1. Introduction
Glaucoma, the most common cause of irreversible blindness worldwide is one of the most heritable of all complex human diseases with an estimated heritability of 70% [Citation1]. Diagnosis of glaucoma relies heavily on clinical biomarkers derived from ocular examination and optic disc parameters such as vertical cup-to-disc ratio (vCDR) and ophthalmic testing including visual field (VF) and optical coherence tomography (OCT). Multiple clinical risk factors have been identified to date including high intraocular pressure (IOP), older age, African or Hispanic ancestry, myopia, thin central corneal thickness and importantly family history. While congenital and childhood glaucoma follow a Mendelian form of inheritance due single gene mutations with large effect size, adult-onset glaucoma, like primary open-angle glaucoma (POAG), tend to follow complex inheritance with contributions from both one’s genetics and environmental exposures. While discovering genes contributing to adult-onset glaucoma has been historically difficult, advancements in genome-wide association studies (GWAS) over the last two decades had allowed for the identification of single nucleotide polymorphisms (SNPs) conferring susceptibility to complex diseases including POAG. GWAS involves scanning the genomes of a large number of individuals to find genetic markers, usually SNPs that are more common in those with a particular disease compared to those without the disease. Since the first published GWAS in 2009 [Citation2], over 300 common variants including some involving genes such as TMCO1, ABCA1, GAS7, ANGPT1, and MYOC, among others, have been linked to an increased risk of developing POAG [Citation3].
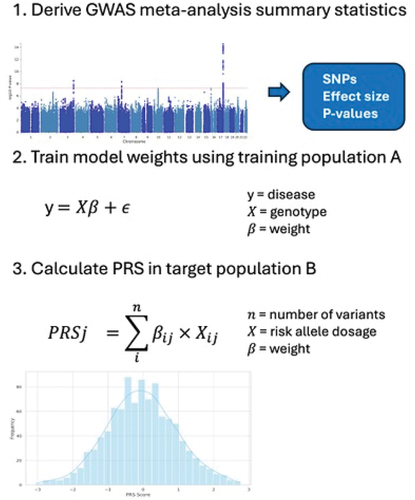
However, the common genetic variants identified from GWAS individually have small effects that contribute incrementally to the disease risk. While the predictive value for each POAG genetic variant is currently too low for clinical use, we can capitalize on the polygenic nature of diseases like POAG. In this context, polygenic risk scores (PRS) have emerged as a valuable tool for capturing the cumulative genetic risk for each individual. PRS is calculated by aggregating the effects of genetic variants identified through GWAS. In the simplest form, genetic risk scores can be constructed by summing risk alleles that meet genome-wide significance in prior GWAS. Polygenic risk scores, however, include variants across the entire genome, including those that many have not reached genome-wide significance in prior GWAS, each weighted by its contribution to disease risk. The standard approach for calculating PRS involves applying a p-value cutoff threshold to GWAS association statistics followed by a ‘pruning’ step that removes highly correlated variants. This approach, while conceptually simple, discards information and limits the predictive value of PRS. More recently, Bayesian statistical methods like LDPred, Lassosum and PRS-CS [Citation4–6] have been developed that jointly model genome-wide markers and account for variant linkage disequilibrium (LD) patterns. These methods derive effects for all variants with weights optimized using an independent training population and subsequently used to calculate genome-wide PRS in the target population (). Recent studies have shown that these genome-wide PRSs have improved prediction accuracy compared to earlier standard approaches for a range of diseases [Citation7,Citation8].
Figure 1. A) Genome-wide association study (GWAS) providing disease association summary statistics. An example Manhattan plot is shown, where the dashed line represents the threshold for genome-wide significance. B) Training (optimization) of the PRS based on GWAS summary association results in a dataset that is independent of the GWAS dataset. C) Calculation of the optimized PRS in target population.
While PRS offer a quantifiable measure of an individual’s genetic predisposition to POAG, it is important to remember that these scores provide relative risk of a disease and not absolute risk. This means that PRS offers a tool to identify individuals who are at higher risk for a disease compared to the non-diseased population and performance is affected by the populations in which the PRS is trained and implemented. Despite these limitations, the use of PRS in clinical settings holds significant potential for early detection, prognostication, and personalized management of POAG. The clinical utility of POAG PRS spans several key areas.
2. Early detection and risk stratification
Prior work demonstrates that higher POAG PRS is associated with disease prevalence. We have shown in the UK biobank, a population-based study, as well as in two clinical biobanks, that prevalence of POAG in the highest decile of genetic risk is 2-6× [Citation9] higher than prevalence of disease in the lowest decile of risk [Citation9]. Additionally, other groups have demonstrated on average 5–7 years earlier age of diagnosis among those with PRS in top 5–10% of risk [Citation10,Citation11]. Together, this work demonstrates that PRS can potentially identify high-risk individuals before symptoms of disease appear. Indeed, using data from the seminal Ocular Hypertension Treatment Study (OHTS) we have shown that 20-year risk of POAG conversion increased nearly 2-fold from 9.52% in the lowest PRS decile to 21.81% in the highest PRS decile [Citation12]. Importantly, early treatment appeared to mitigate the effects of high genetic risk in this population. As early detection and IOP-lowering treatment can significantly delay glaucoma progression and improve outcomes, these early results suggest that individuals with high PRS might benefit from more intensive screening and monitoring strategies.
3. Therapeutic decisions
Not only can PRS be used to identify those at risk for disease, research has shown that it is also associated with disease severity and progression. Using data from the UK Biobank, our group demonstrated a significant association between higher PRS and higher vCDR, higher IOP, lower retinal nerve fiber layer (RNFL) and lower ganglion cell complex (GCC) thickness [Citation9]. Similarly, results from two clinical biobanks show that higher genetic risk is associated with higher IOP, vCDR, thinner retinal nerve fiber layer (RNFL), greater use of IOP lowering medications and higher surgical counts across multiple ancestral groups, all markers of disease severity. Data from the ANRAG cohort supports these findings showing that those with higher PRS having greater likelihood of requiring glaucoma surgery [Citation11]. Importantly, we and others [Citation13] have shown that higher POAG PRS is associated with both structural and functional measures of disease worsening in multiple cohorts based on VF and OCT measures.
These results suggest that PRS can be used to identify individuals at higher risk of aggressive and progressive glaucoma. Those with higher PRS could be monitored more closely and treated earlier and more aggressively to slow disease progression and prevent vision loss and disability. PRS can even conceivably aid in surgical decision-making in order to optimize patient outcomes. Similar approaches have shown utility in cardiovascular disease, where PRS has been used to identify individuals that may benefit from early intervention and lifestyle modification [Citation14]. Similarly, PRS has been instrumental in diabetes care, where it helps in predicting the progression and complications of the disease, thereby guiding treatment decisions [Citation15].
4. Personalized medicine
While a relatively new area of research, genetic and polygenic risk scores can enable the tailoring of medical treatments based on individual genetic risk. Current methods for calculating PRS provide a single score that captures genome-wide genetic information for an individual. Although this is effective for various applications, it can overlook how genetic risk can vary across different biological processes. PRSs based on specific pathways or biological processes may offer a more nuanced approach to predicting glaucoma risk, progression, and treatment response by focusing on the most relevant biological processes for each individual. This could lead to more personalized prevention and therapy. Similarly, PRS that considers clinical subtypes of glaucoma could enhance individual risk and treatment assessments. Such approaches have been tested in cardiovascular disease and oncology where pathway-specific PRS have already shown promise in guiding targeted therapies [Citation16] and risk assessments for hormone-related cancers [Citation17]. The integration of PRS in glaucoma management, therefore, represents an important step toward more individualized care, drawing on successful precedents in other areas of medicine.
5. Healthcare allocation
As a corollary to using PRS for identification of those at high risk of disease, quantification of genetic risk can allow for reduction of unnecessary healthcare utilization. Currently, we lack the ability to differentiate patients at risk of severe or progressive disease from the many glaucoma suspects unlikely to develop vision loss. The inability to predict disease course using basic clinical risk factors such as enlarged vCDR and ocular hypertension results in many unnecessary clinic visits for those who are unlikely to progress causing overwhelmed ophthalmic and glaucoma clinics and significant healthcare costs. For example, in 2009, total glaucoma payments by Medicare alone were estimated at $748 million [Citation18]. Using the OHTS data, we were able to define a statistically optimal POAG PRS threshold to identify individuals who are at low risk of disease onset. Individuals that fell below our PRS threshold showed significantly lower conversion rates over 20 years with this effect being much more pronounced when grouped by the OHTS baseline risk tertiles (based on age, IOP, vCDR, central corneal thickness, and VF pattern standard deviation). For example, among those in the highest risk, OHTS tertile randomized to observation, 23.8% with low PRS vs 61.1% with high PRS progressed to POAG by 20 years. While more research is needed, this finding underscores the importance of combining PRS with known clinical risk factors for better disease prognostication and resource allocation. To this end, research in oncology has demonstrated the utility of PRS for optimization of cancer screening. Incorporation of PRS in screening strategies for colorectal cancer can aid in identifying individuals at lower risk, potentially reducing unnecessary procedures [Citation19]. By focusing resources on higher-risk individuals, PRS can be used to reduce the burden of glaucoma screening and follow-up in lower-risk populations.
6. Practical challenges
Despite current progress, optimal age-specific POAG PRS thresholds are not currently available, limiting clinical impact. Age is an important consideration in establishing thresholds of disease risk especially for younger patients with lower disease prevalence. Determining risk thresholds is a critical step toward integrating genetic risk assessment into clinical practice. The Electronic Medical Records and Genomics (eMERGE) Network [Citation20] is a multicenter effort to integrate PRS into healthcare by assessing genomic risk in diverse populations, aiming to inform new healthcare actions after the return of genome-informed risk assessments. Such integration involves the calibration of genetic ancestry and the development of clinical reporting tools to ensure the PRS’s are appropriately adjusted for individual patient profiles. Incorporating PRS into electronic health records (EHRs) could allow for the seamless incorporation of genetic risk data into treatment algorithms, aiding clinical decision-making. This integration can serve as a blend of primary and secondary prevention, identifying at-risk individuals before disease onset and enabling early intervention strategies. However, there are challenges in implementing PRS in clinical settings, including ensuring the accuracy of PRS calculation through rigorous quality control and addressing issues of non-normal PRS distribution due to population stratification.
7. Ethical considerations
The use of PRS in guiding glaucoma care involves ethical considerations centered around privacy, informed consent, and accessibility. Genetic testing, integral to PRS, raises concerns about the handling of sensitive information and ensuring that patients fully understand the implications of the tests they undergo. For example, false positives could lead to unnecessary clinical actions and emotional distress. Ensuring the accuracy of PRS is crucial to prevent potential harm that could arise from incorrect risk estimations. Moreover, there is an ethical imperative to address disparities in access to genetic testing, which may otherwise exacerbate health inequities [Citation21].
8. Limitations
Currently, the GWAS used for PRS derivation primarily consist of European ancestry individuals. The relative lack of diversity and under-representation of Hispanic/Latino or African ancestry individuals may decrease predictive accuracy and generalizability in these populations, potentially leaking to misinformed clinical decisions. Indeed, there is some evidence that among African descent individuals, PRS developed from a meta-analysis within the same population can be more predictive than a PRS derived from a larger predominantly European cohort [Citation22].
Developing ethnic-specific PRS is necessary to overcome these limitations and ensure that the benefits of genomic medicine are equitably distributed [Citation21]. However, creating such scores is challenging due to the smaller sample sizes available for non-European populations as well as greater genetic diversity within African ancestry populations. Achieving a PRS that is applicable across diverse ethnicities requires concerted efforts to increase the diversity of GWAS cohorts, as well as methodological advancements to account for varied linkage disequilibrium patterns and allele frequencies across populations.
9. Expert opinion
While the integration of PRS into clinical practice is still evolving, recent progress suggests that PRS risk stratification is a significant step toward a personalized approach to glaucoma care. In the glaucoma clinic, risk stratification using PRS presents an opportunity to focus clinical care and healthcare resources on those individuals most likely to develop disease, while at the same time reducing the clinical surveillance of those not likely to clinically progress. The focus on ‘high risk’ individuals will maximize health care costs and will likely prevent or delay blindness in those likely to be affected by progressive disease. There is still more work to be done before we reach this important goal. Establishing PRS thresholds for high and low risk is a critically important step as well as understanding the clinical features of those with high risk, including rates of disease progression. Also important is generalizing current PRSs to include data from more diverse GWAS, allowing for improved outcomes in diverse populations. Additionally, ‘absolute’ risk has not been explored and could be an important factor in evaluating PRS across, rather within, populations. However, current research activity is promising and suggests that PRS risk stratification can become a useful tool in the glaucoma clinic, potentially revolutionizing the management of POAG. Ultimately, PRS risk stratification could facilitate earlier therapeutic interventions and more tailored treatment strategies, with the goal of preserving vision in those most genetically susceptible to this complex disease.
Declaration of interest
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
Reviewer disclosures
Peer reviewers on this manuscript have no relevant financial or other relationships to disclose.
Additional information
Funding
References
- Wang K, Gaitsch H, Poon H, et al. Classification of common human diseases derived from shared genetic and environmental determinants. Nat Genet. 2017;49(9):1319–1325. doi: 10.1038/ng.3931
- Nakano M, Ikeda Y, Taniguchi T, et al. Three susceptible loci associated with primary open-angle glaucoma identified by genome-wide association study in a Japanese population. Proc Natl Acad Sci USA. 2009;106(31):12838–12842. doi: 10.1073/pnas.0906397106
- Han X, Gharahkhani P, Hamel AR, et al. Large-scale multitrait genome-wide association analyses identify hundreds of glaucoma risk loci. Nat Genet. 2023;55(7):1116–1125. doi: 10.1038/s41588-023-01428-5
- Mak TSH, Porsch RM, Choi SW, et al. Polygenic scores via penalized regression on summary statistics. Genet Epidemiol. 2017;41(6):469–480. doi: 10.1002/gepi.22050
- Ge T, Chen C-Y, Ni Y, et al. Polygenic prediction via Bayesian regression and continuous shrinkage priors. Nat Commun. 2019;10(1):1776. doi: 10.1038/s41467-019-09718-5
- Privé F, Arbel J, Vilhjálmsson BJ, et al. LDpred2: better, faster, stronger. Bioinforma Oxf Engl. 2021;36(22–23):5424–5431. doi: 10.1093/bioinformatics/btaa1029
- Moser G, Lee SH, Hayes BJ, et al. Simultaneous discovery, estimation and prediction analysis of complex traits using a bayesian mixture model. PLOS Genet. 2015;11(4):e1004969. doi: 10.1371/journal.pgen.1004969
- Zhou X, Carbonetto P, Stephens M, et al. Polygenic modeling with bayesian sparse linear mixed models. PLOS Genet. 2013;9(2):e1003264. doi: 10.1371/journal.pgen.1003264
- Sekimitsu S, Xiang D, Smith SL, et al. Deep ocular phenotyping across primary open-angle glaucoma genetic burden. JAMA Ophthalmol. 2023;141(9):891–899. doi: 10.1001/jamaophthalmol.2023.3645
- Fan BJ, Bailey JC, Igo RP, et al. Association of a primary open-angle glaucoma genetic risk score with earlier age at diagnosis. JAMA Ophthalmol. 2019;137(10):1190–1194. doi: 10.1001/jamaophthalmol.2019.3109
- Craig JE, Han X, Qassim A, et al. Multitrait analysis of glaucoma identifies new risk loci and enables polygenic prediction of disease susceptibility and progression. Nat Genet. 2020;52(2):160–166. doi: 10.1038/s41588-019-0556-y
- Singh RK, Zhao Y, Elze T, et al. Polygenic risk score improves prediction of primary open angle glaucoma onset in the ocular hypertension treatment study. JAMA Ophthalmol. 2024 Mar 14;Online ahead of print:e240151. doi: 10.1001/jamaophthalmol.2024.0151
- Siggs OM, Qassim A, Han X, et al. Association of high polygenic risk with visual field worsening despite treatment in early primary open-angle glaucoma. JAMA Ophthalmol. 2022;141(1):73–77. doi: 10.1001/jamaophthalmol.2022.4688
- Inouye M, Abraham G, Nelson CP, et al. Genomic risk prediction of coronary artery disease in 480,000 adults: implications for primary prevention. J Am Coll Cardiol. 2018;72(16):1883–1893. doi: 10.1016/j.jacc.2018.07.079
- Udler MS, McCarthy MI, Florez JC, et al. Genetic risk scores for diabetes diagnosis and precision medicine. Endocr Rev. 2019;40(6):1500–1520. doi: 10.1210/er.2019-00088
- Natarajan P, Young R, Stitziel NO, et al. Polygenic risk score identifies subgroup with higher burden of atherosclerosis and greater relative benefit from statin therapy in the primary prevention setting. Circulation. 2017;135(22):2091–2101. doi: 10.1161/CIRCULATIONAHA.116.024436
- Mavaddat N, Michailidou K, Dennis J, et al. Polygenic risk scores for prediction of breast cancer and breast cancer subtypes. Am J Hum Genet. 2019;104(1):21–34. doi: 10.1016/j.ajhg.2018.11.002
- Quigley HA, Cassard SD, Gower EW, et al. The cost of glaucoma care provided to medicare beneficiaries from 2002 to 2009. Ophthalmology. 2013;120(11):2249–2257. doi: 10.1016/j.ophtha.2013.04.027
- Saunders CL, Kilian B, Thompson DJ, et al. External validation of risk prediction models incorporating common genetic variants for incident colorectal cancer using UK Biobank. Cancer Prev Res Phila Pa. 2020;13(6):509–520. doi: 10.1158/1940-6207.CAPR-19-0521
- Lennon N, Kottyan LC, Kachulis C, et al. Selection, optimization and validation of ten chronic disease polygenic risk scores for clinical implementation in diverse US populations. Nature Med. 2024;30(2):480–487. doi: 10.1038/s41591-024-02796-z
- Adeyemo A, Balaconis MK, Darnes DR, et al. Responsible use of polygenic risk scores in the clinic: potential benefits, risks and gaps. Nat Med. 2021;27(11):1876–1884. doi: 10.1038/s41591-021-01549-6
- Verma S, Gudiseva HV, Chavali VRM, et al. A multi-cohort genome-wide association study in African ancestry individuals reveals risk loci for primary open-angle glaucoma. Cell. 2024;187(2):464–480.e10. doi: 10.1016/j.cell.2023.12.006
