ABSTRACT
Discrete-event simulation (DES) is a widely used computational method in health services and health economic studies. This scoping review investigates to what extent authors share DES models and audits if sharing adheres to best practice. The Web of Science, Scopus, PubMed, and ACM Digital Library databases were searched between January 1 2019 till December 31 2022. Cost-effectiveness, health service research and methodology studies in a health context were included. Data extraction and audit were performed by two reviewers. We measured the proportion of literature that shared models; we report analyses by publication type, year of publication, COVID-19 application; and free and open source versus commercial software. Out of the 564 studies included, 47 (8.3%) cited a published computer model, rising to 9.0% in 2022. Studies were more likely to share models if they had been developed using free and open source tools. Studies rarely followed best practice when sharing computer models. Although still in the minority, healthcare DES authors are increasingly sharing their computer model artefacts. Although commercial software dominates the DES literature, free and open source software plays a crucial role in sharing. The DES community can adopt simple best practices to improve the quality of sharing.
1. Introduction
1.1. Healthcare discrete-event simulation
In healthcare, discrete-event simulation (DES) is the most used simulation method for modelling (Philip et al., Citation2022; Roy et al., Citation2021; Salleh et al., Citation2017; Salmon et al., Citation2018). DES has proven useful within the field of health as it can be used to model patient care pathways, optimise health service delivery, investigate health queuing systems, and conduct health technology assessment.
It has been applied to a wide variety of important clinical and health problems such as stroke care (Lahr et al., Citation2020), emergency departments (Mohiuddin et al., Citation2017), chronic obstructive pulmonary disease (Hoogendoorn et al., Citation2021), sexual health (Mohiuddin et al., Citation2020), reducing delayed discharges (Onen-Dumlu et al., Citation2022), mental health waiting times (Noorain et al., Citation2019), critical care (Penn et al., Citation2020), managing health services during the COVID-19 pandemic (Yakutcan et al., Citation2022), and end of life care (Chalk et al., Citation2021). Healthcare DES computer models are often complex research artefacts: they are time-consuming to build, depend on specialist software, and logic may be difficult to describe accurately using words and diagrams alone (Monks et al., Citation2019).
1.2. Published computer models: Study motivation
To enhance transparency of model logic and offer others the potential to understand, learn from, or reuse a model, one option available to authors of healthcare DES studies is to openly publish the computer model. We define a computer model to be either a model written in a specialist simulation software package, or a model written in a general purpose programming language. The computer model is an executable artefact that is an implementation of the study conceptual model (Robinson, Citation2014) and is used for experimentation.
The current extent of model sharing and practice of sharing DES computer models in the healthcare literature is unknown. To understand if and how authors of DES studies are sharing their models, draw lessons, and evaluate if this can be improved to benefit the wider community, we conduct a review of the contemporary DES literature between 2019 and 2022 inclusive. Reviews in other computational fields report that the sharing of model code and files has historically been low (Collberg & Proebsting, Citation2016; Janssen et al., Citation2020; Rahmandad & Sterman, Citation2012; Stodden et al., Citation2018). The closest of these fields to our review in healthcare DES is in the field of Agent (or Individual) Based Simulation (Janssen et al., Citation2020). The study examined 7500 articles reporting agent-based models and found only 11% of articles shared model code, although there was an upward trend: 18% of ABS publications were found to share their model in some form by 2018. This study applies a similar approach to Janssen et al. (Citation2020) in the healthcare DES literature.
2. Sharing of computer models
2.1. Sharing models is a subset of reproducibility
Our focus in this study is on the practice of sharing healthcare DES computer models: to what extent do health researchers openly share their DES computer models, how do they do it, and what actions could the DES community take to improve what is shared? We consider the open publication of models to be a subset of, and complementary to, the broader topic of tackling the reproducibility of computational analyses and modelling. There has been a long-standing effort to provide incentives for authors to make their computational work reproducibile (Ayllón et al., Citation2021; Grimm et al., Citation2010; Heroux, Citation2015; Marco & Janssen, Citation2008; Monks et al., Citation2019; Reinhardt et al., Citation2018; Ruscheinski et al., Citation2020; The Turing Way Community, Citation2022). One of the most well known of these within the modelling and simulation community is the Association of Computing Machinery’s (ACM) Reproducible Computational Results (RCR) initiative (https://www.acm.org/publications/policies/artifact-review-and-badging-current). The RCR is an optional extra peer review process for authors who publish in ACM journals. Computational artefacts, i.e., models or algorithms, are peer reviewed by specialists and author publications are awarded badges based on the results. ACM RCR badges include: artefacts evaluated (as functional or reusable), artefacts available (deposited in a FORCE-11 compliant (Smith et al., Citation2016) archive such as the Open Science Framework) and Results Validated (either using the author provided artefacts or a higher level using independent methods).
Initiatives such as RCR are limited to specific journals, but health researchers publish, and may share DES models, in a wide variety of outlets. For example, mathematical, medical and clinical, Health Economic, health policy, and Operational Research journals, as well as specialist conferences that publish full peer-reviewed papers (such as the Winter Simulation Conference). In these non RCR supported journals, it is unlikely that model artefacts are peer reviewed. Those authors that share models may instead be guided by discipline norms, journal rules, open research guides such as the Turing Way (The Turing Way Community, Citation2022), or one of several DES reporting guidelines (Eddy et al., Citation2012; Monks et al., Citation2019; Zhang et al., Citation2020).
The DES reporting guidelines take different approaches to publication of DES computer models. The International Society for Pharmacoeconomics and Outcomes Research and the Society for Medical Decision Making (ISPOR-SDM; Eddy et al. (Citation2012)) encourage authors to make non-confidential versions of their computer models available to enhance transparency, but state that open models should not be a formal requirement of publication. The task-force note a number of reasons why code might not be able to be shared including intellectual property and cost. The Strengthening the Reporting of Empirical Simulation Studies (STRESS-DES; Monks et al. (Citation2019)) guidelines takes the position that model code is an enhancement to transparency, not a requirement. The STRESS checklist asks for detailed information on the software environment and computing hardware used to execute the model. Section 6 goes further and requires a statement on how the computer model can be accessed. This is intended to prompt authors to think about enhanced transparency, and enhance publication in journals that do not ask for “code and data availability statements”. The reporting checklist developed by Zhang et al. (Citation2020) focuses only on logic and validation reporting and does not prompt users for information on model code.
One exception to the position of publication as an enhancement versus requirement for transparency is perhaps models tackling Covid-19. At the start of the coronavirus pandemic, the lack of transparency and access to epidemiological model code used to inform economic and public health policy contributed to public confusion and polarisation. This has led to some calling for open publication of all model code related to any aspect of COVID-19 (Sills et al., Citation2020).
2.2. State-of the art practices for sharing computer model artefacts
The topic of sharing code, simulated experiments, computer model artefacts, and the reproducibility of published results is a live topic in other computational fields such as neuroscience, life sciences, and ecology (Cadwallader & Hrynaszkiewicz, Citation2022; Eglen et al., Citation2017; Halchenko & Hanke, Citation2015; Heil et al., Citation2021; Krafczyk et al., Citation2021). Outside of the academic literature there are recent community-driven guides, standards, and digital repositories. These include the Turing Way developed by the Alan Turing Institute (The Turing Way Community, Citation2022), the Open Modelling Foundation standards (https://www.openmodelingfoundation.org/), and the ability to deposit models using the Network for Computational Modeling in Social and Ecological Sciences (CoMSES Net; https://www.comses.net/). The state-of-the-art for sharing model artefacts is an emerging and evolving field; although in biostatistics it has been talked about as far back as 2011 (Peng, Citation2011). Although the recent literature is disparate, when brought together, the literature agrees on a number of practices that benefit the ability of others to find, access, reuse, and freely adapt shared model artefacts.
Contemporary sharing of computer model artefacts is best done through a digital open science repository that has FORCE11 compliant citation (Smith et al., Citation2016) and guarantees on persistence of digital artefacts (Lin et al., Citation2020). Examples include Zenodo (https://zenodo.org/); Figshare (https://figshare.com/); the Open Science Framework (https://osf.io/); and CoMSES Net. Deposited models are provided with a permanent Digital Object Identifier (DOI) that can be used to cite the artefact. Researchers should already be familiar with DOIs as they are minted and allocated to published journal articles. An example is 10.1016/j.envsoft.2020.104873 that identifies an article by Janssen et al. (Citation2020). The advantage of this approach is that the exact code that is cited in the journal article is preserved (authors are free to work on new versions of the code). A related concept is that of the Open Researcher and Contributor Identifier (ORCID; Taylor et al. (Citation2017). This is a unique identifier for an individual researcher. A trusted archive will accommodate ORCIDs within the meta-data of a deposited artefact: providing an unambiguous permanent link back to the authors of the artefact. This offers an improvement over email addresses listed with a journal article that may become outdated shortly after publication.
Published models should also be accompanied by an open licence (Eglen et al., Citation2017; Halchenko & Hanke, Citation2015; Heil et al., Citation2021). A licence details how others may use or adapt the artefact(s), as well as re-share any adaptations and credit authors. At a minimum a licence specifies the terms of use for a model, and waives the authors of any liability if the artefact is reused. There are many types of standard licence to choose from. For example, licensers of models might choose between a permissive-type licence (e.g., the MIT; or BSD 2-Clause) or a copyleft type licence (e.g., GNU General Public Licence v3). An alternative that is often used with open data, and open access publication, but also relevant for models are Creative Commons licences such as the CC-BY 4.0 (Taylor et al., Citation2017).
Permissive and copyleft licences are also used by DES packages developed using Free and Open Source Software (FOSS). Note that FOSS here is more than open source code. It grants the freedom for users to reuse, adapt, and distribute copies however they choose. Examples include R Simmer (GPL-2), SimPy (MIT), and JaamSim (Apache 2.0). For an overview of FOSS packages for DES see (Dagkakis & Heavey, Citation2016).
To maximise the chances that another user can execute a computer artefact, a model’s dependencies and the software environment must be specified (Heil et al., Citation2021; Krafczyk et al., Citation2021). This can be challenging: many computational artefacts rely on other software that may be operating system specific. Formal methods exist to manage dependencies. Complexity can range from package managers, such as “conda” or “renv”, to containerisation (where a model, parameters, an operating system, and dependencies are deployed via a container and software such as Docker), to Open Science Gateways that allow remote execution (Taylor et al., Citation2017). Such methods may be best suited to computational artefacts written in codefor example, a simulation package in Python, or R. Models developed in commercial Visual Interactive Modelling packages such as Arena or Simul8 rely on software with strict proprietary licencing stipulations (i.e., paid licences), but the software and operating system versions can be reported within the meta-data of the deposited artefact. Several commercial simulation packages now provide cloud versions of their software where users may upload a computer model and allow others to execute it without installation. However, such tools do not adhere to the guarantees provided by a trusted digital repository such as Zenodo.
Execution of a computer model artefact should be guided by a clear set of instructions: for example, the inclusion of a README file that includes an overview of what a model does, how to execute it, and how to vary parameters (Cadwallader & Hrynaszkiewicz, Citation2022; Eglen et al., Citation2017). Documentation of models developed using code only could be enhanced with notebooks that combine code and explanation (Ayllón et al., Citation2021; The Turing Way Community, Citation2022).
Finally, if coded models are to be trusted, reused, or adapted, then some form of testing and verification should be included with the shared model (The Turing Way Community, Citation2022). Test driven development is one option for the simulation community (Onggo & Karatas, Citation2016).
2.3. Time, effort, and alternatives
The state-of-the-art methods and benefits outlined above do come at the cost of time and effort. Publishing a computer model artefact along with a journal article may also prompt authors to clean up code and models ready for sharing. There is some evidence that the time authors are willing to spend on this varies with experience, with more established authors being willing to spend more time than those with fewer publications (Cadwallader & Hrynaszkiewicz, Citation2022). Authors may choose to adopt one or a combination of the practices recommended by the literature. More complex methods require more effort. For example, in a small trial, the journal Nature Biotechnology reported that it took a median of 9 days for authors to set up an online executable version of their computational artefacts using the containerisation and compute services provided by Code Ocean (Nature Biotechnology, Citation2019). In contrast, depositing code or a model in a trusted digital archive such as Zenodo requires only the time to upload the data and effort to add metadata such as ORCIDs.
A simple alternative option to direct publication of computer models is to use a Data Availability Statement (DAS). A DAS provides a way for authors to describe how others might access the computational artefacts used within the research. For example, “the materials used within this study are available from the corresponding author on reasonable request”. A substantial downside is that DAS statements offering to share are frequently not honoured, even in journals mandating reproducibility standards (Collberg & Proebsting, Citation2016; Gabelica et al., Citation2022; Janssen et al., Citation2020; Stodden et al., Citation2018). In the largest simulation review to date, the study researchers contacted all authors of ABS papers that included a DAS within their paper. They received a response from less than 1% of authors to provide their code; the majority of these indicated that their model is no longer available, or failed to provide a runnable version (Janssen et al., Citation2020). Outside of simulation modelling other disciplines have reported varying results when contacting authors of papers with DAS statements, with positive responses of 7% (Gabelica et al., Citation2022), 33% (Collberg & Proebsting, Citation2016), and 44% (Stodden et al., Citation2018).
3. Review aims
We aim to determine to what extent authors of contemporary DES health studies share computer models, where models are shared, and how it is done.
3.1. Primary research questions
What proportion of DES healthcare studies share code?
How is sharing affected by FOSS, Covid-19, publication type, and year of publication?
What proportion of studies make use of a reporting guideline?
What methods, tools, and resources did authors use to share their computer models and code?
To what extent do the DES health community follow best practice for open science when sharing computer models?
To what extent can the healthcare DES community improve its sharing of computer models?
For our last two research questions we conducted a best practice audit of the shared computer models only.
4. Methods
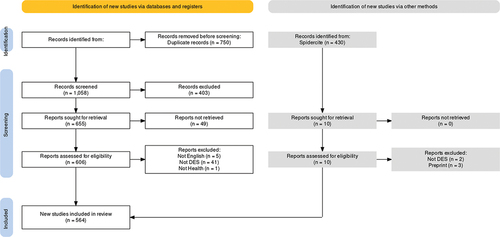
The reporting of our study conforms to the PRISMA reporting guidelines (Page et al., Citation2021).
4.1. Search strategy
The databases Scopus, PubMed Central, ACM digital library, and Web of Science (WoS) were searched to retrieve articles on DES applications in healthcare from 2019 to 2022. Scopus has previously demonstrated wide coverage for systematic reviews of modelling and simulation in multiple domains, including healthcare. We searched PubMed using general terms rather than MeSH (Medical Subject Headings) Terms, which have been found to produce large numbers of irrelevant results for operations research and operations management papers. The PubMed search allowed inclusion of healthcare simulation applications published in healthcare journals. We included WoS as others have shown that it can identify different articles to Scopus (Martín-Martín et al., Citation2018). We used the ALL keyword to search all fields and broaden the search in WoS as much as possible to pick up any papers we may have missed across SCOPUS and PubMed. We included the ACM Digital Library to ensure our search covered Transactions on Modelling and Computer Simulation and ACM healthcare journals.
The key search terms included: “health”, “healthcare”, or “patient”, and “discrete”, “event”, and “simulation”, in the title, abstract, and/or keywords. Search years were limited to 2019–2022, and Scopus terms were limited to articles, excluding letters, notes, editorials, conference reviews, short surveys, data papers, and erratums.
We limited our search years to allow time for the March 2018 publication of the STRESS guidelines to take effect. STRESS includes the requirement to include a code access statement (Section 6). This also follows the publication of an Open Science checklist, and the February 2019 release of The Turing Way, a guide to reproducible research published by the UK’s National Institute for Data Science. We also considered the ACM RCR initiative, and the impact this has had on available DES models. An exploratory search for papers badged as artefacts available within any field containing “discrete event simulation” between January 1 2016 and December 31 2018 returned seven papers, none of which related to healthcare DES.
We also conducted backwards and forwards citation chasing to enhance our search. Backwards citation chasing was conducted in context during data extraction. Where studies cited models in prior work we followed that up and reviewed the paper. Forward citation chasing was conducted using spidercite (https://sr-accelerator.com) on all papers that cited a published model.
We include all search strings used within our online supplementary material (Monks & Harper, Citation2023a)
4.2. Study selection
4.2.1. Identifying duplicate records
We combined all database searches and then removed all duplicate records. Duplicates were matched using a two-step process. First, we used our own software “pydedup” (https://github.com/TomMonks/pydedup) to automate the majority of the process to remove duplicates. All publication titles were stripped of punctuation and whitespace with only unique titles being returned. Second, we imported the remaining records into the reference software Zotero for manual matching of the remaining duplicates. This enabled us to identify the remaining small number of close matches, e.g., records from one database that contained mathematical symbols that had a representation in words from another database. We manually merged the duplicates in this second stage.
4.2.2. Abstract, title, and keyword screening
The titles, abstracts, and keywords of all unique records were then screened by one member of the team using the reference management software Zotero. We tagged each record as a “Yes” or “No”. Excluded records were re-reviewed in a second pass to mitigate the risk of missing key papers.
Articles were included in the second stage of review and data extraction if the title, abstract, or keywords indicated that a healthcare patient-based DES model was used in an applied setting, cost-effectiveness study, or methodology study. We included hybrid DES models, for example, those studies that hybridised agent-based simulation and DES. Where there was any uncertainty, for example, in the simulation method used, we included the paper for full text reviewed in the second stage.
4.3. Data extraction
Where possible we always viewed papers at the publishers’ site so we could identify, download, and access any supplementary material or information that may not be directly included in the article PDF. If an article built on and cited previously published work/models, we followed up the paper in an attempt to complete data extraction. Any uncertainty was managed by dual review, and an additional proportion of papers were reviewed by both authors (23%). A previous review (Janssen et al., Citation2020) handled “model code is available upon request” by emailing authors directly. Given the low response rate found in their study (<1%) we chose not to follow up in this study.
We extracted the following data from each article:
Type of article: journal, conference (e.g., Winter Simulation Conference), or book.
Name of reporting guidelines used (if present).
Name of simulation software, if reported.
If the software was commercially or FOSS licenced.
Methods of sharing (see below).
Standard health or COVID-19 study.
For the method of sharing the computer model, we adopted the sharing classification developed by Janssen et al. (Citation2020):
Archive (Zenodo, Figshare, Open Science Framework (OSF), Dataverse, Datadryd, CoMSES, university archives, etc.);
Repository (Github/Lab/Bitbucket, Sourceforge, Google code, etc.);
Journal supplementary material, personal and organisational (own website, Dropbox, ResearchGate, Google Drive, Amazon CloudFront, etc.);
Platform (e.g., vendor website – e.g., Simul8 Cloud, Anylogic Cloud, etc. This might also include free infrastructure to support FOSS, e.g., StreamLit.io, RShiny.io, Binder, or hybrid free/paid infrastructure like Code Ocean).
4.4. Best practice audit
We split our assessment of the best practice of how models were shared into two groups: models developed using a coding language or framework (e.g., MatLab, R, or Python based) and models developed in commercial off the shelf Visual Interactive Modelling software (VIM; e.g., Arena, Anylogic, or Simio).
We know of no general best practice auditing tools for sharing of simulation code and models. However, general guidance for open science and reproducible research is available from the Turing Way (The Turing Way Community, Citation2022) developed by The Alan Turing Institute: the UK’s official institute for data science and AI. The Turing Way is a data science community project and at the time of writing (2023) has 290 contributors and reviewers. We reviewed the Turing Way checklists for Open Research, Licensing, Reproducible Environments, and Code Testing and selected relevant quality criteria. We enhanced this list by validating it against two further sources of guidance related to open science. First, we adapted the high-level open scholarship recommendations for modelling and simulation (Taylor et al., Citation2017). Second, the Open Modelling Foundation’s (OMF) minimal and ideal reusability standards .Footnote1 The OMF defines reusability as “implicitly includ[ing] usability and focuses on the ability of humans and machines to execute, inspect, and understand the software so that it can be modified, built upon, or incorporated into other software”. These latter sources added two further items not specifically listed in the Turing Ways checklists.
We excluded some Turing checklists from our review as they were not relevant to the quality of model sharing. For example, the Research Data Management checklist is focused on raw data that in a typical DES study would have been used to derive model parameters. Our focus is on the sharing of the model itself and not underlying raw data. Another example is the Turing’s recommendations to publish open notebooks containing all details of experiments. This was on the basis that the modelling and simulation community might adopt a large number of approaches and tools to manage their models and artefacts. Instead, we included a broader item checking for instructions to execute experiments in any format. We also excluded most of the OMF’s ideal reusability standards including the use of containerisation tools (such as Docker or Podman) in order to keep our best practice criteria simple for the Modelling and Simulation (M&S) community.
We emphasise that our aim is to audit the practice of the sharing of DES computer model artefacts, not to test if model artefacts reproduce the results reported within a paper. As such we are not conducting a full ACM RCR style peer review. We note that metrics within our audit overlap with those others have listed as requirements for reproducibility of computational studies (Heil et al., Citation2021; Krafczyk et al., Citation2021; Venkatesh et al., Citation2022). We also note an overlap with the ACM RCR artefact available badge (in terms of archiving in a digital open science respository) and part of the requirements for an artefact evaluated as functional badge (in terms of documentation). We list these in , to illustrate which data were extracted for the coding and VIM groups and detail the provenance of the items.
Table 1. Best practice audit: metrics and sources.
We extracted the type of licence included with each shared model. For example, GPL-3 or MIT. When no licence was included, we recorded this as “None”. When a model was published as journal supplementary material, we assigned the same licence as applied to the paper if it was not explicitly stated. For example, if a paper was published under a CC-BY 4.0 licence and there was no explicit licence attached to supplementary material we assumed the same licence for the model.
4.5. Analysis environment
All analysis code was written in Python 3.9.15. Data cleaning and manipulation were done using pandas (McKinney, Citation2010) and NumPy (van der Walt et al., Citation2011). All charts were produced with MatPlotLib (Hunter, Citation2007). Identification of duplicate references was conducted using “pydedup” (available https://github.com/TomMonks/pydedup). Notebooks were produced using Jupyter Lab v3.5.2 (Kluyver et al., Citation2016). The analyses were run on Intel i9-9900K CPU with 64GB RAM running the Pop!_OS 20.04 Linux. The references were managed via Zotero 6.0.15 (Zotero, Citation2011).
We generated the PRISMA 2020 compliant flow diagram of our scoping review using an online R Shiny app (Haddaway et al., Citation2022)
4.6. Availability of code and data
We have archived the Python code, the main, and the best practice datasets generated in this study (Monks & Harper, Citation2023b). We provide an online supplementary appendix in the form of a Jupyter Book https://tommonks.github.io/des_sharing_lit_review. Live code and data can also be accessed via GitHub https://github.com/TomMonks/des_sharing_lit_review. Instructions are included to allow online execution of the code to reproduce the results in this paper using Binder.
We have also created a Zotero online database that contains the studies that shared computer simulation models https://www.zotero.org/groups/4877863/des_papers_with_code/library
5. Results
We were able to access 564 potentially eligible papers where a DES study was used in a health context. During data extraction, we excluded further 47 studies (see ); the vast majority of these studies were excluded as the full text revealed that DES was not used.
5.1. Study characteristics
Out of the 564 included studies, 431 were journal articles; 109 were full conference papers; and 24 were book chapters. A total of 69 studies investigated COVID-19 using DES. The majority of the studies used commercial off-the-shelf simulation software (n = 340); the top three being ARENA (n = 124), Anylogic (n=78), and Simul8 (n = 51). A total of 60 studies were classified as “unknown” as they did not report the software used. This left a minority of studies (n = 101) using FOSS tools. The most popular FOSS languages were R (n = 50) and Python (n = 33).
5.2. What proportion of studies shared model code?
In total, 47 (8.3%) of the included 564 studies cited an openly available DES computer model that was used to generate their results. Of these 47, the majority (n = 29) of models were developed using FOSS tools. The proportion of computer models published varied by the type of publication. A total of 42 (9.7%) journal articles cited a published DES model; full conference papers and book chapters cited a model in ∼4% of cases. summarises overall findings and is broken down by journal, conference, and book publication of models.
Table 2. Sharing of models by article type.
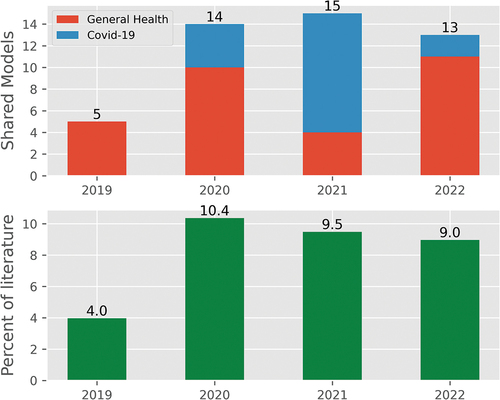
The proportion of the literature that cited an openly available DES computer model increased from 4.0% in 2019 to 9.0% in 2022; the years 2020–2022 were similar in proportion. illustrates the number of shared healthcare DES models by year split into Non-Covid and Covid as well as the proportion of the included studies. The number of DES models published per year was similar (mean = 141.0); with 2021 reporting a small increase in numbers relative to the other three (158), and also the largest number of COVID-19 models (n = 35; the mean number between 2020 and 2022 is 23.0).
Studies were more likely to share computer models if they had been developed using FOSS tools (29 of 101 studies; 28.7%), or if they were tackling COVID-19 (17 of 69 studies; 24.6%).
5.3. What proportion of the studies used a reporting guideline?
In general, the use of reporting guidelines in DES healthcare studies was rare. A total of 72 (12.8%) studies used models published in articles that mentioned a known simulation reporting guideline or general checklist. The most frequently mentioned were one of the ISPOR guidelines (n = 37; 6.6%); followed by the STRESS guidelines (n = 22; 3.9%).
5.4. What methods, tools, and resources did authors use to share their models and code?
summarises the method of model archiving split into models written in a programming language (either FOSS or commercial) versus models written in a VIM package. A total of 31 (66.0%) models were shared as code; while 16 (34.0%) models developed using a VIM were shared.
Table 3. Janssen et al. classification of model archiving.
Model files generated using a VIM were primarily shared as journal supplementary material (n = 4; typically just the model file), or via AnyLogic Cloud (n = 6; we found no evidence of any other simulation vendor cloud platform in the included studies). Code models were often shared via GitHub (n = 19), but also as journal supplementary material (n = 6) and organisational website (n = 4). A total of five models were deposited in a digital open science archive. A small number of authors used multiple methods to archive and share their models. For example, an institutional archive and GitLab (Anagnostou et al., Citation2022).
5.5. To what extent do the DES health community follow best practice for open science when sharing models?
summarises the results of the best practice audit; totals are split by code and VIM-based models. In general, computer models and artefacts were published without a DOI (n = 7); rarely included ORCIDs for authors (n = 6); rarely included an open licence (n = 21); were mostly supported by a README file (n = 28); rarely included documentation detailing how to run the model (n = 15); provided no form of formal or informal dependency management (n = 21); did not include any evidence of model testing (n = 3); were almost all downloadable (n = 38); and rarely executable via a cloud-based platform (n = 10).
Table 4. Best practice audit results.
6. Discussion
6.1. Summary of main results
We found that a minority (n = 47; 8.3%) of the 564 papers reviewed shared a simulation model either as a code or a visual interactive simulation file. The trend is that sharing has increased during the study period: increasing from 4% in 2019 to 8%–10% between 2020 and 2022. Studies published in journal articles were also more likely to share their computer model (9.7%) compared to full conference papers (4%). This might reflect that conference papers were work in progress and authors were not ready to share their work. However, we note that one model shared via a Winter Simulation Conference paper rated very high in our best practice audit (Anagnostou et al., Citation2022). Regardless of the way the split is calculated, sharing of health DES models does not match the findings from Agent-Based Simulation where 18% of studies shared models in 2018 (Janssen et al., Citation2020). It appears that 4 years later DES models in health are shared far less often than ABS models in general.
Of the 47 models shared the majority were implemented using a FOSS simulation tool such as R Simmer. This is perhaps not surprising given the freedom FOSS licences grant authors and other researchers or healthcare services that may opt to reuse the work. We also found that only a minority (∼25%) of models aiming to support health services to care for patients during the COVID-19 pandemic were shared. It would appear that healthcare DES fell short of the sharing standard other fields had called for during the pandemic (Sills et al., Citation2020). One positive is that COVID-19 models were shared more often than computer models in general healthcare settings.
Over 65% (31 out of 47) of the DES models shared via a publication were developed using a code-based simulation package. Sharing was most often done by GitHub. Others have stated that open science archives are the gold standard for research artefact preservation (Heil et al., Citation2021; Janssen et al., Citation2020; Krafczyk et al., Citation2021). An open science archive can mint a DOI and provide long-term storage guarantees. Disadvantage of GitHub (and other cloud-based version control tools) is that there are no guarantees on how long code will remain stored, and it is unclear which version of the code was used in a publication. We only found two instances of code being managed on both GitHub/Lab and deposited in an open science archive. Models built using commercial VIM software were shared less often (16 out 47 models). The majority of these were attached to journal supplementary material or via AnyLogic Cloud. The latter of these approaches will likely lead to broken links in the future due to changes in commercially provided infrastructure and licencing.
Although FOSS simulation packages made up the majority of the shared/archived models, we identified that it is still commercial off-the-shelf simulation packages that dominate the academic literature overall. We confirmed a prior result (Vázquez-Serrano et al., Citation2021) that Arena was the most used package in healthcare DES, with AnyLogic and Simul8 also being in regular use. Definitively explaining why model sharing is most associated with FOSS packages is not possible at this time. We can confirm that this is not related to the effort needed to perform basic archiving. If we take a digital open science repository such as Zenodo, we note that there is no additional work to archive a model file built in a commercial package to one coded in Python. Our speculations are that this is (i) related to the general philosophy of freedom built into FOSS tools. That is, FOSS tools attract certain types of users and these users are more likely to share their model artefacts. (ii) FOSS tools are more likely to be code based (although not exclusively) and that lends itself to managing code via repositories such as GitHub.
Keeping to the topic of simulation software, we found that 11% of studies failed to report what simulation software was used at all. This result was also observed by Vázquez-Serrano et al. (Citation2021) and by Brailsford et al. (Citation2019) in their exhaustive review of hybrid simulation models. Our result is somewhat more striking than these previous reviews as we focused our review post publication of the Strengthening the Reporting of Empirical Simulation Studies checklist that recommends authors report software used. An explanation is that our review found that the uptake of reporting guidelines within healthcare DES was low. Cost-effectiveness studies modelling individual patient trajectories using DES were the most likely to include some mention or cite using reporting guidelines, typically one of the ISPOR publications.
6.2. Summary of best practice audit results
The best practice audit revealed that the tools and methods of sharing DES models could be greatly – and in many cases, easily – improved within health and medicine.
6.2.1. Open scholarship
We found that very few models were deposited in an open science repository such as Zenodo, OSF, or even an academic institution. This meant that these model artefacts had no guarantees of long-term storage and could not be easily cited. In the case of FOSS or code-based models, authors primarily opted for linking to code version control repository such as GitHub or GitLab. While this allowed authors to share their code, it is possible that these peer-reviewed links will be invalid in future years. We also found instances where model binary files used within commercial simulation software such as Anylogic were inappropriately committed to GitHub instead of an open science repository.
Similarly, the review found limited use of ORCIDs in repositories, archives, or platforms that shared DES models. This meant there was no robust way for the models artefacts to be linked to a researcher and their portfolios.
The majority of DES models were shared without an open licence. If we split the models into code-based and VIM-based simulation software, we found that VIM models were the least likely to include a clear open licence. The VIM models that did have an open licence were typically journal supplementary material and by default adopted the licence applied to the journal article as a whole. Without a licence authors retain exclusive copyright to their research artefacts. This means that other researchers and potential users can view the code/model but not reuse it or adapt it.
6.2.2. Tools to facilitate reuse of models
In general, we found that only a minority of DES models are stored with any form of clear instruction to run the model. This was less likely for VIM models (20%) than code-based models (40%). As we did not have licenced copies of all the commercial software used, it is possible that instructions were contained within the models themselves. If this is indeed the case, the authors did not describe this within papers or repositories.
Dependency management was in general of a poor quality or not used at all. Only 23% of code-based simulation models had any formal dependency management, while VIM-based models fared slightly better informally by stating the version of the software used. This latter result is perhaps due to the simplicity of stating which version of the commercial “of the shelf” DES software you are using versus a complex software environment for code-based DES model. However, we note that in many cases authors did not even state the version of R or Python they were using.
Surprisingly, we found that several of the VIM-based DES models shared were not downloadable – even when an author stated it could be downloaded. These were all hosted on cloud-based services where the model was interactive and executable to some extent. We found that some models shared through the same platforms were downloadable, so a plausible explanation is that this was just a setting that was missed by study authors.
Although testing and model verification is standard practice and covered by many simulation text books, we found very little evidence of model testing both of models written in a coding language and commercial software. Given the other findings, it is not surprising that there was limited evidence of testing among the shared models, although this does not mean that testing activities did not take place. An explanation could be that model testing had been completed informally and incrementally as models were coded.
6.3. Additional observations on sharing
We have four additional areas of learning from our review that are beneficial to report.
6.3.1. Poor quality logic diagrams
We found peer reviewed articles that contained poor quality and low-resolution images of model logic. For example, we found several studies where the ARENA simulation software was used and model documentation was based on screenshots of the software as opposed to carefully designed diagrams. These images were often of no use to reporting and sharing: resolution was too low and text was unreadable.
6.3.2. Data availability statements
Where journals mandated, study authors provided a data availability statement within their publication. A generic example of this is “The data used within this study” or “the model inputs” are “available upon reasonable request”. Such statements ignored the availability of the model itself. It is unclear if such an omission is intentional, not considered relevant, or not considered at all due to standard practice in the authors’ home discipline. What is clear is that journal editors and reviewers do not consider models as “data” and as such are not enforcing details on access.
6.3.3. Open science journals and reporting guidelines
Due to journal policy, we expected articles published in open science championing journals such as PLOS1 and BioMedCentral to enforce links to model artefacts. However, this was not always the case. In some cases, we found no model, or an online appendix with a table of input data. We also found that these journals did not enforce reporting guidelines. It may be that authors, reviewers, and editors do not consider these requirements to extend to computational artefacts such as DES models.
6.3.4. Time spent to set up sharing
The majority of DES model sharing we found did not score well in our best practice audit. One explanation is that sharing of the model was only considered at the end of the simulation study or model write-up with minimal time dedicated to the mechanism and quality of sharing. Evidence to support this statement can be taken from the GitHub repositories with a single or small number of commits (updates to code) – a sign that model code was only uploaded at the end of a project. Often without any documentation, licence, instructions to run, information on software dependencies, or link to a paper describing the work.
6.4. Implications
6.4.1. Reviewers
Outside of journals that offer formal RCR style review initiatives, reviewers can still improve the quality of DES model sharing within health and medicine by encouraging authors to follow best practice. Naturally the reviewers of clinical studies, methodology, and health service problems using DES models will have different technical background and knowledge. We also acknowledge that this is an optional activity for reviewers. We therefore propose two levels of light touch review that a study referee may consider when authors of healthcare DES studies cite a shared version of the computer model.
6.4.1.1. Open scholarship review
Non-technical reviewers should focus on the open scholarship of the DES model. Others have recommended that code should enhance study reporting and not be a requirement (Eddy et al., Citation2012; Monks et al., Citation2019). We agree with this position and, if already not included, an open scholarship review could begin by asking authors to include and cite a reporting checklist such as STRESS-DES (Monks et al., Citation2019) to improve the documentation and explanation of models and data.
Our results show that reviewers can improve on this existing base level of open scholarship by encouraging authors to deposit their computer model in an open science archive (for example, their home institution or external archive such as Zenodo or OSF). Reviewers should favour this option over and above journal supplementary material, which is unreliable. We found one instance where authors claimed a model is available as supplementary material, but the journal did not provide a link. Open science repositories mint a DOI, for citation and findability, and provide guarantees on persistence long term. Reviewers can boost the findability of the models by asking authors to include their ORCID details as metadata wherever the model is deposited.
Where necessary, reviewers should also ask authors to provide an open licence with their model. In our study we found that authors provided a mix of open licences. Appropriate choices used by studies included in our review include the MIT licence, GPL-3, and BSD-3. The appropriate licence will depend on the project and what software has been used. Reviewers could signpost authors to journal guidelines (if applicable) or licence guides such as https://choosealicense.com/to support decisions.
Lastly, reviewers should ask authors to provide basic steps to use and run their DES models. This does not need to include a detailed peer review, but instructions should be visible for open scholarship.
The vast majority of DES health models we found were published outside of ACM journals, and, at the time of writing, authors did not have an opportunity to apply for an RCR review, or receive a badge. Although no badge will be issued, a reviewer making our simple open scholarship recommendations can support DES healthcare authors to meet the requirements of the ACM RCR artefact available badge and begin to work towards meeting the requirements of an RCR artefact evaluated functional badge.
6.4.1.2. Longevity review
Given our best practice study findings, more technical reviewers should still prioritise our recommendations for open scholarship. Long term maintainability and reuse of code-based models can be maximised through the use of formal dependency management tools. The exact nature of these tools depends on the language used. R and Python were common tools used for DES in our study. Tools that resolve and install dependencies such as renv and conda can be used here, respectively. For DES models developed in commercial software, reporting guidelines that include items for software should be followed. For example, The Strengthening the Reporting of Empirical Simulation Studies for DES (STRESS-DES) includes a section on software environment.
A longevity review does not guarantee the ability to replicate the results in a paper, but we argue that it improves the chance of artefacts being functional for others to execute with minimal reviewer effort. For example, instead of directly testing model dependencies, reviewers could ask authors to “confirm the method that has been used to manage software dependencies in your study”.
6.4.2. Researchers
Researchers who wish to share their model artefacts will want to minimise time and effort. To mitigate end-of-study time constraints (e.g., due to funding or need to publish results) and increase sharing quality, health researchers should consider open working practices from day 1 of their study (Harper, & Monks, Citation2023) For researchers using FOSS tools to build models, this could be used to set up a GitHub repository (or other cloud version control) workflow. Many guides exist and are free to access, for example, from Software Carpentry (https://swcarpentry.github.io/git-novice/). Repositories can be initially setup as private if needed. Raw study data and patient records should not be stored online. However, aggregate simulation model parameters (either real or synthetic), such as those for statistical distributions, could be included. Authors may wish to consult our best practice audit criteria to support their sharing, for example, to include an open licence and deposit in an open science repository in order to mint a DOI.
We also note that many DES studies are conducted using propriety software, e.g., Arena, with a minority using FOSS packages such as SimPy. While training and time is a consideration in all studies, we encourage researchers who wish to share their models to consider a FOSS simulation tool to enable health services such as the NHS to easily take up and use their work without financial or nuanced licencing blocks. If the upward trend of model sharing continues, there will be more example models for researchers to learn from to build their own.
6.4.3. Funders
The results of our study complement the findings of the UK’s Goldacre review into using health data to support research and analysis (Goldacre & Morely, Citation2022), i.e., funders should incentivise researchers to share their DES models. We echo Goldacre’s call for “UKRI and/or NIHR to launch an open funding call specifically for open code projects in health data science” (Goldacre & Morely, Citation2022). These specific funders are UK based, but could equally apply internationally. One addition to Goldacre’s recommendations, in the context of our DES findings, is that funders could encourage or require researchers to make use of well-maintained FOSS simulation tools that facilitate open working and reuse. This latter requirement could assist in raising the proportion of COVID-19 DES studies where computer models are shared. Lastly, given that time, effort, quality, and knowledge are all issues in sharing DES model artefacts, funders should support work that has the potential for new innovation. This might be to improve quality by standardising computational notebooks such as that proposed by TRACE (Ayllón et al., Citation2021).
6.5. Strengths and limitations
To our knowledge, our study is the first to evaluate the quality of sharing in healthcare DES. A strength of our work is that our best practice audit is simple, effective, and quick to apply to new studies. Given its simplicity and the large amount of guidance on open science and coding available for free online, it is important to recognise that it does show clear deficiencies in the last 4 years of the literature. For example, in open licencing, model testing (an expectation spelled out in any simulation textbook), and long-term storage and access.
Another strength is that we have followed open working practices in this study. All code and data are available, openly licenced, and deposited in Zenodo. We have also taken the additional step of providing an online companion book where code can be run to reproduce the results of the study. This is also deposited in Zenodo to ensure long-term availability.
Our study cannot make any definitive statements about the reproducibility of the studies that shared their code versus those that used a reporting guideline versus those that did neither. Our aim instead was to focus on the practice of sharing and the current deficiencies as defined by gold standard guides such as the Turing Way and Open Modelling Initiative.
Our findings are based on information we found in the publication. We recognise that the model code may have been published online but not mentioned in the article. We feel our approach is the most appropriate as articles are the primary means by which researchers find research studies and their artefacts.
In order to scope computer model and code sharing practices in the DES healthcare literature, we attempted to conduct a broad and inclusive search of well-known databases. While we cannot be certain that we have found all shared computer models, we can attempt to compare our included studies to those included in previous healthcare DES reviews. A direct comparison of the number of studies included with other healthcare DES reviews is difficult due to differing aims, search terms, inclusion criteria, and reporting. We can say that our review included more than double the number of health DES articles of a 2021 review covering 1994 to 2021 (n = 231; Vázquez-Serrano et al., Citation2021). For the years that overlapped (2019–2021) we cross-checked our included studies with those included in the 2021 study (Vázquez-Serrano et al., Citation2021) and found no additional DES articles. Our results also compare favourably to an earlier 2018 article covering healthcare DES between 1997 and 2016 (n = 211; Zhang, Citation2018). This study used very similar search terms to our own but only included WoS and PubMed in their databases, was unable to cross-check with other reviews, and did not conduct backwards and forwards citation chasing.
6.6. Conclusions
Although still in the minority, there is evidence that healthcare DES authors are increasingly sharing their model artefacts. This presents opportunities for others, including early career researchers, and health service practitioners to reuse, learn from, and reproduce results from DES papers. The current trend of sharing DES models is far from perfect. There are many (simple) best practices the community can adopt, such as the use of trusted archives, and documentation, to improve its sharing. Reviewers, researchers, and funders all have a role to incentivise and support authors to do so. A potential barrier to sharing remains time; if sharing of models is going to happen, then planning for this from the start of a project potentially spreads the effort across the study duration. It seems essential that new methods for preparing and sharing high-quality simulations are developed. Finally, although commercial software dominates the DES literature, free and open source software plays a crucial role in sharing and could be considered by authors looking to share models.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
Notes
References
- Anagnostou, A., Groen, D., Taylor, S. J., Suleimenova, D., Abubakar, N., Saha, A., Mintram, K., Ghorbani, M., Daroge, H., Islam, T., Xue, Y., Okine, E., & Anokye, N. (2022). FACS-CHARM: A hybrid agent-based and discrete-event simulation approach for COVID-19 management at regional level. In 2022 winter simulation conference (wsc), Singapore (pp. 1223–1234).
- Ayllón, D., Railsback, S. F., Gallagher, C., Augusiak, J., Baveco, H., Berger, U., Charles, S., Martin, R., Focks, A., Galic, N., Liu, C., van Loon, E. E., Nabe-Nielsen, J., Piou, C., Polhill, J. G, Preuss, T. G, Radchuk, V., Schmolke, A., Stadnicka-Michalak, J., Thorbek, P., & Grimm, V. (2021, February). Keeping modelling notebooks with TRACE: Good for you and good for environmental research and management support. Environmental Modelling & Software, 136, 104932. Retrieved April 25, 2023, from https://www.sciencedirect.com/science/article/pii/S1364815220309890
- Brailsford, S. C., Eldabi, T., Kunc, M., Mustafee, N., & Osorio, A. F. (2019, November). Hybrid simulation modelling in operational research: A state-of-the-art review. European Journal of Operational Research, 278(3), 721–737. Retrieved April 03, 2023, from https://www.sciencedirect.com/science/article/pii/S0377221718308786
- Cadwallader, L., & Hrynaszkiewicz, I. (2022, August). A survey of researchers’ code sharing and code reuse practices, and assessment of interactive notebook prototypes. PeerJ, 10, e13933. Retrieved April 25, 2023, from https://www.ncbi.nlm.nih.gov/pmc/articles/PMC9406794/
- Chalk, D., Robbins, S., Kandasamy, R., Rush, K., Aggarwal, A., Sullivan, R., & Chamberlain, C. (2021). Modelling palliative and end-of-life resource requirements during covid-19: Implications for quality care. British Medical Journal Open, 11(5), e043795. https://bmjopen.bmj.com/content/11/5/e043795
- Collberg, C., & Proebsting, T. A. (2016, February). Repeatability in computer systems research. Communications of the ACM, 59(3), 62–69. https://doi.org/10.1145/2812803
- Dagkakis, G., & Heavey, C. (2016). A review of open source discrete event simulation software for operations research. Journal of Simulation, 10(3), 193–206. https://doi.org/10.1057/jos.2015.9
- Eddy, D. M., Hollingworth, W., Caro, J. J., Tsevat, J., McDonald, K. M., & Wong, J. B. (2012). Model transparency and validation: A report of the ISPOR-SMDM modeling good research practices task force-7. Medical Decision Making, 32(5), 733–743. PMID: 22990088. https://doi.org/10.1177/0272989X12454579.
- Eglen, S. J., Marwick, B., Halchenko, Y. O., Hanke, M., Sufi, S., Gleeson, P., Poline, J.-B., Silver, R. A., Davison, A. P., Lanyon, L., Abrams, M., Wachtler, T., Willshaw, D. J., Pouzat, C., & Poline, J.-B. (2017, June). Toward standard practices for sharing computer code and programs in neuroscience. Nature Neuroscience, 20(6), 770–773. (Number: 6 Publisher: Nature Publishing Group) Retrieved April 25, 2023, from https://www.nature.com/articles/nn.4550
- Gabelica, M., Bojčić, R., & Puljak, L. (2022, October). Many researchers were not compliant with their published data sharing statement: A mixed-methods study. Journal of Clinical Epidemiology, 150, 33–41. Retrieved April 25, 2023, from https://www.sciencedirect.com/science/article/pii/S089543562200141X
- Goldacre, B., & Morely, J. (2022). Better, broader, safer: Using health data for research and analysis. A review commissioned by the secretary of state for health and social care. Department of Health and Social Care. https://www.gov.uk/government/publications/better-broader-safer-using-health-data-for-research-and-analysis
- Grimm, V., Berger, U., DeAngelis, D. L., Polhill, J. G., Giske, J., & Railsback, S. F. (2010, November). The ODD protocol: A review and first update. Ecological Modelling, 221(23), 2760–2768. Retrieved June 02, 2023, from https://www.sciencedirect.com/science/article/pii/S030438001000414X
- Haddaway, N. R., Page, M. J., Pritchard, C. C., & McGuinness, L. A. (2022, June). PRISMA2020: An R package and shiny app for producing PRISMA 2020-compliant flow diagrams, with interactivity for optimised digital transparency and open synthesis. Campbell Systematic Reviews, 18(2), e1230. (Publisher: John Wiley & Sons, Ltd) Retrieved May 06, 2022, from https://doi.org/10.1002/cl2.1230
- Halchenko, Y. O., & Hanke, M. (2015). Four aspects to make science open “by design” and not as an after-thought. GigaScience, 4(1), 31. https://doi.org/10.1186/s13742-015-0072-7
- Harper, A., & Monks, T. (2023, March). A framework to share healthcare simulations on the web using free and open source tools and Python. In Proceedings of SW21 The OR Society Simulation Workshop (pp. 250–260). Operational Research Society. Retreived June 11, 2023, from https://www.theorsociety.com/media/7313/doiorg1036819sw23030.pdf
- Heil, B. J., Hoffman, M. M., Markowetz, F., Lee, S.-I., Greene, C. S., & Hicks, S. C. (2021, October). Reproducibility standards for machine learning in the life sciences. Nature Methods, 18(10), 1132–1135. (Number: 10 Publisher: Nature Publishing Group) Retrieved April 25, 2023, from https://www.nature.com/articles/s41592-021-01256-7
- Heroux, M. A. (2015, June). Editorial: ACM TOMS replicated computational results initiative. ACM Transactions on Mathematical Software, 41(3), 5. 13:1–13 Retrieved June 02, 2023, from https://dl.acm.org/doi/10.1145/2743015
- Hoogendoorn, M., Ramos, I. C., Soulard, S., Cook, J., Soini, E., Paulsson, E., & van Mölken, M. R. (2021). Cost-effectiveness of the fixed-dose combination tiotropium/olodaterol versus tiotropium monotherapy or a fixed-dose combination of long-acting β2-agonist/inhaled corticosteroid for COPD in Finland, Sweden and the Netherlands: A model-based study. British Medical Journal Open, 11(8), e049675. https://bmjopen.bmj.com/content/11/8/e049675
- Hunter, J. D. (2007). Matplotlib: A 2d graphics environment. Computing in Science & Engineering, 9(3), 90–95. https://doi.org/10.1109/MCSE.2007.55
- Janssen, M. A., Pritchard, C., & Lee, A. (2020). On code sharing and model documentation of published individual and agent-based models. Environmental Modelling & Software, 134, 104873. https://doi.org/10.1016/j.envsoft.2020.104873
- Kluyver, T., Ragan-Kelley, B., Pérez, F., Granger, B., Bussonnier, M., Frederic, J., Kelley, K., Hamrick, J., Grout, J., Corlay, S., Ivanov, P., Avila, D, Abdalla, S., Willing, C., Jupyter, &Jupyter development team. (2016). Jupyter notebooks – a publishing format for reproducible computational workflows. In F. Loizides & B. Scmidt (Eds.), IOS press (pp. 87–90). Retrieved July 04, 2022, from https://eprints.soton.ac.uk/403913/
- Krafczyk, M. S., Shi, A., Bhaskar, A., Marinov, D., & Stodden, V. (2021). Learning from reproducing computational results: Introducing three principles and the reproduction package. Philosophical Transactions Series A, Mathematical, Physical, and Engineering Sciences, 379(2197), 20200069. Retrieved April 03, 2023, from https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8059663/
- Lahr, M. M. H., van der Zee, D.-J., Luijckx, G.-J., & Buskens, E. (2020). Optimising acute stroke care organisation: A simulation study to assess the potential to increase intravenous thrombolysis rates and patient gains. British Medical Journal Open, 10(1), e032780. https://bmjopen.bmj.com/content/10/1/e032780
- Lin, D., Crabtree, J., Dillo, I., Downs, R. R., Edmunds, R., Giaretta, D., De Giusti, M., L’Hours, H., Hugo, W., Jenkyns, R., Khodiyar, V., Martone, M. E., Mokrane, M., Navale, V., Petters, J., Sierman, B., Sokolova, D. V., Stockhause, M., & Westbrook, J. (2020, May). The TRUST principles for digital repositories. Scientific Data, 7(1), 144. (Number: 1 Publisher: Nature Publishing Group) Retrieved April 25, 2023, from https://www.nature.com/articles/s41597-020-0486-7
- Marco, A., & Janssen, L. N. A. (2008, March). Towards a community framework for agent-based modelling. JASSS). [Text.Article]. Retrieved June 02, 2023, from https://jasss.soc.surrey.ac.uk/11/2/6.html
- Martín-Martín, A., Orduna-Malea, E., Thelwall, M., & Delgado López-Cózar, E. (2018). Google Scholar, Web of Science, and Scopus: A systematic comparison of citations in 252 subject categories. Journal of Informetrics, 12(4), 1160–1177. https://www.sciencedirect.com/science/article/pii/S1751157718303249
- McKinney, W. (2010). Data structures for statistical computing in Python. In S. van der Walt, & J. Millman, Proceedings of the 9th Python in Science Conference (pp. 56–61). https://doi.org/10.25080/Majora-92bf1922-00a
- Mohiuddin, S., Busby, J., Savović, J., Richards, A., Northstone, K., Hollingworth, W., Donovan, J. L., & Vasilakis, C. (2017). Patient flow within UK emergency departments: A systematic review of the use of computer simulation modelling methods. British Medical Journal Open, 7(5), e015007. https://bmjopen.bmj.com/content/7/5/e015007
- Mohiuddin, S., Gardiner, R., Crofts, M., Muir, P., Steer, J., Turner, J., Wheeler, H., Hollingworth, W., & Horner, P. J. (2020). Modelling patient flows and resource use within a sexual health clinic through discrete event simulation to inform service redesign. British Medical Journal Open, 10(7), e037084. https://bmjopen.bmj.com/content/10/7/e037084
- Monks, T., Currie, C. S., Onggo, B. S., Robinson, S., Kunc, M., & Taylor, S. J. (2019). Strengthening the reporting of empirical simulation studies: Introducing the stress guidelines. Journal of Simulation, 13(1), 55–67. https://doi.org/10.1080/17477778.2018.1442155
- Monks, T., & Harper, A. (2023a, June). Database search strings for computer model and code sharing practices in healthcare discrete- event simulation: A systematic scoping review. Zenodo. https://doi.org/10.5281/zenodo.8046898
- Monks, T., & Harper, A. (2023b, June). Supplementary material v1.0.0: Model and code sharing practices in healthcare discrete-event simulation: A systematic scoping review. Zenodo. https://doi.org/10.5281/zenodo.8054806
- Nature Biotechnology. (2019 May). Changing coding culture. Nature Biotechnology, 37(5), 485–485. (Number: 5 Publisher: Nature Publishing Group) Retrieved April 04, 2023, from https://www.nature.com/articles/s41587-019-0136-9
- Noorain, S., Kotiadis, K., & Scaparra, M. P. (2019). Application of discrete-event simulation for planning and operations issues in mental healthcare. In 2019 winter simulation conference (wsc), National Harbor, MD, USA (p. 1184–1195).
- Onen-Dumlu, Z., Harper, A. L., Forte, P. G., Powell, A. L., Pitt, M., Vasilakis, C., Wood, R. M., & Pietrantonio, F. (2022, 06). Optimising the balance of acute and intermediate care capacity for the complex discharge pathway: Computer modelling study during COVID-19 recovery in england. PLOS ONE, 17(6), 1–16. https://doi.org/10.1371/journal.pone.0268837
- Onggo, B. S., & Karatas, M. (2016). Test-driven simulation modelling: A case study using agent-based maritime search-operation simulation. European Journal of Operational Research, 254(2), 517–531. https://www.sciencedirect.com/science/article/pii/S0377221716301965
- Page, M. J., McKenzie, J. E., Bossuyt, P. M., Boutron, I., Hoffmann, T. C., Mulrow, C. D., Shamseer, L., Tetzlaff, J. M., Akl, E. A., Brennan, S. U., Chou, R., Glanville, J., Grimshaw, J. M., Lalu, M. M., Li, T., Loder, E. W., Mayo-Wilson, E., McDonald, S., McGuinness, L. A., Stewart, L. A., … Moher, D. (2021). The prisma 2020 statement: An updated guideline for reporting systematic reviews. BMJ, 372. https://www.bmj.com/content/372/bmj.n71
- Peng, R. (2011). Reproducible research in computational science. Science, 334(6060), 1226–1227. Retrieved April 25, 2023, from https://www.science.org/doi/10.1126/science.1213847
- Penn, M., Monks, T., Kazmierska, A., & Alkoheji, M. (2020). Towards generic modelling of hospital wards: Reuse and redevelopment of simple models. Journal of Simulation, 14(2), 107–118. https://doi.org/10.1080/17477778.2019.1664264.
- Philip, A. M., Prasannavenkatesan, S., & Mustafee, N. (2022). Simulation modelling of hospital outpatient department: A review of the literature and bibliometric analysis. SIMULATION, 99(6), 573–597. https://doi.org/10.1177/00375497221139282
- Rahmandad, H., & Sterman, J. D. (2012). Reporting guidelines for simulation-based research in social sciences. System Dynamics Review, 28(4), 396–411. https://onlinelibrary.wiley.com/doi/abs/10.1002/sdr.1481
- Reinhardt, O., Rucheinski, A., & Uhrmacher, A. M. (2018, December). ODD+P: COMPLEMENTING the ODD PROTOCOL with PROVENANCE INFORMATION. In 2018 Winter Simulation Conference (WSC), Gothenburg, Sweden (pp. 727–738).
- Robinson, S. (2014). Simulation: The practice of model development and use. Palgrave Macmillan.
- Roy, S., Venkatesan, S. P., & Goh, M. (2021). Healthcare services: A systematic review of patient-centric logistics issues using simulation. Journal of the Operational Research Society, 72(10), 2342–2364. https://doi.org/10.1080/01605682.2020.1790306.
- Ruscheinski, A., Warnke, T., & Uhrmacher, A. M. (2020, June). Artifact-based workflows for supporting simulation studies. IEEE Transactions on Knowledge and Data Engineering, 32(6), 1064–1078. (Conference Name: IEEE Transactions on Knowledge and Data Engineering) https://doi.org/10.1109/TKDE.2019.2899840
- Salleh, S., Thokala, P., Brennan, A., Hughes, R., & Booth, A. (2017). Simulation modelling in healthcare: An umbrella review of systematic literature reviews. PharmacoEconomics, 35(9), 937–949. https://doi.org/10.1007/s40273-017-0523-3
- Salmon, A., Rachuba, S., Briscoe, S., & Pitt, M. (2018). A structured literature review of simulation modelling applied to emergency departments: Current patterns and emerging trends. Operations Research for Health Care, 19, 1–13. https://www.sciencedirect.com/science/article/pii/S2211692317301042
- Sills, J., Barton, C. M., Alberti, M., Ames, D., Atkinson, J.-A., Bales, J., Burke, E., Chen, M., Diallo, S. Y., Earn, D. J. D., Fath, B., Feng, Z., Gibbons, H., Hammond, R., Heffernan, J., Houser, H., Hovmand, S. P., Kopainsky, B., Mabry, P. L., Mair, C., … Tucker, G. (2020). Call for transparency of COVID-19 models. Science, 368(6490), 482–483. https://www.science.org/doi/abs/10.1126/science.abb8637
- Smith, A. M., Katz, D. S., Niemeyer, K. E., & FORCE11 Software Citation Working Group. (2016 September). Software citation principles. Peer Journal Computer Science, 2, e86. Retrieved April 25, 2023, from https://peerj.com/articles/cs-86
- Stodden, V., Seiler, J., & Ma, Z. (2018). An empirical analysis of journal policy effectiveness for computational reproducibility. Proceedings of the National Academy of Sciences, 115(11), 2584–2589. https://doi.org/10.1073/pnas.1708290115
- Taylor, S. J., Anagnostou, A., Fabiyi, A., Currie, C., Monks, T., Barbera, R., & Becker, B. (2017). Open science: Approaches and benefits for modeling & simulation. In 2017 winter simulation conference (wsc), Las Vegas, NV, USA (pp. 535–549).
- The Turing Way Community. (2022 July) The Turing Way: A handbook for reproducible, ethical and collaborative research. Zenodo. https://doi.org/10.5281/zenodo.7470333
- van der Walt, S., Colbert, S. C., & Varoquaux, G. (2011). The numpy array: A structure for efficient numerical computation. Computing in Science & Engineering, 13(2), 22–30. https://doi.org/10.1109/MCSE.2011.37
- Vázquez-Serrano, J. I., Peimbert-García, R. E., & Cárdenas-Barrón, L. E. (2021, January). Discrete-event simulation modeling in healthcare: A comprehensive review. International Journal of Environmental Research and Public Health, 18(22), 12262. (Number: 22 Publisher: Multidisciplinary Digital Publishing Institute) Retrieved April 03, 2023, from https://www.mdpi.com/1660-4601/18/22/12262
- Venkatesh, K., Santomartino, S. M., Sulam, J., & Yi, P. H. (2022, August). Code and data sharing practices in the radiology artificial intelligence literature: A meta-research study. Radiology: Artificial Intelligence, 4(5), e220081. Retrieved January 04, 2023 from https://www.ncbi.nlm.nih.gov/pmc/articles/PMC9530751/
- Yakutcan, U., Hurst, J. R., Lebcir, R., & Demir, E. (2022). Assessing the impact of COVID-19 measures on COPD management and patients: A simulation-based decision support tool for COPD services in the uk. British Medical Journal Open, 12(10), e062305. https://bmjopen.bmj.com/content/12/10/e062305
- Zhang, X. (2018, September). Application of discrete event simulation in health care: A systematic review. BMC Health Services Research, 18(1), 687. Retrieved April 03, 2023, from https://doi.org/10.1186/s12913-018-3456-4
- Zhang, X., Lhachimi, S. K., & Rogowski, W. H. (2020). Reporting quality of discrete event simulations in healthcare—results from a generic reporting checklist. Value in Health, 23(4), 506–514. https://www.sciencedirect.com/science/article/pii/S1098301520300401
- Zotero. (2011). Zotero. GitHub Repository. https://github.com/zotero/zotero