ABSTRACT
Introduction
The potential of virtual reality (VR) to contribute to drug design and development has been recognized for many years. A recent advance is to use VR not only to visualize and interact with molecules, but also to interact with molecular dynamics simulations ‘on the fly’ (interactive molecular dynamics in VR, IMD-VR), which is useful for flexible docking and examining binding processes and conformational changes.
Areas Covered
The authors use the term ‘interactive VR’ to refer to software where interactivity is an inherent part of the user VR experience e.g. in making structural modifications or interacting with a physically rigorous molecular dynamics (MD) simulation, as opposed to using VR controllers to rotate and translate the molecule for enhanced visualization. Here, they describe these methods and their application to problems relevant to drug discovery, highlighting the possibilities that they offer in this arena.
Expert opinion
The ease of viewing and manipulating molecular structures and dynamics, using accessible VR hardware, and the ability to modify structures on the fly (e.g. adding or deleting atoms) – and for groups of researchers to work together in the same virtual environment – makes modern interactive VR a valuable tool to add to the armory of drug design and development methods.
1. Introduction
Human perception, intuition, creativity, and expertise are central to computer-aided drug discovery and design (CADD) [Citation1–5]. This is likely to remain so for the foreseeable future, despite recent significant and rapid advances in machine learning and artificial intelligence. The three-dimensional (3D) structure of molecules is essential to their function. An increasing aim of drug designs is to ‘escape from flatland’, i.e. to design and develop molecules with more complex 3D structure, which are found higher up the drug development pathway and tend to exhibit better solubility [Citation6] and lower toxicity [Citation7]. The human mind has evolved to visualize and understand 3D space, and to interact with and manipulate objects in this type of environment. In psychologist and computer scientist J. C. R. Licklider’s 1960 essay ‘Man-computer symbiosis’ [Citation8], he postulates that humans and computers will develop a symbiotic relationship, where the different strengths will complement each other, e.g. human spatial reasoning and computer speed and accuracy. Virtual reality (VR) provides a means of bringing together human intuition with computational power and resources, and can be an effective interface between them.
Many CADD applications that involve molecular models allow modification of structures, and display structures or simulations of 3D biomolecules via two-dimensional (2D) interfaces such as a monitor, using controls such as a mouse, making it difficult to fully utilize human 3D intelligence. VR provides a platform for 3D visualization of complex biomolecular structures and their dynamics. With the addition of interactivity via using human hands and fine motor control to directly manipulate objects, VR provides ways to interact, such as modifying structures ‘on the fly’ (e.g. chemical changes) or directing simulations toward a solution visible to a human. Interactive VR is intuitive to use and enables humans to focus on the areas of the drug discovery process that benefit from human knowledge and perception, e.g. chemical intuition, visualizing chirality, predicting how proteins and ligands fit together, the effects of a conformational change, or which alterations to the chemical structure might improve affinity or specificity. This has significant potential tospeed drug design and development.
VR is the use of artificial sense stimuli and can extend to manual interaction with a computer program to ‘trick’ the human mind into both perceiving a virtual world and feeling embodied in that world [Citation9]. It can allow for the addition of extra modalities, such as depth, touch, and sound, to enhance human interaction with computer software. To be fully immersed in VR is to feel disconnected from real-world stimuli, and to feel completely engaged with the simulated world [Citation10]. Although VR has been around for many years [Citation11–16], recent improvements to hardware and software have now made VR a technology that is ready for widespread scientific use. Advances include low cost, high resolution, fast refresh screens (for visualizing VR e.g. through lenses in a VR headset), fast graphics processing units (GPUs), high-level 3D graphics engines, and scientific VR software. Some examples of using VR in science and engineering for 3D visualization are: virtual restoration of archeological finds [Citation17]; virtual exploration and cartography [Citation18]; viewing the sea bed [Citation19]; safety training in chemical manufacturing [Citation20]; sport psychology [Citation21]; telepresence for clinicians [Citation22]; teaching anatomy [Citation23,Citation24]; investigating the molecular structures related to the SARS-CoV-2 virus [Citation25] to name a few.
The promise of VR in drug discovery has been recognized for many years [Citation26–28], and is now beginninig to be realised. VR offers several potential benefits over traditional molecular visualization tools, and over traditional interfaces for interacting with biomolecular simulations and molecular modeling. Firstly, VR allows the researcher to visualize drug molecules and their macromolecular targets in full 3D, which allows more detailed perception and deeper understanding of these complex systems, and so can informs design and modification of ligands in the process of structure-based design and development [Citation29,Citation30]. Secondly, VR allows interaction with molecules, primarily through VR controllers. The controllers can be thought of as a kind of ‘virtual pair of hands,’ allowing the user to grasp parts of a molecule or molecules as easily as if they were tangible, real-world objects. Thirdly, recent developments of interactive VR permit users to interact with a running molecular dynamics (MD) simulation the atomic level [Citation31], allowing them to manipulate the system, modifying its structure and interactions ‘on the fly’ (). Finally, some VR software allows multiple users to occupy the same virtual space [Citation30,Citation31] for collaboration and, e.g. for teaching (collaborative VR). In other words, modern VR technology (hardware and software) is not merely an update with better graphics: the addition of molecular motion via MD simulations, the ability to directly manipulate those simulations, to work together virtually, and to create and modify molecular structure from within the program turns VR from a visualization method to a research tool in its own right.
Figure 1. Interactivity for a protein-ligand system in the VR software Narupa IMD [Citation31,Citation32]. A) A protein-drug system (influenza neuraminidase complexed with the neuraminidase inhibitor oseltamivir) shown in the interactive molecular dynamics in virtual reality (IMD-VR) software Narupa IMD, where all atoms are rendered with the default ball and stick renderer. The VR controllers are also rendered in VR, where the orange circle represents the point of interaction with the controller (like a cursor for a mouse). B) Rendering of all atoms changed from ball and stick to lines and a different color scheme. C) The protein is here rendered in the ‘cartoon’ renderer and colored in rainbow, where red represents the N-terminal of the protein and violet represents the C-terminal. The drug is here rendered in the ‘cpm’ renderer and with atomistic colors to differentiate it from the protein. D) By grabbing single atoms of the drug molecule using each VR controller, the user can apply a force to coax the drug out of the active site. The force from the VR controllers is shown by the two light yellow sine waves between the controller and the drug molecule. E) The drug has now started to dissociate from the protein because of the interaction by the user in VR. F) A view of a VR user in real life, where the monitor is showing what the user sees in VR.
![Figure 1. Interactivity for a protein-ligand system in the VR software Narupa IMD [Citation31,Citation32]. A) A protein-drug system (influenza neuraminidase complexed with the neuraminidase inhibitor oseltamivir) shown in the interactive molecular dynamics in virtual reality (IMD-VR) software Narupa IMD, where all atoms are rendered with the default ball and stick renderer. The VR controllers are also rendered in VR, where the orange circle represents the point of interaction with the controller (like a cursor for a mouse). B) Rendering of all atoms changed from ball and stick to lines and a different color scheme. C) The protein is here rendered in the ‘cartoon’ renderer and colored in rainbow, where red represents the N-terminal of the protein and violet represents the C-terminal. The drug is here rendered in the ‘cpm’ renderer and with atomistic colors to differentiate it from the protein. D) By grabbing single atoms of the drug molecule using each VR controller, the user can apply a force to coax the drug out of the active site. The force from the VR controllers is shown by the two light yellow sine waves between the controller and the drug molecule. E) The drug has now started to dissociate from the protein because of the interaction by the user in VR. F) A view of a VR user in real life, where the monitor is showing what the user sees in VR.](/cms/asset/1618e02c-a5c8-484d-b8a3-2ff95c78460e/iedc_a_2079632_f0001_oc.jpg)
We use the phrase ‘interactive VR’ to refer to programs that allow the user to interact with and change structure of a molecular system, i.e. not programs where interactivity is limited to using the controllers to rotate, zoom, and translate the 3D structure to improve visualization. In this review, we discuss current VR hardware and some relevant VR software for structure-based drug design. We give examples of how interactive VR is currently being used to visualize and manipulate biomolecules, addressing features that are useful in terms of CADD. This is a rapidly developing field, and we highlight some pertinent recent applications, noting that this is not an exhaustive list of all VR-based activities in drug discovery; it provides a snapshot of capabilities of some current hardware and software.
2. VR hardware and software
There are many different types of VR hardware currently available, offering varying levels of immersion and interactivity. Low-cost solutions such as cardboard viewers that hold a smartphone in front of the user’s eyes offer a low-cost way of experiencing VR [Citation32–34], however, the field of view is restricted in this case, and there is limited interactivity. At the expensive end of the spectrum lies the Cave Automatic Virtual Environment (CAVE), a room-size virtual environment first developed 30 years ago that tracks the user via CAVE controllers and glasses [Citation35] (there are also state of the art VR systems that track the user’s entire body from head to feet such as Vicon Origin [Citation36], but these are primarily used for entertainment applications so are not discussed here). A CAVE consists of multiple screens that project the simulated space onto either 4 or 6 walls. The user(s) experience(s) VR through stereoscopic LCD shutter glasses that display a 3D image and can also interact using CAVE compatible wireless controllers [Citation37]. CAVE set-ups have been used for drug discovery in the past, however they are expensive, costing hundreds of thousands of dollars for a high-end set-up [Citation38,Citation39], and can have problems with immersion if the glasses do not cover the peripheral vision as well as problems with ventilation and claustrophobia [Citation40]. Head-mounted displays (HMD), such as the Oculus Quest 2, HTC Vive Pro, and the Valve Index, are generally used with the VR software discussed in this review. The advantages of using HMDs are that: (i) they are now relatively cheap (between $300-$1,600 USD at the time of writing for all the necessary VR equipment, not including the computer) [Citation41–43]; (ii) they are portable and lightweight; (iii) users experience a wide field of view (ranging between 90 and 130 degrees in the horizontal dimension); (iv) they have good spatiotemporal resolution; and (v) the experience can be fully immersive.
Typically, HMDs allow either 3- or 6- degrees of freedom (DoF). HMDs with 3-DoF allow tracking of rotational motion about the x, y, and z axes (known as pitch, yaw, and roll), but do not track translational movement. Examples of 3-DoF HMDs include the Oculus Go, Samsung Gear VR, and Google Daydream. This is the simplest kind of tracking and is achieved by sensors built into the headset. HMDs that allow 6-DoF track both rotational and translational motion in the x, y, and z directions. 6-DoF HMDs may require external trackers positioned within the room, while the newer HMDs, such as the Oculus Quest 2 and Oculus Rift S, are capable of 6-DoF through sensors and image processing within the headset. This kind of tracking approach seems to be the most common with the latest HMDs, suggesting this is where the industry is heading. The advantage of utilizing 6-DoF in VR is that the user can exploit their full range of motion to explore the simulated environment, by walking, crouching, and bending to achieve the best view.
There are several articles that discuss and compare the specifications of the most common HMDs in detail, including resolution, refresh rate, and pixel density [Citation44,Citation45]. For biomolecular modeling and manipulation, a commodity HMD is often the most affordable and practical solution; current examples at the time of writing include the HTC Vive Pro [Citation46], Valve Index [Citation47], and Oculus Quest 2 [Citation48]. Facebook recently renamed itself to Meta [Citation49], after the ‘metaverse’ (first coined in the novel Snow Crash [Citation50] as an online VR world accessible and manipulable by users), highlighting an emphasis on their VR range, Oculus. Valve [Citation47] owns the gaming platform Steam™ and is therefore likely to maintain their headset line so as to be able to continue with their core business, and HTC has been in the area for a long time having made collaborations with large corporations. All three of these headsets are reasonable choices, however we recommend that the user checks whether their HMD is compatible with the interactive VR program they wish to use. Different programs mentioned in this review offer different capabilities. Some allow topological changes to the protein or ligand, some offer different renderers, some allow interaction with a physically rigorous MD simulation directly, from within the VR environment.
3. Tools for interactive VR: controllers, gloves, and hands
There is a natural desire to reach out and touch simulated objects when they are presented in an immersive setting. VR controllers, such as those which can be bought as a set with HMDs, provide the user with a virtual ‘pair of hands.’ The user can interactively and intuitively grasp and manipulate structures as if they were physical objects in the real world. This allows a more extensive exploration of structures than manipulation on a 2D screen [Citation51] as it is facilitated by the 6 degrees of rotational and translational freedom and the depth perception afforded by VR (although it should be noted that attempts to improve visualization of 3D molecules have been made without the use of VR, e.g. stereo viewers and 3D printing of molecules [Citation52,Citation53]). Compared to using a mouse to interact with the simulation, which only permits a single interaction at a time via a mouse click, some VR software allow interaction from two controllers per user simultaneously, resulting in greater control over the simulation. This can be important when exploring large molecules or studying an area of the system that may be non-trivial to navigate e.g. a buried binding cavity.
However, there is still the limitation that a VR controller does not have the same range of motion as a real pair of hands, provided by the flexibility of human thumbs, fingers, and wrists. The development of VR gloves allows the incorporation of the complex motions that hands are capable of. There have been some advances in VR gloves specifically for molecular manipulation () [Citation54].
Figure 2. VR Gloves and their application [Citation54]. (A) Pinch sensing gloves. The tracker is placed on top of the glove and the interaction occurs through a pinching motion between either the thumb and index finger, or thumb and middle finger. (B) Two users in VR interacting with a real-time MD simulation of a polyalanine peptide. A pinching motion allows the users to grab an atom and interactively maneuver the protein around the VR environment. Image reproduced from [Citation54] licensed under CC BY-SA.
![Figure 2. VR Gloves and their application [Citation54]. (A) Pinch sensing gloves. The tracker is placed on top of the glove and the interaction occurs through a pinching motion between either the thumb and index finger, or thumb and middle finger. (B) Two users in VR interacting with a real-time MD simulation of a polyalanine peptide. A pinching motion allows the users to grab an atom and interactively maneuver the protein around the VR environment. Image reproduced from [Citation54] licensed under CC BY-SA.](/cms/asset/8b1bd4ec-d8e7-48c3-9583-e7ac13163fa2/iedc_a_2079632_f0002_oc.jpg)
Hand tracking is an alternative approach. Molecular Rift [Citation55] is an example of VR software that is focused on manipulation of biomolecules in 3D space using hand tracking rather than VR controllers. Their gesture recognition software [Citation56] allows basic structure manipulation such as rotation and translation of the molecule through a series of predetermined hand gestures. While hand tracking has the potential to be the most instinctive way for humans to manipulate molecules in VR – it removes the need for using specific hardware – a focus group of Molecular Rift users found learning the gestures to be a lengthy process as some were not immediately obvious [Citation55].
An exciting direction is to incorporate haptic feedback (i.e. the modality of touch) into the virtual world [Citation31,Citation57,Citation58]. Specialist haptic systems have already been developed [Citation59–65], however they can be very expensive or not easily available to the consumer. Haptic feedback in VR has been explored for use in surgical simulations [Citation66–68], where a prospective surgeon can, for example, feel the resistance of body tissue beneath their simulated surgical tool. As this kind of physical sensation is well defined, it is relatively easy to mimic in a VR setting. Haptic feedback in terms of molecular simulation presents a different type of challenge [Citation69–71] because there is no reference for what kind of feedback is ‘realistic’ or intuitively useful when manipulating these objects. On the other hand, there is the possibility of exploiting a phenomenon known as ‘pseudo-haptics’ to ‘feel’ molecules in VR. This phenomenon is where the user experiences the illusion of feeling things within the simulation [Citation71]; there is no actual physical force. A recent experiment showed that users of the interactive VR software Narupa IMD (see below) could distinguish molecular properties using this pseudo-haptic feedback [Citation72]. The study involved users interacting with an interactive molecular dynamics (IMD) simulation, where three Buckminster fullerene (C60) molecules were simulated in the same virtual space, all with different bond force constants. The force constants affected how rigid the molecule appeared in VR: a large force constant meant the molecule was stiff and inelastic, and a small force constant meant the molecule was more flexible. Most participants were able to notice a difference between how the molecules behaved in the simulation, and correctly rank the C60 molecules according to ‘elasticity’ (bond force constants). There is potential for the sensation of pseudo-haptic feedback to be exploited for drug design. For example, an IMD-VR user could estimate the barrier to dissociation of a drug from a protein based on how ‘easy’ or ‘difficult’ it feels to remove the drug from the binding site.
4. Applications of VR relevant to drug discovery
Although this review focuses on interactive VR, non-interactive VR of biomolecular structures clearly has uses in drug discovery, as noted above. VR provides depth perception which improves how the molecules are perceived in 3D over 2D screens and stereoscopic representations. As the screens are directly in front of your eyes, VR offers the advantage of being able to view molecules as simply as moving your head and with an increased field of view (compared to a screen or using stereoglasses), making the technology excellent as a graphical display.
The developers of ProteinVR [Citation29] (an example of a protein viewed in ProteinVR is shown in (.4) tested how visualizing a protein-ligand complex in ProteinVR compares to the 2D molecular viewer, VMD [Citation73]. The complex chosen (T. brucei RNA editing ligase 1, known as REL1, and V2, a naphthalene-based inhibitor) has a binding pocket that lies in a deep, narrow cavity. The structure of the complex used had been generated using the automated docking program AutoDock Vina [Citation74]. The users found that, by viewing the molecule in VR, it allowed intuitive exploration of the narrow binding pocket cavity by the user moving their head [Citation29]. They hypothesized that the AutoDock Vina pose may be incorrect and suggested interactions which could be more favorable to binding [Citation29]. This is an excellent demonstration of how users can utilize their spatial awareness in VR to note irregularities in the structure that might otherwise go unnoticed on a 2D screen.
Figure 3. Scenes from VR software for molecular modeling. 1) A scene from the interactive molecular dynamics tutorial from the Nanome software [Citation30], where the user is inspecting the first frame of a trajectory. 2) UnityMol software [Citation103] showing a cartoon and hyperball representation of the influenza neuraminidase protein (PDB 3TI6). 3) Hydrogen bonding (rendered as white glowing interactions) between a 20 amino acid long polypeptide in the software, Peppy [Citation88]. 4) The SARS-CoV Mpro (PDB 2Q6G) in a rainbow cartoon representation using the web-based VR software ProteinVR [Citation29]. Images created by the author using interactive VR software Nanome [Citation30], UnityMol [Citation103], Peppy [Citation88], and ProteinVR [Citation29].
![Figure 3. Scenes from VR software for molecular modeling. 1) A scene from the interactive molecular dynamics tutorial from the Nanome software [Citation30], where the user is inspecting the first frame of a trajectory. 2) UnityMol software [Citation103] showing a cartoon and hyperball representation of the influenza neuraminidase protein (PDB 3TI6). 3) Hydrogen bonding (rendered as white glowing interactions) between a 20 amino acid long polypeptide in the software, Peppy [Citation88]. 4) The SARS-CoV Mpro (PDB 2Q6G) in a rainbow cartoon representation using the web-based VR software ProteinVR [Citation29]. Images created by the author using interactive VR software Nanome [Citation30], UnityMol [Citation103], Peppy [Citation88], and ProteinVR [Citation29].](/cms/asset/82351b45-c7af-4679-8616-d559fd3b4435/iedc_a_2079632_f0003_oc.jpg)
Molecules are fundamentally dynamic [Citation75], (and protein dynamics is often important for function) and therefore it can be important to consider molecular motion when designing drugs, e.g. to examine how a protein and drug candidate bind and interact with each other over time. MD simulations [Citation76–79] are increasingly widely used in drug design and development. Popular programs currently used for visualizing molecular dynamics trajectories include VMD [Citation73], Pymol [Citation80], and Chimera [Citation81]. However, these typically project 3D motion onto a 2D screen, so information is lost. Interactive VR software such as Nanome [Citation30] and ChimeraX [Citation82,Citation83] can be used to visualize MD trajectories (Figure 3.1). Features such as the ability to play, pause, rewind, fast-forward, slow down, and speed up the simulation, which are available in traditional molecular viewers, are also available in these VR software packages.
As with standard screen-based GUIs, it is useful to visualize molecular properties and interactions such as hydrogen bonds and the electrostatic potential of molecules in VR. Interactions such as hydrogen bonds, salt bridges, and charge-dipole interactions contribute greatly to protein structure and stability, and ligand binding [Citation84,Citation85]. Drug binding relies on favorable interactions between the drug and the receptor, therefore viewing and understanding these interactions is crucial to drug design and development [Citation86,Citation87]. Many interactive VR programs such as Molecular Rift [Citation55], Nanome [Citation30], and Peppy [Citation88] include a toggle for switching hydrogen bond visualization on and off. By viewing these interactions in 3D, the user can utilize their own depth perception to inspect the distances and angles between interacting groups. This is valuable for suggesting where modifications could be made to the drug or protein to generate additional hydrogen bonds. Molecular Rift and Peppy offer a striking visualization of this type of interaction, portraying the contact between the donor and acceptor atoms as a bright, dynamic cloud (Figure 3.3). In contrast to a flat dashed line, which is often used to indicate a hydrogen bond in 2D molecular visualization software, this makes it obvious to the user when a hydrogen bond is present because it is visually distinct from the rest of the biomolecule. Nanome (version 1.22.1) also provides the option to measure the distance and angle manually in VR, by clicking on the measuring tool and physically selecting atoms of interest. The value of the distance or angle is then displayed above the selected atoms. Finally, the ability to simply move your head and inspect the molecule from a different angle may be helpful when looking at hydrogen bonds (or other non-covalent interactions) between atoms buried below the surface of the molecule, where the structure is more labyrinthine. Recently, interactive VR (specifically using Nanome) was used to generate structural analogues of a small molecule with the aim of creating a SARS-CoV-2 main protease (Mpro) inhibitor [Citation89]. The SARS-CoV-2 Mpro is a promising drug target for combating COVID-19, and it is indicative of recent progress that interactive VR is being used for drug design and modification purposes for this and other COVID targets.
In recent years, the pharmaceutical company Novartis has recognized the potential for interactive VR for early-stage drug research [Citation90]. Using Oculus and Nanome, scientists at Novartis are utilizing interactive VR to investigate structural insights for potential drugs to combat COVID-19. US biotechnology company Nimbus Therapeutics have also deployed Nanome for drug discovery, stating that they expect the technology to ‘save tens of thousands of dollars per year’ and drastically accelerate the drug design process [Citation91]. In December 2021, Roivant Discovery, the drug discovery division of biotechnology company Roivant Sciences, deployed the largest platform of Nanome to date [Citation92]. By employing this interactive VR technology, dozens of scientists across the USA can collaboratively modify and process molecular data, allowing more informed decisions to be made and, ultimately, speed up the drug discovery process. Nanome has also been used for research into psychedelics by biotechnology start-up Psilera Inc., where scientists are investigating how natural psychoactive compounds may treat mood disorders and neurodegenerative diseases [Citation93]. Nanome was founded in 2015, and is currently working with more than 15 biopharmaceutical companies in the US [Citation92]. A recent partnership with Japanese information and communications technology company Fujitsu is allowing Nanome to provide their platform to Japanese pharmaceutical companies [Citation94]. This impressive interest from the pharmaceutical industry indicates the potential that interactive VR has for early-stage drug development.
Nanome is also being used in education. Correspondence between the authors and Prof. Shozeb Haider at the University College London (UCL) School of Pharmacy helpfully revealed that he and colleagues at UCL are using Nanome for teaching and research, and have found interactive VR to be a ‘very easy and intuitive [tool] to teach students how structure-based drug design’ and that feedback from students has been ‘overwhelmingly positive’ (2022 e-mail from Prof. Haider to the authors; unreferenced). He also stated that ‘Multi-site discussions have been quite productive in our research group during the lockdown’ (2022 e-mail from Prof. Haider to the authors; unreferenced), highlighting the importance of the collaborative aspect of interactive VR.
In addition to visualizing electrostatic interactions, it may also be important to view the overall electrostatic potential of the molecule(s), as this can aid predictions in how and where a drug may bind to a receptor [Citation95]. The visualization of electrostatic potentials using computer graphics was introduced by Weiner et al. in 1982 [Citation96], who proposed that this kind of visualization would be beneficial for drug design. Indeed, the visualization of electrostatic surfaces has proven to be useful for investigating protein-ligand binding for CADD [Citation97–99]. Software such as adaptive Poisson-Boltzmann solver (APBS) [Citation100,Citation101] and PDB2PQR [Citation102] facilitate electrostatic calculations of biomolecules for visualization.
The developers behind UnityMol (example of interface shown in Figure 3.2) have created a VR platform for simultaneous calculation and visualization of electrostatic properties of molecules, named UnityMol-APBS [Citation103]. The advantages that this programme has over APBS with a 2D viewer is that the user (i) has increased depth perception and field of view for examining the molecule, and (ii) can prepare APBS input files from within the virtual interface, using a graphical user interface (GUI) to send the input files directly to APBS tools. By simply reaching out, selecting the atoms of interest, and then clicking a button in the GUI, the user can seamlessly perform calculations within VR using the interface with APBS. The combination of the virtual interface with the APBS toolkit significantly reduces the time needed to manually prepare input files, which can often be verbose and rely on the user having prior knowledge of the software’s language and formatting style. Here, the need for learning a completely new software program is removed and replaced by a few easy gestures.
To investigate the utility of VR for exploring electrostatic surface potentials, the UnityMol-APBS developers examined a particular enzyme (Torpedo californica acetylcholinesterase, known to operate quickly due to its electrostatic properties) and a substrate molecule [Citation103]. In VR, with a large field of view and ability to translate, rotate, and scale the size of the molecule, it was immediately clear from the cluster of animated electrostatic field lines – which represent the electrostatic potential gradient – where the active site was situated. The immersive nature of VR also provides a sense of spatial awareness, meaning the user can grab the substrate and translate it as a rigid body through the binding cavity. In this way, users can get a sense of how feasible the pathway is. It should be noted that, interactive VR is being used as a tool for drug research and development; that is not to say that the same results could not be obtained without the interactive VR element. Rather, pharmaceutical companies and research groups are using interactive VR to aid the drug development process, particularly in the early stages, and see value in this approach.
5. On-the-fly modification
Drug design has been described as both a science and an art [Citation104], and therefore, creativity and ingenuity are integral to success. A study investigating the relationship between creativity and factors relating to creativity (such as flow of work and attention) of individuals tasked with a clothing design challenge found (via EEG measurements of brainwaves) showed that a VR environment allows participants to focus more on the challenge at hand. Users reported that the ability to walk about and observe their creation from different angles inspired new ideas for improvement, compared to designing their product with a pencil and paper [Citation105,Citation106]. Furthermore, the ability to interact with complex 3D structures in VR inspires creative discussion around what interactions contribute to drug binding, and what could be altered to improve these interactions [Citation30]. Many programs used in CADD are focused on building and structural modification of small drug candidates and proteins. As the orientation of functional groups can be vital in terms of drugs binding, performing complex 3D modeling on a flat screen could lead to some error. Performing these modifications ‘on-the-fly’ in a 3D space could allow immediate answers to hypotheses and thus drive intellectual discussion.
The function of many proteins relies on the structure they adopt. The capacity to perform structural modifications of proteins and ligands could be enormously powerful [Citation107]. The interactive VR chemistry educational tool Peppy allows such modifications: a cohort of undergraduate chemists used it to perform a range of 3D modeling tasks: from creating short chain polypeptides to constructing α-helices and β-sheets with complex hydrogen bonding networks to teach them the fundamentals of protein structure [Citation88]. This study showed that students using interactive VR were enthusiastic and creative when building and scrutinizing polypeptides. Some students also reported that interacting with polypeptides in VR cleared up previous misconceptions they had from learning protein structure in 2D [Citation88]. The developers of Peppy state that, while their primary aim is teaching secondary structure of proteins to undergraduate students, they also recognize its potential use in research [Citation88]. They also highlight that the goal of designing a tool capable of on-the-fly modifications and mutations was to encourage engagement and creativity, as well as inspiring a deeper exploration of biomolecular structures than a 2D builder would allow. A limitation of this software is that it does not currently support larger proteins (currently, the maximum is 32 residues) [Citation88]. Nanome allows modifications of much bigger proteins [Citation30].
The ability to make on-the-fly modifications to both the internal structure (such as rotating functional groups) and the chemical structure (such as mutating a functional group) allows rapid testing of hypotheses and inspires creative thinking, as previous studies of GUIs etc have shown [Citation105,Citation106]. Nanome allows building and modifying drug-like molecules within a VR environment [Citation30]. Researchers were able to build a small molecule inhibitor from scratch in the active site of an RIP2 kinase (PDB 5W5O), achieving an RMSD of 1.8 Å with respect to the crystal structure [Citation30] (around 2 Å is considered by docking programmes to be a sensible cutoff for finding the ‘correct’ pose [Citation108]). Furthermore, starting from the crystal structure, users were tasked with making chemical alterations to the drug, where the objective was to form additional hydrogen bonds with the enzyme to achieve a more tightly bound pose [Citation30]. This shows that, not only were users of Nanome able to produce sensible drug-protein structures similar to crystal structures, but they could also make predictions for more favorable interactions and trial them directly [Citation30]. Nanome also provides an energy minimization feature that can remove human induced artifacts, such as unrealistically long chemical bonds, and return a more energetically reasonable structure. In another test case, a VR session was held where four Nanome users (chemists) were tasked with pointing out sites for macrocyclization of an active starting compound [Citation30]. The aim was to preserve activity of the compound while improving physicochemical properties. Within minutes the users had become accustomed to the software and were discussing how they could alter the structure they were given. The group were successful in creating structures with improved physicochemical properties, whilst retaining activity.
DNA nanostructures pose an interesting avenue for drug development as they can be used for targeted drug delivery. Vivern (Visulization, Interaction, and Virtual EnviRoNment) provides an interactive VR platform for users to build and analyze DNA nanostructures [Citation109], including many on-the-fly modification tools, such as a ‘DNA un-twister’ and a ‘Magic Scale Lens’ that allows atomic level inspection of the DNA structures. In a recent user study, experts from the DNA nanotechnology field were tasked with using Vivern to build DN superstructures from scratch and analyzing existing structures [Citation109]. The experts found that, from within the VR environment, it is much easier to spot mistakes and rectify them, which can of course be crucial from a drug design perspective.
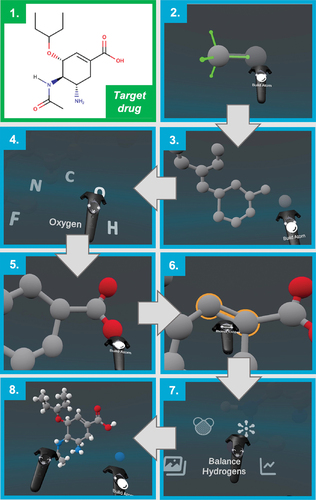
The interactive VR tool Narupa Builder (), an application of the interactive VR framework Narupa, also allows the creation and manipulation of molecules. shows a drug structure (oseltamivir carboxylate, an influenza neuraminidase inhibitor) and the steps taken by a user to create the drug in an interactive VR environment. While this small drug was generated atom by atom, Narupa Builder also has a built-in library that contains common molecular fragments, such as amino acids and aromatic rings, that allow the rapid building of larger molecules. There is also an energy minimization feature, like Nanome, and the ability to export the resulting structure to a MOL2 format. The Narupa Builder is freely available for download on Windows via https://irl.itch.io/narupa-builder.
Figure 4. Constructing oseltamivir using the Narupa Builder. 1) The structure of the target drug (oseltamivir carboxylate, commonly known as Tamiflu, an influenza drug) to be built using Narupa Builder. 2) Building the first carbon atoms of the 6 membered ring. The green axes represent the possible sites for a new atom to be bonded to the carbon. 3) The carbon backbone of the drug, without any hydrogens. 4) Changing from a carbon atom to an oxygen atom to be added to the drug. 5) Creating the carboxylate functional group. 6) Changing the bond order between two carbon atoms from a single to a double bond. 7) Finally, adding hydrogens to all the possible sites of hydrogenation. 8) The resulting structure.
6. Interactive molecular dynamics in virtual reality (IMD-VR)
One of the most exciting recent developments in this area is the combination of interactive molecular dynamics simulations with virtual reality (IMD-VR) [Citation31], allowing researchers to literally reach into their MD simulations and orchestrate the pathway of molecular motion [Citation110]. IMD [Citation111–116] has some similarities to steered MD (SMD) [Citation117], which uses small forces applied over a long simulation time to simulate atomic force microscopy (AFM) experiments or biological structural change. IMD differs as large forces are applied by the user, and the simulations are very fast to run at the cost of a decrease in accuracy [Citation118]. In IMD, the user steers the system to drive rare/high barrier events such as protein conformational changes and ligand binding. Various scientific problems have been tackled with IMD including: ligand binding [Citation119], protein engineering [Citation120], polymers [Citation112], colloids [Citation113], transport through membrane proteins [Citation121], fitting X-ray crystallography [Citation116], education [Citation122,Citation123] and even small ~(40 atoms) ab initio quantum mechanical calculations [Citation124]. However, performing complex 3D tasks with a 2D interface can be challenging. Although IMD was around during the early VR era [Citation112], the breakthrough year was 2001 when Stone, Gullingsrud, and Schulten [Citation111] interfaced the graphical display capabilities of VMD with the molecular mechanics/dynamics engine NAMD [Citation125] and contemporary others did IMD in Java [Citation113]. Interactivity was recognized as important, and since interaction with the simulation is via applied forces, further work in this area concentrated on adding haptic interfaces [Citation111,Citation118,Citation120]: for example, force-feedback gloves combined with MD simulations via IMD were used in VR to design proteins for their stiffness and mechanical properties [Citation120]. Some groups combined IMD, VR, and haptics [Citation119,Citation121], however, such frameworks only allowed one user at a time to interact with the simulation.
The IMD-VR software Narupa IMD [Citation126] allows users to interact with MD simulations (utilizing the OpenMM physics engine [Citation127]) on-the-fly in a fully immersive VR space. The purpose of OpenMM here is to propagate the MD; that is, the user provides force that moves the atoms, and OpenMM numerically integrates Newton’s equations of motion and provides the new positions of the atoms. For every timestep, this process is repeated to provide a trajectory that an IMD-VR user has effectively manipulated. Thhe user can interact directly with molecules in VR, manually applying forces that change the simulation. It should be noted that other force engines can be used with Narupa IMD. For example, electronic structure programs can be used for studies of chemical reactivity. In a recent human-computer interaction (HCI) study [Citation51], participants were tasked with performing a range of molecular manipulation assignments (threading methane through a nanotube, changing helical screw sense, and tying a knot in a protein) using either a mouse and keyboard, a touchscreen, or IMD-VR. The participants were able to complete these complex 3D tasks faster using IMD-VR and with a higher success rate. IMD-VR has also been used as an undergraduate chemistry teaching tool, where students were tasked with performing small chemical rearrangements and docking tasks [Citation128]. Feedback from students suggested that visualizing molecules in an interactive VR setting is more engaging than the traditional computer and mouse set-up they had previously used, and allows them to grasp chemistry concepts more quickly.
Automated docking of small molecules, fragments, and potential drugs is a popular CADD method due to its capacity to rapidly predict binding poses of small molecules to a target receptor molecule [Citation129–132]. This provides insight into both the structural orientations (binding poses) of the small molecules as well as the corresponding binding affinities. Docking flexible molecules still poses a challenge for CADD, and human intervention is often required. Deeks et al. investigated the use of Narupa IMD for flexible, human driven, protein-ligand docking [Citation133]. The experimental protocol involved testing whether expert and novice Narupa IMD users were able to interactively guide ligands (benzamidine, oseltamivir, and amprenavir) in and out of the binding pockets of three viral enzymes (trypsin, neuraminidase, and HIV-1 protease, respectively), and recreate their respective crystallographic protein-ligand binding poses (within 2.15 Å RMSD of the crystal structure) in 5–10 minutes of real time. A bound structure from the interactive MD was then extracted and a short amount of MD was performed to test whether the structures created with IMD-VR were stable. By utilizing human ability at 3D spatial manipulation, after only a short amount of time in VR (40 minutes, including time spent learning how to operate the VR equipment), non-expert users (most of whom were also not experts in drug docking) were able to produce docked structures of these three viral enzymes with FDA approved drugs [Citation133].
In a separate study [Citation134], expert Narupa IMD users generated structures of a small molecule inhibitor and an 11-amino acid oligopeptide substrate complexed to the SARS-CoV-2 main protease (Mpro), an enzyme involved in replication of the virus that causes COVID-19 [Citation135]. Various protocols were tested during the IMD-VR docking process, including running the simulation with and without backbone restraints on the enzyme. The expert users were able to produce structures that replicate experimentally observed crystal structures [Citation134]. The results emphasized the importance of formation of important hydrogen bonds and led to the recommendation to focus on forming such interactions to create stable bound structures. The expert users were also able to interactively dock an inhibitor and a substrate to an apo form of the Mpro, following the same procedure of forming key hydrogen bonds [Citation134]. The docked structures generated from apo Mpro remained stable in MD. A further study that compared docked structures of peptide-Mpro complexes generated from AutoDock CrankPep (a popular automated docking program), Pymol, and IMD-VR showed that bound structures from all three methods were in good agreement with each other [Citation136], which was encouraging.
Narupa IMD can save frames of the trajectory generated by the user. For example, if a user were to interactively grab a drug bound to an enzyme and pull it out into the solvent space, the user can save the frames of this trajectory using OpenMM, just like any other standard MD engine would allow. In a matter of minutes of real time, users can simulate a rare event such as drug unbinding [Citation133,Citation134,Citation136]. Capturing these rare events may otherwise require a large amount of computational time and resources, as well as expert knowledge of the biological system prior to simulation. IMD-VR can allow users to sample a substantial range of the rugged energy landscape of a protein in an intuitive manner, using human spatial and chemical intuition to guide movements. IMD-VR allows manipulations on the atomic scale in order to observe how molecular systems evolve in real-time. This could provide an interesting way to explore the effects of a perturbation: e.g. when a drug binds to a protein, users can observe how the protein responds. It should be noted that the forces that a user exerts on the simulation may be high (this is a non-equilibrium simulation) and may cause significant perturbation. This should be considered when generating IMD-VR trajectories.
Currently, one important limitation of Narupa IMD is the number of atoms in the system that can be simulated and rendered at an appropriate frame rate (which of course depends on the available hardware). While it is possible to visualize several hundred thousand atoms for a static structure (where no MD engine is used, just observing the structure, e.g. an LSD molecule bound to a membrane-embedded HT2B human serotonin receptor using Narupa, see https://vimeo.com/420035802), dynamically rendering the positions of all these atoms as an MD simulation propagates is not currently feasible due to limitations of processing power. Clearly, this depends on the hardware being used for the simulation. aAdvances in hardware will improve issues with framerates. Similarly, the limitation of processing power and number of atoms also means that explicit solvent simulations are not yet generally practical using Narupa IMD. In all of the Narupa IMD applications referenced here [Citation51,Citation128,Citation133,Citation134,Citation136], implicit solvent was used in order to ensure the framerate was reasonable.
IMD-VR has great potential in drug design and development. As discussed above, the ability to observe how a molecule responds to a perturbation could be useful in drug discovery, in understanding lgand binding affinity and kinetics. For example, drug bindinig to a protein may require a confromational change such as loop opening: such prrocesses can be driven intuitively by the user using IMD-VR to generate binding pathways. Being able to simply reach out and create binding poses and pathways of drug binding on-the-fly, using human spatial and chemical intuition, will help in drug development. Also transformative is the demonstrated potential for groups of researchers to work together virtually on molecular design and modeling problems.
7. Conclusions
Interactive VR software will not replace other CADD methods; many of the features described in this review are already available in non-VR biomolecular manipulation software and have been useful for a long time. Instead, interactive VR offers an alternative way of modeling and manipulating biomolecules and can be used in conjunction with other CADD methods. For example, it may be useful to use VR to visualize a docked structure created from automated docking, employing the depth perception and immersion to examine results and deepen understanding of the structure. Additionally, the interactive nature of the VR allows exploration of the structure in an intuitive manner, using simple gestures to grab and manipulate the system easily.
We encourage readers to try out these VR programs themselves. The ease of use of modern interactive VR programs, the immersion that they allow in 3D molecular space, and the ability to ‘touch’ molecules cannot be described adequately here; it is necessary to experience them. Despite the wealth of anecdotal evidence of VR tools improvement over comparable non-VR visualization and manipulation programs [Citation93,Citation137,Citation138], there is a pressing need for well designed and rigorously tested user studies and HCI experiments to quantify this improvement. In this review we have only mentioned VR; as headsets shrink in size, weight and cost, the related technology of augmented reality, AR, (VR with an optical pass-through such that the user sees both the virtual and real worlds simultaneously) will grow in popularity. It is easy to see how both collaborative VR and AR could be invaluable tools for teaching and research collaboration, e.g. for distance learning. The only modalities discussed at length here were vision, manual interactivity, and touch; there are others that can be added to VR. For example, InteraChem [Citation122] is an interactive VR program built on Narupa IMD-VR for use as a teaching tool. It includes an interesting modality of atomic ‘happiness’ that encodes the energetic feasibility of a particular bonding arrangement and it is represented by emojis drawn on atoms. This approach co-opts the part of the brain that deals with human social interaction and makes high-level concepts incredibly easy to understand. Temperature feedback (of controllers or gloves) has also been suggested as a possible method of information communication in IMD [Citation120]. Color is part of how we see the world and can be used to encode information easily and intuitively. VR also includes the ability to play sounds and here extra information can be passed to the user. There is room for significant creativity in including these aspects here. How does the sound of a molecule’s vibrations change as it goes through a transition state? Getting good answers to these and similar questions, and developing the user experience, will allow interactive VR to be even more intuitive for human users. The easier the technology is to use, the more it can harness human creativity efficiently.
8. Expert opinion
Interactive VR is an emerging area that offers tremendous potential in drug design and development. State-of-the-art VR software and hardware can harness interactivity to manipulate and modify biomolecules on-the-fly, using human chemical and spatial intuition in practical and accessible ways. While humans cannot defeat computers in terms of speed and efficiency for producing results, human perception remains a vital part of drug design and discovery. The ability to be present within a molecular simulation and interact with it holds great promise.
Sophisticated VR hardware is now widely available and relatively not very expensive. Currently, this includes HMDs and controllers. More ‘natural’ (i.e. hand-like) controllers such VR gloves are emerging and allow more tactile exploration of biomolecular structures. It is also possible to harness senses other than just sight, e.g. touch: haptic feedback could be potentially very useful for drug design e.g. feel the energy change and identify the bottleneck to pulling a drug out of a binding site. Other human senses such as sound could be utilized to aid in exploring molecular systems, adding new ways to convey information about a model and its responses, and indeed this is being investigated [Citation139–141].
A particularly interesting emerging frontier is IMD in VR. While IMD is by no means a new concept, the concept of humans interacting with real time MD simulations in a VR is still relatively new. Molecules are inherently dynamic, and molecular motion is important to consider when modeling biomolecules. sSome dynamics important to drug design occur on the order of nanoseconds to microseconds, e.g. loop opening and drug binding, it may be useful to coax the system to explore areas of high energy using IMD-VR that may not otherwise be reached using standard MD simulations, i.e. using IMD-VR to generate rare but important states such as transition states for binding, or to drive conformational changes. While this kind of biased MD is not exclusive to VR (there are many non-VR enhanced sampling techniques available such as metadynamics, umbrella sampling, the string method, etc.), being able to physically reach out and maneuver molecules to find binding modes and pathways for drugs holds immense potential.
Finally, the collaborative nature of interactive VR means that many users can occupy the same virtual space and even interact with the simulated molecules together. This means that scientists across the globe can inhabit the same simulation and observe the same molecular phenomena. This could be useful for sharing new drug candidates or protein structures with colleagues, rapidly and intuitively, with full appreciation of 3-D molecular structure.
Interactive VR is a rapidly developing area. The VR programs for molecular modeling mentioned in this review have been published within the last 6 years, with the majority being published in the last 3 years. VR software and hardware will continue to evolve rapidly. In the next ten years, it is likely that a VR setup will be a staple for drug designers, structural biologists, and medicinal and computational chemists. VR will also allow research teams to collaborate virtually. With the ease of viewing and manipulating 3D static and dynamical molecular properties, the ability to interact MD simulations, and the ability to change structures on the fly, interactive VR will be an increasingly important tool in the armory of computational drug design and development.
Article highlights
Interactive VR here refers to software where the user can alter the molecular structure of the system in VR, i.e. not just using controllers to interactively change the view of the system by e.g. translating or rotating the object.
Interactive VR users can interact with physically rigorous molecular dynamics simulations to steer the simulation, by simply reaching out, taking hold of the atoms, and guiding them around the simulation space, e.g. to open or close protein loops, and flexibly dock drugs to protein targets.
Head mounted displays (HMDs) are the best VR hardware to use for interactive biomolecular modelling and manipulation.
Interactive VR also allows new ways for drug designers to collaborate virtually, e.g. using the cloud.
This box summarizes key points contained in the article.
Reviewer disclosures
Peer reviewers on this manuscript have no relevant financial or other relationships to disclose.
Additional information
Funding
References
- Gund P, Andose JD, Rhodes JB, et al. Three-dimensional molecular modeling and drug design. Science. 1980;208(4451):1425–1431.
- Marshall GR. Computer-aided drug design. Annu Rev Pharmacol Toxicol. 1987;27(1):193–213.
- Van Drie JH. Computer-aided drug design: the next 20 years. J Comput Aided Mol Des. 2007;21(10):591–601.
- Macalino SJY, Gosu V, Hong S, et al. Role of computer-aided drug design in modern drug discovery. Arch Pharm Res. 2015;38(9):1686–1701.
- Jorgensen WL. The many roles of computation in drug discovery. Science. 2004;303(5665):1813–1818.
- Lovering F, Bikker J, Humblet C. Escape from flatland: increasing saturation as an approach to improving clinical success. J Med Chem. 2009;52(21):6752–6756.
- Lovering F. Escape from Flatland 2: complexity and promiscuity. Med Chem Comm. 2013;4(3):515–519.
- Licklider JC. Man-computer symbiosis. IRE Trans Human Factors Electron. 1960;1:4–11.
- Wright WG. Using virtual reality to augment perception, enhance sensorimotor adaptation, and change our minds. Front Syst Neurosci. 2014;8:56.
- Biocca F, Delaney B. Immersive virtual reality technology. Communication in the Age of Virtual Reality. 1995;15(32):10–5555.
- Sutherland IE, editor. A head-mounted three dimensional display. Proceedings of the December 9-11, 1968, Fall Joint Computer Conference, Part I, San Francisco, California; 1968.
- Krueger MW, Gionfriddo T, Hinrichsen K, editors. VIDEOPLACE—an artificial reality. Proceedings of the SIGCHI Conference on Human Factors in Computing Systems, San Francisco, California; 1985.
- Lanier J. Virtual reality: the promise of the future. Interact Learn Int. 1992;8(4):275–279.
- Fisher SS, McGreevy M, Humphries J, et al., editors. Virtual environment display system. Proceedings of the 1986 workshop on Interactive 3D graphics, Chapel Hill, North Carolina USA; 1987.
- Lippman A. Movie-maps: an application of the optical videodisc to computer graphics. Acm Siggraph Comput Graphics. 1980;14(3):32–42.
- Sutherland I. The ultimate display. 1965
- Shakya S. Virtual restoration of damaged archeological artifacts obtained from expeditions using 3D visualization. J Innovative Image Process (JIIP). 2019;1(2):102–110.
- Lütjens M, Kersten TP, Dorschel B, et al. Virtual reality in cartography: immersive 3D visualization of the arctic clyde inlet (Canada) using digital elevation models and bathymetric data. Multimodal Technol Inter. 2019;3(1):9.
- Li R. Dynamic three-dimensional visualization system of sea area flow field based on virtual reality technology. Ccamlr Sci. 2019;26(1):23–29.
- Poyade M, Eaglesham C, Trench J, et al. A transferable psychological evaluation of virtual reality applied to safety training in chemical manufacturing. ACS Chem Health Saf. 2021;28(1):55–65.
- Bird JM. The use of virtual reality head-mounted displays within applied sport psychology. J Sport Psychol Action. 2020;11(2):115–128.
- Hilty DM, Randhawa K, Maheu MM, et al. A review of telepresence, virtual reality, and augmented reality applied to clinical care. J Technol Behav Sci. 2020;5(2):178–205.
- Moro C, Štromberga Z, Raikos A, et al. The effectiveness of virtual and augmented reality in health sciences and medical anatomy. Anat Sci Educ. 2017;10(6):549–559.
- Desselle MR, Brown RA, James AR, et al. Augmented and virtual reality in surgery. Comput Sci Eng. 2020;22(3):18–26.
- Calvelo M, Á P, Garcia-Fandino R. An immersive journey to the molecular structure of SARS-CoV-2: virtual reality in COVID-19. Comput Struct Biotechnol J. 2020;18:2621–2628
- Tse C-M, Li H, Leung K-S , et al., editors. Interactive drug design in virtual reality. 15th International Conference on Information Visualisation, London, UK; 2011: IEEE.
- Zonta N, Brancale A. Virtual reality applications in antiviral drug design. Antiviral Res. 2009;82(2):A74.
- Kleinberg ML, Wanke LA. New approaches and technologies in drug design and discovery. Am J Health Syst Pharm. 1995;52(12):1323–1336.
- Cassidy KC, Šefčík J, Raghav Y, et al., ProteinVR: web-based molecular visualization in virtual reality. PLoS Comput Biol. 2020;16(3): e1007747
- Kingsley LJ, Brunet V, Lelais G, et al. Development of a virtual reality platform for effective communication of structural data in drug discovery. J Mol Graphics Modell. 2019;89:234–241
- O’Connor MB, Bennie SJ, Deeks HM, et al., Interactive molecular dynamics in virtual reality from quantum chemistry to drug binding: an open-source multi-person framework. J Chem Phys. 2019;150(22): 220901
- Paper VR: VR storm studio; 2021 [2021 18 October]. Available from: https://vrstorm.hu/en/paper-vr/
- Samsung gear VR: samsung; 2021 [2021 18 October]. Available from: https://www.samsung.com/global/galaxy/gear-vr/
- Google cardboard: google; 2021 [18/January/2021]. Available from: https://arvr.google.com/cardboard/
- Cruz-Neira C, Sandin DJ, DeFanti TA, et al. The CAVE: audio visual experience automatic virtual environment. Commun ACM. 1992;35(6):64–73.
- Origin by vicon: vicon; [31/January/2022]. Available from: https://www.vicon.com/applications/location-based-virtual-reality
- Liu X-H, Wang T, Lin J-P, et al. Using virtual reality for drug discovery: a promising new outlet for novel leads. Expert Opin Drug Discov. 2018;13(12):1103–1114.
- Gaudiosi J. Dassault Systèmes Uses HTC vive to replace cave virtual reality tech. Fortune. 2016.
- Arcane. Immersive projection-based virtual reality (VR) for everyone, mobile and tailor-made 2022 [31/January/2022]. Available from: https://arcanetech.io/produit/vr-cave/
- Mestre DR. CAVE versus head-mounted displays: ongoing thoughts. Electron Imaging. 2017;2017(3):31–35.
- Robertson A. Oculus rift S review: a swan song for first-generation VR: The Verge; 2019 2022 January 31. Available from: https://www.theverge.com/2019/4/30/18523941/oculus-rift-s-review-vr-headset-price-specs-features
- Valve. Valve Index VR Kit Steam2022 [31/January/2022]. Available from: https://store.steampowered.com/sub/354231
- Vive. Vive pro series vive2022 [31/January/2022]. Available from: https://www.vive.com/uk/product/#pro%20series
- Angelov V, Petkov E, Shipkovenski G, et al., editors. Modern virtual reality headsets. 2020 international congress on human-computer interaction, Optimization and Robotic Applications (HORA). 2020: IEEE
- Mehrfard A, Fotouhi J, Taylor G, et al. A comparative analysis of virtual reality head-mounted display systems. arXiv preprint arXiv. 2019;191202913.
- HTC Vive Pro: Vive; 2021 [2021 13 September]. Available from: https://www.vive.com/uk/product/
- Valve Index: Valve Software; 2021 [2021 13 September]. Available from: https://www.valvesoftware.com/en/index
- Oculus Quest 2: Oculus VR; 2021 2021 September 13. Available from: https://www.oculus.com/quest-2/
- Introducing meta: a social technology company: meta; 2021 [2021 December 11]. Available from: https://about.fb.com/news/2021/10/facebook-company-is-now-meta/
- Stephenson N. Snow crash. United States: Bantam Books; 1992.
- O’Connor M, Deeks HM, Dawn E, et al., Sampling molecular conformations and dynamics in a multiuser virtual reality framework. Sci Adv. 2018;4(6): eaat2731.
- Hušák M, editor The use of stereoscopic visualization in chemistry and structural biology. Stereoscopic displays and virtual reality systems XIII. International Society for Optics and Photonics; 2006.
- Zhu X, Li H, Huang L, et al. 3D printing promotes the development of drugs. Biomed Pharmacother. 2020;131:110644.
- Freire R, Glowacki BR, Williams RR, et al. Omg-vr: open-source mudra gloves for manipulating molecular simulations in vr. arXiv preprint arXiv. 2019;190103532.
- Norrby M, Grebner C, Eriksson J, et al., Molecular rift: virtual reality for drug designers. J Chem Inf Model. 2015;55(11): 2475–2484.
- Microsoft kinect: microsoft; 2021 [ 29/10/21]. Available from: https://developer.microsoft.com/en-us/windows/kinect/
- Stone RJ, editor Haptic feedback: a brief history from telepresence to virtual reality. International Workshop on Haptic Human-Computer Interaction, Glasgow, UK. Springer; 2000.
- Burdea GC Force and touch feedback for virtual reality. 1996
- Chinello F, Malvezzi M, Prattichizzo D, et al. A modular wearable finger interface for cutaneous and kinesthetic interaction: control and evaluation. IEEE Trans Ind Electron. 2019;67(1):706–716.
- Whitmire E, Benko H, Holz C, et al., editors. Haptic revolver: touch, shear, texture, and shape rendering on a reconfigurable virtual reality controller. Proceedings of the 2018 CHI Conference on Human Factors in Computing Systems, Montreal, QC, Canada; 2018.
- Al Maimani A, Roudaut A, editors. Frozen suit: designing a changeable stiffness suit and its application to haptic games. Proceedings of the 2017 CHI Conference on Human Factors in Computing Systems, Denver, Colorado, USA; 2017.
- Al-Sada M, Jiang K, Ranade S, et al. HapticSnakes: multi-haptic feedback wearable robots for immersive virtual reality. Virtual Reality. 2020;24(2):191–209.
- Spagnoletti G, Meli L, Baldi TL, et al., editors. Rendering of pressure and textures using wearable haptics in immersive vr environments. 2018 IEEE Conference on Virtual Reality and 3D User Interfaces (VR), Reutlingen, Germany; 2018: IEEE.
- Pacchierotti C, Sinclair S, Solazzi M, et al. Wearable haptic systems for the fingertip and the hand: taxonomy, review, and perspectives. IEEE Trans Haptics. 2017;10(4):580–600.
- HaptX Gloves DK2: haptX Inc.; 2021 [ 29/10/21]. Available from: https://haptx.com/virtual-reality/
- Kreimeier J, Hammer S, Friedmann D, et al., editors. Evaluation of different types of haptic feedback influencing the task-based presence and performance in virtual reality. Proceedings of the 12th ACM International Conference on PErvasive Technologies Related to Assistive Environments, Rhodes, Greece; 2019.
- Alaker M, Wynn GR, Arulampalam T. Virtual reality training in laparoscopic surgery: a systematic review & meta-analysis. Int J Surg. 2016;29:85–94.
- Vaughan N, Dubey VN, Wainwright TW, et al. A review of virtual reality based training simulators for orthopaedic surgery. Med Eng Phys. 2016;38(2):59–71.
- Pusch A, Lécuyer A, editors. Pseudo-haptics: from the theoretical foundations to practical system design guidelines. Proceedings of the 13th international conference on multimodal interfaces, New York, USA; 2011.
- Lécuyer A, Coquillart S, Kheddar A, et al., editors. Pseudo-haptic feedback: can isometric input devices simulate force feedback? Proceedings IEEE Virtual Reality 2000 (Cat. No. 00CB37048), New Jersey, USA; 2000: IEEE.
- Lécuyer A. Simulating haptic feedback using vision: a survey of research and applications of pseudo-haptic feedback. Presence: Teleoperators and Virtual Environments. 2009;18(1):39–53.
- Roebuck Williams R, Varcoe X, Glowacki BR, et al., editors. Subtle sensing: detecting differences in the flexibility of virtually simulated molecular objects. Extended Abstracts of the 2020 CHI Conference on Human Factors in Computing Systems, Hawaii, USA; 2020
- Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14(1):33–38.
- Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31(2):455–461.
- Karplus M, Kuriyan J. Molecular dynamics and protein function. Proc Nat Acad Sci. 2005;102(19):6679–6685.
- Hernández-Rodríguez M, C Rosales-Hernández M, E Mendieta-Wejebe J, et al. Current tools and methods in molecular dynamics (MD) simulations for drug design. Curr Med Chem. 2016;23(34):3909–3924.
- Durrant JD, McCammon JA. Molecular dynamics simulations and drug discovery. BMC Biol. 2011;9(1):1–9.
- Liu X, Shi D, Zhou S, et al. Molecular dynamics simulations and novel drug discovery. Expert Opin Drug Discov. 2018;13(1):23–37.
- De Vivo M, Masetti M, Bottegoni G, et al. Role of molecular dynamics and related methods in drug discovery. J Med Chem. 2016;59(9):4035–4061.
- DeLano WL. Pymol: an open-source molecular graphics tool. CCP4 Newsletter on Protein Crystallography. 2002;40(1):82–92.
- Pettersen EF, Goddard TD, Huang CC, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612.
- Goddard TD, Huang CC, Meng EC, et al., UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 2018;27(1): 14–25.
- Pettersen EF, Goddard TD, Huang CC, et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 2021;30(1):70–82.
- Radić Z, Kirchhoff PD, Quinn DM, et al. Electrostatic influence on the kinetics of ligand binding to acetylcholinesterase: distinctions between active center ligands and fasciculin. J Biol Chem. 1997;272(37):23265–23277.
- Zhou H-X, Pang X. Electrostatic interactions in protein structure, folding, binding, and condensation. Chem Rev. 2018;118(4):1691–1741.
- Náray‐szabó G. Electrostatics in computer‐aided drug design. Int J Quantum Chem. 1989;36(S16):87–99.
- Rathi PC, Ludlow RF, Verdonk ML. Practical high-quality electrostatic potential surfaces for drug discovery using a graph-convolutional deep neural network. J Med Chem. 2019;63(16):8778–8790.
- Doak DG, Denyer GS, Gerrard JA, et al., Peppy: a virtual reality environment for exploring the principles of polypeptide structure. Protein Sci. 2020;29(1): 157–168.
- Kneller DW, Li H, Galanie S, et al., Structural, electronic, and electrostatic determinants for inhibitor binding to subsites s1 and s2 in sars-Cov-2 main protease. J Med Chem. 2021;64(23): 17366–17383.
- Collaborating for coronavirus drug discovery California: oculus; cited 2022 Apr 13]. Available from 2022 Apr 13: https://business.oculus.com/case-studies/novartis/?locale=en_GB
- Accelerating drug discovery to create a healthier future California: Oculus; cited 2022 Apr 13]. Available from 2022 Apr 13: https://business.oculus.com/case-studies/nimbus/?locale=en_GB
- Roivant in largest ever deployment of Nanome VR drug discovery software London UK: VRWorldTech; cited 2022 Apr 13]. Available from 2022 Apr 13: https://vrworldtech.com/2021/12/17/roivant-in-largest-ever-deployment-of-nanome-vr-drug-discovery-software/
- Castellanos S. Virtual reality puts drug researchers inside the molecules they study. Wall Street Journal. Sep 7. 2021. [cited 25 05 2022]. https://www.wsj.com/articles/virtual-reality-puts-drug-researchers-inside-the-molecules-they-study-11631023212
- Grier F Nanome partners with Fujitsu, San Diego: San Diego Business Journal 2020 cited 2022 Apr 13]. Available from 2022 Apr 13: https://www.sdbj.com/news/2020/sep/15/nanome-partners-fujitsu/
- Fried SD, Boxer SG. Electric fields and enzyme catalysis. Annu Rev Biochem. 2017;86:387–415.
- Weiner PK, Langridge R, Blaney JM, et al. Electrostatic potential molecular surfaces. Proc Nat Acad Sci. 1982;79(12):3754–3758.
- Bauer MR, Mackey MD. Electrostatic complementarity as a fast and effective tool to optimize binding and selectivity of protein–ligand complexes. J Med Chem. 2019;62(6):3036–3050.
- Nakamura H, Komatsu K, Nakagawa S, et al. Visualization of electrostatic recognition by enzymes for their ligands and cofactors. J Mol Graph. 1985;3(1):2–11.
- Keil M, Marhofer RJ, Rohwer A, et al. Molecular visualization in the rational drug design process. Front Biosci. 2009;14:2559–2583.
- Jurrus E, Engel D, Star K, et al. Improvements to the APBS biomolecular solvation software suite. Protein Sci. 2018;27(1):112–128.
- Unni S, Huang Y, Hanson RM, et al. Web servers and services for electrostatics calculations with APBS and PDB2PQR. J Comput Chem. 2011;32(7):1488–1491.
- Dolinsky TJ, Nielsen JE, McCammon JA, et al. PDB2PQR: an automated pipeline for the setup of Poisson–Boltzmann electrostatics calculations. Nucleic Acids Res. 2004;32(suppl_2):W665–W667.
- Laureanti J, Brandi J, Offor E, et al., Visualizing biomolecular electrostatics in virtual reality with unitymol‐APBS. Protein Sci. 2020;29(1): 237–246.
- Loging WT. The art and science of the drug discovery pipeline: history of drug discovery. Bioinformatics and Computational Biology in Drug Discovery and Development. 2016;1.
- Chang Y-S, Chou C-H, Chuang M-J, et al. Effects of virtual reality on creative design performance and creative experiential learning. Interact Learn Environ. 2020; 28:1–16.
- Yang X, Lin L, Cheng P-Y, et al. Examining creativity through a virtual reality support system. Edu Technol Res Develop. 2018;66(5):1231–1254.
- Berg JM, Tymoczko JL, Stryer L. Biochemistry. New York: WH Freeman; 2002.
- Pagadala NS, Syed K, Tuszynski J. Software for molecular docking: a review. Biophys Rev. 2017;9(2):91–102.
- Kutak D, Selzer MN, Byska J, et al. Vivern A virtual environment for multiscale visualization and modeling of DNA nanostructures. 2021. https://ieeexplore.ieee.org/document/9523759
- Juárez-Jiménez J, Tew P, Llabres S, et al. A virtual reality ensemble molecular dynamics workflow to study complex conformational changes in proteins. 2020.
- Stone JE, Gullingsrud J, Schulten K, editors. A system for interactive molecular dynamics simulation. Proceedings of the 2001 symposium on Interactive 3D graphics, New York, USA; 2001
- Rapaport D. Interactive molecular dynamics. Phys A Stat Mech Appli. 1997;240(1–2):246–254.
- Vormoor O. Quick and easy interactive molecular dynamics using Java3D. Computing Sci Eng. 2001;3(5):98–104.
- Knoll P, Mirzaei S. Development of an interactive molecular dynamics simulation software package. Rev Sci Instrum. 2003;74(4):2483–2487.
- Schroeder DV. Interactive molecular dynamics. Am J Phys. 2015;83(3):210–218.
- Croll TI, Andersen GR. Re-evaluation of low-resolution crystal structures via interactive molecular-dynamics flexible fitting (iMDFF): a case study in complement C4. Acta Crystallograph Sect D: Struct Biol. 2016;72(9):1006–1016.
- Izrailev S, Stepaniants S, Isralewitz B, et al. Steered molecular dynamics. Computational molecular dynamics: challenges, methods, ideas. Springer; 1999. p. 39–65.
- Grayson P, Tajkhorshid E, Schulten K. Mechanisms of selectivity in channels and enzymes studied with interactive molecular dynamics. Biophys J. 2003;85(1):36–48.
- Ai Z, Fröhlich T, editors Molecular dynamics simulation in virtual environments. Computer graphics forum. Wiley Online Library; 1998.
- Hamdi M, Ferreira A, Sharma G, et al. Prototyping bio-nanorobots using molecular dynamics simulation and virtual reality. Microelectronics J. 2008;39(2):190–201.
- Dreher M, Piuzzi M, Turki A, et al. Interactive molecular dynamics: scaling up to large systems. Procedia Comput Sci. 2013;18:20–29.
- Seritan S, Wang Y, Ford JE, et al., InteraChem: virtual reality visualizer for reactive interactive molecular dynamics. J Chem Educ. 2021;98(11): 3486–3492.
- Ferrell JB, Campbell JP, McCarthy DR, et al. Chemical exploration with virtual reality in organic teaching laboratories. J Chem Educ. 2019;96(9):1961–1966.
- Luehr N, Jin AG, Martínez TJ. Ab initio interactive molecular dynamics on graphical processing units (GPUs). J Chem Theory Comput. 2015;11(10):4536–4544.
- Phillips JC, Braun R, Wang W, et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26(16):1781–1802.
- Jamieson-Binnie AD, O’Connor MB, Barnoud J, et al. Narupa iMD: a VR-enabled multiplayer framework for streaming interactive molecular simulations. ACM SIGGRAPH 2020 Immersive Pavilion. 2020;1–2.
- Eastman P, Swails J, Chodera JD, et al. OpenMM 7: rapid development of high performance algorithms for molecular dynamics. PLoS Comput Biol. 2017;13(7):e1005659.
- Bennie SJ, Ranaghan KE, Deeks H, et al., Teaching enzyme catalysis using interactive molecular dynamics in virtual reality. J Chem Educ. 2019;96(11): 2488–2496.
- Schneider G, Böhm H-J. Virtual screening and fast automated docking methods. Drug Discov Today. 2002;7:64–70.
- Goodsell DS, Morris GM, Olson AJ. Automated docking of flexible ligands: applications of autodock. J Mol Recog. 1996;9(1):1–5.
- Blaney JM, Dixon JS. A good ligand is hard to find: automated docking methods. Perspect Drug Discovery Des. 1993;1(2):301–319.
- Goodsell DS, Olson AJ. Automated docking of substrates to proteins by simulated annealing. Proteins Struct Funct Bioinf. 1990;8(3):195–202.
- Deeks HM, Walters RK, Hare SR, et al., Interactive molecular dynamics in virtual reality for accurate flexible protein-ligand docking. Plos one. 2020;15(3): e0228461.
- Deeks HM, Walters RK, Barnoud J, et al., Interactive molecular dynamics in virtual reality is an effective tool for flexible substrate and inhibitor docking to the SARS-CoV-2 main protease. J Chem Inf Model. 2020;60(12): 5803–5814.
- Zhang L, Lin D, Sun X, et al. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368(6489):409–412.
- Chan HH, Moesser MA, Walters RK, et al. Discovery of SARS-Cov-2 mpro peptide inhibitors from modelling substrate and ligand binding. Chemical science. 2021;12.
- Discovery C4X. The 4Sight project: using VR in the drug discovery space: immerse UK; [31/January/2022]. Available from: https://www.immerseuk.org/case-study/c4x-discovery/
- Dutton GDail. DEMO: Nanome Triggers Deep Drug Development Insights Via Virtual Molecule Design. BioSpace. 2021. [cited 2022 May 25]. https://www.biospace.com/article/demo-nanome-inc-triggers-deep-drug-development-insights-via-virtual-molecule-design-/
- Mitchell TJ, Jones AJ, O’Connor MB, et al., editors. Towards molecular musical instruments: interactive sonifications of 17-alanine, graphene and carbon nanotubes. Proceedings of the 15th International Conference on Audio Mostly, New York, USA; 2020.
- Arbon RE, Jones AJ, Bratholm LA, et al. Sonifying stochastic walks on biomolecular energy landscapes. arXiv preprint arXiv. 2018;180305805.
- Shannon RJ, Deeks HM, Burfoot E, et al. Exploring human-guided strategies for reaction network exploration: interactive molecular dynamics in virtual reality as a tool for citizen scientists. J Chem Phys. 2021;155(15):154106.
