?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
C-X-C motif chemokine ligand 8 (CXCL8) gene, a crucial cytokine with roles in the immune system, functions in the inflammatory response by promoting the activation and migration of neutrophils. Previous studies have reported that the CXCL8 gene resides within the quantitative trait locus (QTL). In this report, the porcine CXCL8 gene was selected to investigate its effect on serum immune traits. CXCL8 mRNA expression was also detected by quantitative real-time polymerase chain reaction (PCR). A SNP (NC_010450.4:g.69934997T > C, rs81218904) of the CXCL8 gene was identified by direct nucleotide sequencing and the SNP was genotyped by MALDI-TOF MS in the three pig populations, which consisted of 300 piglets distributed in three pig breeds, Landrace (68 piglets), Large White (158 piglets) and Songliao Black (74 piglets). An in-depth analysis indicated that there was a significant correlation between the SNP and the serum CXCL8 level (20 days) (14 days after vaccination) (p < .05). Our results suggest that the CXCL8 gene may be served as a genetic marker with effects on serum CXCL8 level in the pig disease resistance breeding.
Genomic variant identification and tissues expression analysis of porcine CXCL8 gene.
The SNP in porcine CXCL8 gene was significantly associated with serum CXCL8 level in three pig populations.
SNP genotyping and association analysis results provided significant evidence for association on serum cytokine trait of porcine CXCL8 gene.
Highlights
Keywords:
Introduction
Cytokines are a group of essential proteins that play decisive roles in regulating immunological processes and inflammatory reactions. As a pre-inflammatory cytokine, C-X-C motif chemokine ligand 8 (CXCL8) is a member of the chemokine family and also named interleukin 8 (IL8), which is known for its leukocyte and lymphocytee chemotactic properties and it plays vital roles in inflammation, immunological reactions, and organism defence (Heinzmann et al. Citation2004). CXCL8 is predominantly produced by monocytes, macrophages and endothelial cells. The main role of CXCL8 is to initiate and increase the inflammatory response caused by pathogens by promoting the activation, migration, adhesion and phagocytosis of neutrophils from the peripheral blood to the tissues (Velloso et al. Citation2005). CXCL8 also has chemotactic activity against T cells and basophils (Jundi and Greene Citation2015).
Previous studies have shown that the inflammatory cytokine CXCL8 may play a role in promoting tumour resistance by enhancing the immunosuppressive microenvironment (David et al. Citation2016). CXCL8 genetic polymorphisms are involved in several human inflammatory diseases, such as chronic periodontitis (Zhang et al. Citation2014), malaria (Mahanta et al. Citation2014) and bacterial meningitis (Fontes et al. Citation2015). Furthermore, CXCL8 gene polymorphisms significantly associate with the somatic cell score trait in Chinese Holstein dairy cows (Chen et al. Citation2015). In duck, the CXCL8 gene enhances the immune response to avian influenza vaccine (Ruan et al. Citation2014).
In pigs, cytokines play central roles in the immunological response. The genetic mechanisms of cytokine action seem to be an attractive strategy and may provide guidelines for establishing effective control measures against infection (Vincent et al. Citation2006). The genomic structure of the porcine CXCL8 gene has been described (Shimanuki et al. Citation2002). The pig CXCL8 gene is located on chromosome 8 and the full length genomic sequence is 3220 bp. It consists of 4 exons and 3 introns. Its transcript is 1488 bp, translating into a protein of 103 amino acid residues. In addition, the CXCL8 gene is located near our reported QTL for interferon-gamma to interleukin 10 ratio in pig chromosome 8 (Animal QTLdb, http://www.animalgenome.org/cgi-bin/QTLdb/SS/qdetails?QTL_ID=23468). These findings strongly suggest that CXCL8 is a promising positional and functional candidate gene for selected immune traits in pig.
Despite the key role of the CXCL8 gene in the pathophysiology of inflammation and immunological processes, a study of the relationship between the variants in the porcine CXCL8 gene and immune traits has not been reported. To achieve a better understanding of the role of the porcine CXCL8 gene in regulating immunological processes and to identify potential variants of the CXCL8 gene associating with the serum CXCL8 level in the pig, we first identified the variant and then performed genotype-phenotype association analysis between the identified SNP and the serum CXCL8 level in three pig populations. We also examined its mRNA expression by quantitative real time polymerase chain reaction (PCR).
Materials and methods
Animal population
The animal population consisted of 300 piglets distributed in four Landrace boar families (12 sows and 68 piglets), six Large White boar families (28 sows and 158 piglets) and four Songliao Black boar families (13 sows and 74 piglets). All piglets were raised under identical standard indoor conditions at the experimental farm of the Institute of Animal Sciences, Chinese Academy of Agricultural Sciences, Beijing, China. All piglets were vaccinated with the Classical Swine Fever (CSF) live vaccine at 21 days of age. The CSF vaccine was safe and used to induce ‘stress’, which allowed us to quantify the serum CXCL8 level.
Blood was obtained from each piglet one day before the vaccination (20 days) and 2 weeks after the vaccination (14 days after vaccination). Ear tissue was also obtained from each piglet for DNA extraction. Seven tissues including spleen, stomach, lung, liver, kidney, heart and muscle of three Large White pigs were collected at the 35th day after slaughter within 30 min, then immediately frozen in liquid nitrogen and stored at −80 °C for expression analysis.
Measurement of the serum CXCL8 level
Sera from the three pig populations were stored at -20 °C. The serum CXCL8 level was measured with a commercial ELISA Test Kit (Biosource, Carlsbad, CA, USA). The kit contained an antibody-coated 96-well test plate, standards of known CXCL8 concentrations, a standard diluent buffer, wash buffers, a biotinylated monoclonal antibody specific to CXCL8, a streptavidin-HRP diluent solution and a stop solution. Each sample was run in triplicate. According to the manufacturer’s instructions, samples were randomly arranged in the test plate and the CXCL8 concentration was calculated based on a standard curve.
Genomic DNA isolation and total RNA extraction
Genomic DNA was extracted from ear tissues using phenol/chloroform and ethanol precipitation (Sambrook et al. Citation1989). DNA quality was measured by spectrophotometry using a NanoDrop™ 2000 Spectrophotometer (Thermo Fisher Scientific, Weehawken, NJ, USA) and electrophoresis using a 1% agarose gel. The total RNA was isolated from different tissues using Trizol Reagent (Invitrogen, Carlsbad, CA, USA) and quantified by using the NanoDropTM 2000 spectrophotometer. RNA integrity was assessed by 1% agarose gel electrophoresis before the first-strand cDNA was synthesised. RNA was purified and reversely transcribed into cDNA using the PrimerScript® RT Reagent Kit with gDNA Eraser (TaKaRa Biotechnology Co., Ltd, Dalian, China), according to the manufacturer’s instructions.
Tissues expression analysis of the porcine CXCL8 gene
The CXCL8 cDNA sequence was obtained from the Ensembl pig database (Sscrofa 10.2, Gene ID: ENSSSCT00000009807). The CXCL8 mRNA level in seven different tissues of three 35-old-day Large White pigs was investigated by quantitative real-time PCR using the LightCycler® 480 II System (Roche Diagnostics GmbH, Mannheim, Germany). The reaction contained 10 μL of 2× SYBR green I mixture, 10 pM each of the forward and reverse primers and 20 ng of cDNA in a final volume of 20 μL. A specific primer set to examine the mRNA expression pattern of the CXCL8 gene was also designed to eliminate potential confounding results from genomic DNA contamination (Supplementary Table S1). The GAPDH (glyceraldehyde-3-dehydrogenase) gene was used as an internal reference gene for normalisation and the primers were as follows: forward (F): 5'-GTCCACTGGTGTCTTCACGA-3′, reverse (R): 5′-GCTGACGATCTTGAGGGAGT-3′ (GenBank Accession No.: AF017079). The reaction conditions were as follows: an initial denaturation at 95 °C for 10 min, 45 cycles at 95 °C for 10 s, 60 °C for 10 s and 72 °C for 10 s. Each reaction was carried out in triplicate and normalised to the GAPDH gene using the 2_△△Ct method (Livak and Schmittgen Citation2001).
SNP identification and genotyping
Five PCR primer sets were designed based on the porcine CXCL8 genomic sequence available in the Sscrofa 10.2 primary assembly (Ensembl Gene ID: ENSSSCG00000008953) to amplify all exons as well as adjacent partial introns (Supplementary Table S1). DNA samples of 30 Large White piglets were selected randomly to construct a DNA pool with equal DNA concentration of 50 ng/μL for each individual. PCR was carried out in a 25-μL volume containing 50 ng pooled DNA, 2.5 μL of 10× PCR buffer, 5 mM of dNTPs, 10 pmol of forward and reverse primer, 0.625 U Taq DNA polymerase (Takara Biotechnology Co., Ltd. Dalian, China) and ddH2O. The reaction conditions were as follows: an initial denaturation at 94 °C for 5 min, followed by 34 cycles at 94 °C for 30 s, 51.6–59.5 °C for 40 s, 72 °C for 40 s, and a final extension at 72 °C for 10 min. PCR fragments were purified with a Gel Extraction Mini Kit (Beijing Tiangen Biotechnology, Beijing, China) and then sequenced using an ABI3730xl DNA Analyzer (Applied Biosystems, Foster, CA, USA). SNPs were detected using Chromas (version 2.3.1) (Lynnon, San Ramon, CA, USA) and DNAMAN (version 6.0) software (Technelysium, South Brisbane, Australia). Matrix-Assisted Laser Desorption/Ionization Time of Flight Mass Spectrometry (MALDI-TOF MS) (Squenom MassARRAY®, Bioyong Technologies Inc. Beijing, China) was carried out to genotype the identified SNP in the pigs.
Association analysis
Association analysis between the SNP genotypes and the serum CXCL8 concentration were examined by fitting the following mixed models using SAS software (Version 9.2, SAS, Carey, NC, USA):
where y was the vector of phenotypic values of serum CXCL8 level analysed;
was the overall mean; β was the vector of fixed effects, including the breed with three levels, sex with two levels and ELISA plate effect with five levels; v was the vector of random litter effects; m was the vector of the SNP genotypes with three levels; a was the vector of the residual polygenetic effects with
, A was the numerator relationship matrix;. X, T and Z were the incidence matrices for β, v and a, respectively; b was the regression coefficient of phenotypes on SNP genotypes; and e was the vector of residual errors with
.
Results and discussion
Tissues expression of the porcine CXCL8 gene
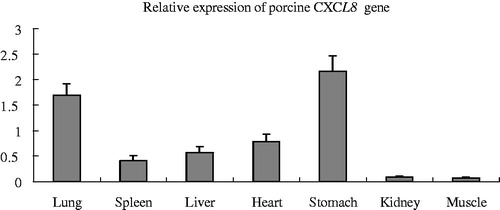
The quantitative real-time quantitative PCR results showed that CXCL8 was expressed in all analysed tissues, except skeletal muscle, with the highest expression level in the stomach, followed by the lung and the lowest expression level in the heart, liver, spleen, kidney and muscle (). Our results were consistent with the gene expression annotation using porcine Affymetrix expression array (Snowball) analysis in the BioGPS dataset (http://biogps.org/). In this study, CXCL8 expression was also highest in the stomach, but lowest in the kidney and skeletal muscle. The reason was likely due to the fact that the kidney and skeletal muscle are not tissues involved in immune-related processes.
Association analysis of the porcine CXCL8 gene
A SNP (NC_010450.4:g.69934997T > C, rs81218904) of CXCL8 gene was detected and was a synonymous and then the SNP was genotyped in 300 piglets by applying MALDI-TOF MS assay. Genetic variation analysis also demonstrated that allelic frequencies were not significantly different among the three pig breeds (p > .05) (). The allele T was dominant in the three pig breeds. By contrast, the allele C had a lower frequency and the CC genotype was lower in Landrace, Large White and Songliao Black pigs than in the TT genotype.
Table 1. Genotype frequencies and allelic frequencies of the SNP (NC_10450.4:g.69934997T > C, rs81218904) the CXCL8 gene determined by MALDI-TOFMS method in the three pig populations.
Based on the aforementioned work, an association study was further performed to determine whether the SNP influenced the serum CXCL8 level in pigs. The results showed that the SNP (NC_010450.4:g.69934997T > C, rs81218904) caused a significant difference in the serum CXCL8 level at both time points (20 days and 14 days after vaccination) (p < .05) (). Multiple comparison tests results showed that the serum CXCL8 level was higher in pigs with genotype TT than in those with genotype CC (p < .05) ().
Table 2. Association analysis and multiple tests of the SNP (NC_010450.4:g.69934997T > C, rs81218904) of the CXCL8 gene with the serum CXCL8 level in the three pig populations.
Chemotactic cytokines are important mediators of immunological responses, inflammation and cancer development. CXCL8 is a chemokine that belongs to the alpha chemokine family, which is produced by various cell types to recruit leukocytes to sites of infection or tissue injury. In humans, the serum CXCL8 level is a valuable biological marker for health and disease status. Akiba et al. (Citation2001) found that the serum CXCL8 level was markedly elevated in most patients with hepatocellular carcinoma compared with healthy subjects and a high serum CXCL8 level significantly correlated with a more aggressive tumour behaviour in patients with resectable hepatocellular carcinoma. The serum CXCL8 protein level was significantly higher in septic patients than in healthy patients (Hu et al. Citation2016). Similarly, the serum CXCL8 level in individuals with a Helicobacter pylori infection also associated strongly with a degree of gastritis (Siregar et al. Citation2015). In addition, the CXCL8 level could act as a prognostic marker for patients with hepatitis B virus-associated hepatocellular carcinoma treated with transarterial chemoembolization (Kim et al. Citation2015) and also as a possible infection site marker in preterm deliveries (Bogavac and Brkić Citation2009). In this study, our results were quite interesting. We showed that the serum CXCL8 level in the Songliao Black pig was the lowest before vaccination, but relatively higher after vaccination, which could be explained by the fact that this breed may have a relatively better immune response capacity than the other two Western commercial pig breeds. These differences of the serum cytokine level among individuals under the same conditions implied that genetic basis of varieties might be a potential influence factor for cytokine expression.
As cytokines play an essential role in the immune system, genes involved in immunological responses may be important candidate genes for selection. Human CXCL8 may behave as a novel messenger to cross-link inflammation and tumour epithelial-mesenchymal transition via autocrine and paracrine pathways (Long et al. Citation2016). The genetic polymorphisms of the human CXCL8 gene significantly associated with cystic fibrosis (Furlan et al. Citation2016) and glioma risk (Atalay et al. Citation2016). Thus, it appears that CXCL8 could be a potential risk factor and may represent a feasible approach to control disease infection.
Interestingly enough, we also found a correlation between the genetic polymorphism and the serum CXCL8 level in this study. Similar results were found in Chinese patients with sepsis and the different genotypes within three SNPs are more susceptible to sepsis (Hu et al. Citation2016). In another study involving a Chinese Holstein population, CXCL8 gene polymorphisms significantly correlated with the somatic cell score trait and different genotypes had significantly different mRNA expression levels (Chen et al. Citation2015).
Discovering new loci or causal genes for disease resistance and characterising their genetic mechanisms in the pig should contribute to both pig health and productivity. Our association analysis results provided a straightforward insight that the CXCL8 gene, which has effects on serum CXCL8 level in the three pig populations and it could serve as a promising candidate gene for detecting the cytokine level during pig breeding. However, the number of pigs analysed in our study was restricted. Further studies will be required to confirm the relationship between the SNP and the serum CXCL8 level in larger pig populations before it can be used for selection.
Conclusion
In summary, one SNP of the CXCL8 gene was identified and it significantly associated with the serum CXCL8 level (20 days) (14 days after vaccination) in the three pig populations. Our results indicated that CXCL8 could be a promising candidate gene for the serum CXCL8 level and the locus could be a useful genetic marker for selection in pig disease resistance breeding programmes.
Ethical approval
In this study, the whole study protocols for collection of the samples of experimental individuals and phenotypic detection were reviewed and approved by the Animal Welfare Committee of Nanjing Agricultural University (Permit No. DK612).
Disclosure statement
The authors have declared that no competing interests exist.
Additional information
Funding
References
- Akiba J, Yano H, Ogasawara S, Higaki K, Kojiro M. 2001. Expression and function of interleukin-8 in human hepatocellular carcinoma. Int J Oncol. 18:257–264.
- Atalay A, Arıkan S, Ozturk O, Öncü M, Tasli ML, Duygulu S, Atalay EO. 2016. The IL-8 gene polymorphisms in Behçet's disease observed in Denizli province of Turkey. Immunol Invest. 45:298–311.
- Bogavac MA, Brkić S. 2009. Serum proinflammatory cytokine-interleukin-8 as possible infection site marker in preterm deliveries. J Perinat Med. 37:707–708.
- Chen R, Wang Z, Yang Z, Zhu X, Ji D, Mao Y. 2015. Association of IL8 -105G/A with mastitis somatic cell score in Chinese Holstein dairy cows . Anim Biotechnol. 26:143–147.
- David JM, Dominguez C, Hamilton DH, Palena C. 2016. The IL-8/IL-8R axis: a double agent in tumor immune resistance. Vaccines (Basel). 4:22.
- Fontes FL, de Araújo LF, Coutinho LG, Leib SL, Agnez-Lima LF. 2015. Genetic polymorphisms associated with the inflammatory response in bacterial meningitis. BMC Med Genet. 16:70
- Furlan LL, Marson FA, Ribeiro JD, Bertuzzo CS, Salomão Junior JB, Souza DR. 2016. IL8 gene as modifier of cystic fibrosis: unraveling the factors which influence clinical variability. Hum Genet. 135:881–894.
- Heinzmann A, Ahlert I, Kurz T, Berner R, Deichmann KA. 2004. Association study suggests opposite effects of polymorphisms within IL8 on bronchial asthma and respiratory syncytial virus bronchiolitis. J Allergy Clin Immunol. 114:671–676.
- Hu D, Wang H, Huang X, Jiang Y, Qin Y, Xiong B, Qin G, Sooranna SR, Pinhu L. 2016. Investigation of association between IL-8 serum levels and IL8 polymorphisms in Chinese patients with sepsis. Gene. 594:165–170.
- Jundi K, Greene CM. 2015. Transcription of interleukin-8: how altered regulation can affect cystic fibrosis lung disease. Biomolecules. 5:1386–1398.
- Kim SS, Cho HJ, Won JH, Bae JI, Kang DR, Lee JD, Shin SJ, Lee KM, Yoo BM, Kim JK, et al. 2015. Interleukin-8 level as a prognostic marker in patients with hepatitis B virus-associated hepatocellular carcinoma treated with transarterial chemoembolization. Cytokine. 76:449–457.
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 25:402–408.
- Long X, Ye Y, Zhang L, Liu P, Yu W, Wei F, Ren X, Yu JIL. 2016. IL-8, a novel messenger to cross-link inflammation and tumor EMT via autocrine and paracrine pathways (Review) . Int J Oncol. 48:5–12.
- Mahanta A, Kakati S, Baruah S. 2014. The association of IL-8-251T/A polymorphism with complicated malaria in Karbi Anglong district of Assam . Cytokine. 65:210–216.
- Ruan Y, Ji X, Wen M, Zhu X, Fu X. 2014. Interleukin 8 enhances the immune response of ducks to avian influenza vaccine. Acta Virol. 58:356–358.
- Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual. 2nd ed. New York, NY, USA: Cold Spring Harbor Laboratory Press.
- Shimanuki S, Kobayashi E, Awata T. 2002. Genomic structure of the porcine interleukin 8 gene and development of a microsatellite marker within intron 1. Anim Genet. 33:470–471.
- Siregar GA, Halim S, Sitepu VR. 2015. Serum TNF-a, IL-8, VEGF levels in Helicobacter pylori infection and their association with degree of gastritis. Acta Med Indones. 47:120–126.
- Velloso LFM, Silva Filho LVF, Miyoshi MH, Rozov T. 2005. Bronchopulmonary dysplasia. J Pediatr (Rio J). 81:99–110.
- Vincent A, Thacker B, Halbur P, Rothschild M, Thacker E. 2006. An investigation of susceptibility to porcine reproductive and respiratory syndrome virus between two genetically diverse commercial lines of pigs. J Anim Sci. 84:49–57.
- Zhang N, Xu Y, Zhang B, Zhang T, Yang H, Zhang B, Feng Z, Zhong D. 2014. Analysis of interleukin-8 gene variants reveals their relative importance as genetic susceptibility factors for chronic periodontitis in the Han population. PLoS One. 9:e104436.