clin Pharmacol Ther. 2015;99:285–297.
- Constandt H. Disruptive change by linked data and semantic technologies in healthcare and life sciences. Drug Discovery World. 2013;14:72–76.
- Han Z-J, Xue -W-W, Tao L, et al. Identification of novel immune-relevant drug target genes for Alzheimer’s Disease by combining ontology inference with network analysis. CNS Neurosci Ther [Internet]. 2018 [cited 2018 Nov 7]. Available from: https://onlinelibrary.wiley.com/doi/full/10.1111/cns.13051.
- Tetko IV. Internet in drug design and discovery. Open Appl Inf J. 2008;2:18–21.
- Katsila T, Matsoukas M-T. How far have we come with contextual data integration in drug discovery? Expert Opin Drug Discov. 2018;13:791–794.
- Neumann EK, Miller E, Wilbanks J. What the semantic web could do for the life sciences. Drug Discovery Today: BIOSILICO. 2004;2:228–236.
- Frey JG. The value of the Semantic web in the laboratory. Drug Discov Today. 2009;14:552–561.
- Wild DJ, Ding Y, Sheth AP, et al. Systems chemical biology and the Semantic web: what they mean for the future of drug discovery research. Drug Discov Today. 2012;17:469–474.
- Chen H, Engkvist O, Wang Y, et al. The rise of deep learning in drug discovery. Drug Discov Today. 2018;23:1241–1250.
- Lavecchia A. Machine-learning approaches in drug discovery: methods and applications. Drug Discov Today. 2015;20:318–331.
- Zhang L, Tan J, Han D, et al. From machine learning to deep learning: progress in machine intelligence for rational drug discovery. Drug Discov Today. 2017;22:1680–1685.
- Machado CM, Rebholz-Schuhmann D, Freitas AT, et al. The semantic web in translational medicine: current applications and future directions. Brief Bioinform. 2015;16:89–103.
- Berners-Lee T, Hendler J, Lassila O. The Semantic web. Sci Am. 2001;284:34–43.
- Shadbolt N, Berners-Lee T, Hall W. The Semantic web revisited. IEEE Intell Syst. 2006;21:96–101.
- RDF - Semantic Web Standards [Internet]. [cited 2018 Nov 22]. Available from: https://www.w3.org/RDF/.
- R2RML: RDB to RDF Mapping Language [Internet]. [cited 2019 Feb 14]. Available from: https://www.w3.org/TR/r2rml/.
- Wilkinson MD, Dumontier M, Aalbersberg IJ, et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci Data. 2016;3:160018.
- OWL - Semantic Web Standards [Internet]. [cited 2018 Nov 22]. Available from: https://www.w3.org/OWL/.
- RDFS - Semantic Web Standards [Internet]. [cited 2018 Nov 22]. Available from: https://www.w3.org/2001/sw/wiki/RDFS.
- OWL 2 Web Ontology Language Document Overview. 2nd ed [Internet]. [cited 2018 Nov 22. Available from. https://www.w3.org/TR/owl2-overview/
- SWRL: A Semantic Web Rule Language Combining OWL and RuleML [Internet]. [cited 2018 Dec 18]. Available from: https://www.w3.org/Submission/SWRL/.
- Chichester C, Digles D, Siebes R, et al. Drug discovery FAQs: workflows for answering multidomain drug discovery questions. Drug Discov Today. 2015;20:399–405.
- Li K, Rodosthenous RS, Kashanchi F, et al. Advances, challenges, and opportunities in extracellular RNA biology: insights from the NIH exRNA Strategic Workshop. JCI Insight [Internet]. [cited 2019 Feb 10];3. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5928855/
- JSON-LD - JSON for Linking Data [Internet]. [cited 2018 Dec 7]. Available from: https://json-ld.org/.
- HTML Microdata [Internet]. [cited 2018 Dec 7]. Available from: https://www.w3.org/TR/microdata/.
- Djokic-Petrovic M, Cvjetkovic V, Yang J, et al. PIBAS FedSPARQL: a web-based platform for integration and exploration of bioinformatics datasets. J Biomed Semantics [Internet]. 2017 [cited 2019 Feb 15];8. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5607505/
- Horn Rules Semantics - W3C RIF-WG Wiki [Internet]. [cited 2019 Feb 14]. Available from: https://www.w3.org/2005/rules/wg/wiki/Horn_Rules_Semantics.html.
- SPIN - SPARQL Syntax [Internet]. [cited 2019 Feb 14]. Available from: https://www.w3.org/Submission/2011/SUBM-spin-sparql-20110222/.
- Shapes Constraint Language (SHACL) [Internet]. [cited 2019 Feb 14]. Available from: https://www.w3.org/TR/shacl/.
- RDF 1.1 Concepts and Abstract Syntax [Internet]. [cited 2018 Nov 22]. Available from: https://www.w3.org/TR/rdf11-concepts/.
- SPARQL 1.1 Overview [Internet]. [cited 2018 Nov 22]. Available from: https://www.w3.org/TR/2013/REC-sparql11-overview-20130321/.
- Semantic Web and Semantic Technology Trends in 2018 [Internet]. DATAVERSITY. 2017 [cited 2018 Nov 22]. Available from: http://www.dataversity.net/semantic-technology-semantic-web-trends-2018/.
- Introducing the Knowledge Graph: things, not strings [Internet]. Official google blog. [cited 2018 Nov 23]. Available from: https://googleblog.blogspot.com/2012/05/introducing-knowledge-graph-things-not.html.
- Borkum MI, Frey JG. Usage and applications of Semantic web techniques and technologies to support chemistry research. J Cheminform. 2014;6:18.
- SKOS Simple Knowledge Organization System Primer [Internet]. [cited 2018 Nov 22]. Available from: https://www.w3.org/TR/skos-primer/.
- Weibel S. The Dublin core: a simple content description model for electronic resources. Bull Am Soc Inf Sci Technol. 1997;24:9–11.
- Chen B, Dong X, Jiao D, et al. Chem2Bio2RDF: a semantic framework for linking and data mining chemogenomic and systems chemical biology data. BMC Bioinformatics. 2010;11:255.
- Chen H, Xie G. The use of web ontology languages and other semantic web tools in drug discovery. Expert Opin Drug Discov. 2010;5:413–423.
- Abeyruwan S, Vempati UD, Küçük-McGinty H, et al. Evolving bioassay ontology (BAO): modularization, integration and applications. J Biomed Semantics. 2014;5:S5.
- Lin Y, Mehta S, Küçük-McGinty H, et al. Drug target ontology to classify and integrate drug discovery data. J Biomed Semantics Internet. 2017;8:50. Available from: https://jbiomedsem.biomedcentral.com/articles/10.1186/s13326-017-0161-x.
- Sahoo SS, Valdez J, Kim M, et al. ProvCaRe: characterizing scientific reproducibility of biomedical research studies using semantic provenance metadata. Int J Med Inform [Internet]. 2018. [cited 2018 Nov 7]. Available from: http://www.sciencedirect.com/science/article/pii/S1386505618302697
- Ontotext’s Partnership with DrugBank to Open New Perspectives in Pharma Research [Internet]. Ontotext. [cited 2019 Feb 12]. Available from: https://www.ontotext.com/company/news/ontotexts-partnership-drugbank/.
- Gardner SP. Ontologies in drug discovery. Drug Discov Today. 2005;2:235–240.
- Vazquez-Naya J M, Martinez-Romero M, Porto-Pazos A B, et al. Ontologies of drug discovery and design for neurology, cardiology and oncology. Curr Pharm Des. 2010;16:2724–2736.
- Tao C, He Y, Arabandi SA. 2013 workshop: vaccine and drug ontology studies (VDOS 2013). J Biomed Semantics. 2014;5:16.
- Hanna J, Joseph E, Brochhausen M, et al. Building a drug ontology based on RxNorm and other sources. J Biomed Semantics. 2013;4:44.
- He Y, Cowell L, Diehl AD, et al. VO: vaccine Ontology. Nat Precedings [Internet]. 2009. [cited 2019 Feb 4]. Available from: http://precedings.nature.com/documents/3553/version/1
- Press TM. Building ontologies with basic formal ontology [Internet]. The MIT Press; [ cited 2019 Feb 4]. Available from: http://mitpress.mit.edu/books/building-ontologies-basic-formal-ontology
- Shen F, Lee Y. Knowledge discovery from biomedical ontologies in cross domains. PLOS ONE. 2016;11:e0160005.
- Gene Ontology: tool for the unification of biology. Nature Genetics [Internet]. [cited 2019 Feb 4]. Available from: https://www.nature.com/articles/ng0500_25.
- Foulger RE, Osumi-Sutherland D, McIntosh BK, et al. Representing virus-host interactions and other multi-organism processes in the Gene Ontology. BMC Microbiol [Internet]. 2015 [cited 2018 Nov 7];15. Available from: http://www.biomedcentral.com/1471-2180/15/146
- BioAssay Ontology; The BioAssay Ontology (BAO) describes chemical biology screening assays and their results including high-throughput screening (HTS) data for the purpose of categorizing assays and data analysis.BioAssay Ontology; The BioAssay Ontology (BAO) describes chemical biology screening assays and their results including high-throughput screening (HTS) data for the purpose of categorizing assays and data analysis [Internet]. [cited 2018 Nov 22]. Available from: http://bioassayontology.org/.
- Visser U, Abeyruwan S, Vempati U, et al. BioAssay Ontology (BAO): a semantic description of bioassays and high-throughput screening results. BMC Bioinformatics. 2011;12:257.
- Clark AM, Litterman NK, Kranz JE, et al. BioAssay templates for the semantic web. PeerJ Comput Sci. 2016;2:e61.
- Slenter DN, Kutmon M, Hanspers K, et al. WikiPathways: a multifaceted pathway database bridging metabolomics to other omics research. Nucleic Acids Res. 2018;46:D661–D667.
- Vrandečić D, Krötzsch M. Wikidata: a free collaborative knowledgebase. Commun ACM. 2014;57:78–85.
- Guha RV, Brickley D, Macbeth S. Schema.org: evolution of structured data on the web. Commun ACM. 2016;59:44–51.
- Ekins S, Mietchen D, Coffee M, et al. Open drug discovery for the Zika virus. F1000Res [Internet]. 2016 [cited 2018 Dec 7];5. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4841202/.
- Marshall LJ, Austin CP, Casey W, et al. Recommendations toward a human pathway-based approach to disease research. Drug Discov Today. 2018;23:1824–1832.
- Tang J, Tanoli Z-R, Ravikumar B, et al. Drug target commons: a community effort to build a consensus knowledge base for drug-target interactions. Cell Chem Biol. 2018;25:224–229.e2.
- Wild D Making semantics work in drug discovery [Internet]. [cited 2018 Dec 18]. Available from: http://scimaps.org/exhibit/images/130325/overview-semantics-wild.pdf.
- Semantic search (invited talk) - IEEE Conference Publication [Internet]. [cited 2018 Dec 12]. Available from: https://ieeexplore.ieee.org/abstract/document/8258348.
- Ekseth OK, Meyer JC, Hvasshovd SO A new database for drug discovery through application of data-integration and semantics. 2018 IEEE 12th International Conference on Semantic Computing (ICSC), Laguna Hills, CA; 2018. p. 403–410.
- Gray AJG, Groth P, Loizou A, et al. Applying linked data approaches to pharmacology: architectural decisions and implementation. Semant Web. 2014;5:101–113.
- Williams AJ, Harland L, Groth P, et al. Open PHACTS: semantic interoperability for drug discovery. Drug Discov Today. 2012;17:1188–1198.
- Dunlop I, Ramgolam R, Pettifer S, et al. Open PHACTS explorer bringing the web to the semantic web. 2013.
- Degtyarenko K, de Matos P, Ennis M, et al. ChEBI: a database and ontology for chemical entities of biological interest. Nucleic Acids Res. 2008;36:D344–D350.
- Miller RA, Woollard P, Willighagen EL, et al. Explicit interaction information from WikiPathways in RDF facilitates drug discovery in the Open PHACTS Discovery Platform. F1000 Res. 2018;7:75.
- López-Massaguer O, Sanz F, Pastor M. An automated tool for obtaining QSAR-ready series of compounds using semantic web technologies. Bioinformatics. 2018;34:131–133.
- Data-driven drug development. Scientific computing world [Internet]. [cited 2019 Feb 12]. Available from: https://www.scientific-computing.com/lig2019/data-driven-drug-development.
- Chen L, Chu C, Lu J, et al. Gene ontology and KEGG pathway enrichment analysis of a drug target-based classification system. PLOS ONE. 2015;10:e0126492.
- Xu J, Hagler A. Chemoinformatics and drug discovery. Molecules. 2002;7:566–600.
- Ferrero E, Agarwal P. Connecting genetics and gene expression data for target prioritisation and drug repositioning. BioData Min. 2018;11:7.
- Andronis C, Sharma A, Virvilis V, et al. Literature mining, ontologies and information visualization for drug repurposing. Brief Bioinform. 2011;12:357–368.
- Chen B, Ding Y, Wild DJ. Assessing drug target association using semantic linked data. PLoS Comput Biol. 2012;8:e1002574.
- Ristoski P, Paulheim H. Semantic Web in data mining and knowledge discovery: A comprehensive survey. J Web Semant. 2016;36:1–22.
- Paulheim H, Ristoski P, Mitichkin E, et al. Data Mining with Background Knowledge from the Web:14.
- Malas TB, Kudrin R, Starikov S, et al. Semantic Knowledge Graph Network Features for Drug Repurposing. 2017.
- Sang S, Yang Z, Wang L, et al. SemaTyP: a knowledge graph based literature mining method for drug discovery. BMC Bioinformatics. 2018;19:193.
- Passi A, Rajput NK, Wild DJ, et al. RepTB: a gene ontology based drug repurposing approach for tuberculosis. J Cheminform. 2018;10:24.
- Cheng F, Liu C, Jiang J, et al. Prediction of drug-target interactions and drug repositioning via network-based inference. PLoS Comput Biol. 2012;8:e1002503.
- Fu G, Ding Y, Seal A, et al. Predicting drug target interactions using meta-path-based semantic network analysis. BMC Bioinformatics [Internet]. 2016 [cited 2018 Nov 7];17] Available from: http://bmcbioinformatics.biomedcentral.com/articles/10.1186/s12859-016-1005-x
- Olier I, Sadawi N, Bickerton GR, et al. Meta-QSAR: a large-scale application of meta-learning to drug design and discovery. Mach Learn. 2018;107:285–311.
- Herrero-Zazo M, Segura-Bedmar I, Hastings J, et al. DINTO: using OWL ontologies and SWRL rules to infer drug–drug interactions and their mechanisms. J Chem Inf Model. 2015;55:1698–1707.
- HiLIT | Lifesciences & Healthcare Platform | Vitrana [Internet]. [cited 2019 Feb 12]. Available from: http://www.vitrana.com/products/hilit/.
- Analytics C Clarivate analytics launches life science alliances program to accelerate innovation [Internet]. [cited 2019 Feb 12]. Available from: https://www.prnewswire.com/in/news-releases/clarivate-analytics-launches-life-science-alliances-program-to-accelerate-innovation-893047805.html.
- Zheng P, Zeng B, Liu M, et al. The gut microbiome from patients with schizophrenia modulates the glutamate-glutamine-GABA cycle and schizophrenia-relevant behaviors in mice. Sci Adv. 2019;5:eaau8317.
- Hu N Home [Internet]. NuMedii. [cited 2019 Feb 12]. Available from: http://numedii.com/.
- Dastgheib S, Webb C, Duan Q, et al. Accelerating Drug Discovery in Rare and Complex Diseases:2.
- Marcos E, Zhao B, He Y. The ontology of vaccine adverse events (OVAE) and its usage in representing and analyzing adverse events associated with US-licensed human vaccines. J Biomed Semantics. 2013;4:40.
- OAE: The Ontology of Adverse Events | Journal of Biomedical Semantics | Full Text [Internet]. [cited 2018 Dec 12]. Available from: https://jbiomedsem.biomedcentral.com/articles/10.1186/2041-1480-5-29.
- Semantic Web, and Other Technologies to Watch: January 2007 (1) [Internet]. [cited 2018 Dec 11]. Available from: https://www.w3.org/2007/Talks/0130-sb-W3CTechSemWeb/#(24).