Abstract
microRNAs (miRNAs) are small non-coding RNAs that regulate various biological processes by targeting mRNAs and affecting their stability or translation. miR-122 is a major miRNA in mammalian liver but its function in poultry liver has not been studied. Therefore, the objective of the current study was to determine the function of miR-122 in the chicken liver. Through bioinformatics, we found P4HA1 was a candidate target of miR-122. P4HA1 mRNA and protein expression were increased in hepatocytes when miR-122 was knocked down by locked nucleic acid (LNA)-antisense-miR-122 (LNA-122). Reporter gene analyses confirmed that P4HA1 was a target of miR-122. Because P4HA1 is known to play an important role in liver metabolism, these findings suggest that miR-122 may be involved in liver metabolism in the chicken by regulating the P4HA1 expression.
The predicted target site of miR-122 in chicken P4HA1 is conserved among species.
P4HA1 mRNA and protein are upregulated in miR-122 knockdown chicken hepatocytes.
miR-122 targets P4HA1 in chicken.
Highlights
Introduction
microRNAs (miRNAs), a class of small non-coding RNAs of ∼22 nucleotides in length, regulate gene expression by interacting with the 3′ untranslated region (UTR) of target mRNAs (Bartel Citation2004). The interaction through an imperfect complementarity between miRNA and target mRNA causes translational repression or mRNA cleavage in animals (Caenorhabditis elegans, mouse) (Lee et al. Citation1993; Yekta et al. Citation2004). miRNAs play roles in many biological processes, including growth, development and metabolism (Bartel Citation2004; Wienholds and Plasterk Citation2005).
miR-122 is a tissue-specific and abundant miRNA in the liver (human, mouse) (Lagos-Quintana et al. Citation2002; Chang et al. Citation2003). miR-122 is an important regulator of metabolism in the liver. The expression level of miR-122 is lower in minipigs fed a high-cholesterol diet than in those fed a standard diet, implying a potential role of miR-122 in lipid metabolism (Cirera et al. Citation2010). CAT-1 is an important amino acid metabolic regulator and a target gene of miR-122. miR-122 may affect protein metabolism by regulating the expression of CAT-1 (human) (Chang et al. Citation2004). The metabolic functions of miR-122 have been studied by miR-122 knockdown. When miR-122 is inhibited by AMO or LNA in animals, cholesterol levels decrease (African green monkey, mouse) (Krutzfeldt et al. Citation2005; Esau et al. Citation2006; Elmen, Lindow, Schutz, et al. Citation2008; Elmen, Lindow, Silahtaroglu, et al. Citation2008), and the expression levels of genes associated with the maintenance of liver phenotype increase (mouse) (Krutzfeldt, et al. Citation2005; Elmen, Lindow, Silahtaroglu, et al. Citation2008). When miR-122 is knocked down in hepatocytes, hepatic fatty acid and cholesterol synthesis rates decrease, and fatty acid oxidation increases (mouse) (Esau et al. Citation2006), suggesting an important role of miR-122 in lipid metabolism in liver. miR-122 also plays roles in some other bioprocesses in the liver, such as circadian rhythm (mouse) (Gatfield et al. Citation2009), hepatocyte differentiation (mouse) (Laudadio et al. Citation2012), and liver development (mouse) (Xu et al. Citation2010).
Some metabolic process are different between birds and mammals; for example, lipid synthesis occurs mostly in adipose tissue in mammals, but it takes place exclusively in the liver in birds. Studies of miR-122 have been mainly performed in mammals. To the best of our knowledge, no studies have been conducted to understand the function of miR-122 in the chicken liver. Therefore, the objective of the current study was to identify the target genes of miR-122 in the chicken liver. Through bioinformatics, our laboratory found P4HA1 was a candidate target of miR-122. And through RNA-seq, we found P4HA1 mRNA expression was increased in miR-122-knocked down chicken hepatocytes (p < .05) (Wang et al. Citation2015). In this paper, we provide experimental evidence confirming that miR-122 targets P4HA1 and regulates its expression in chicken hepatocytes.
Materials and methods
Computational prediction of miR-122 target genes
The 3′ UTR sequences of Gallus gallus were downloaded from the 3′ UTR database (http://utrdb.ba.itb.cnr.it/) (Mignone et al. Citation2004). The target genes of miR-122 and the binding sites in the 3′ UTR of targets were predicted using previously described methods (Wang et al. Citation2013). The target sites in the 3′ UTR of selected genes were further analysed by a BLAST search in GenBank (http://www.ncbi.nlm.nih.gov), and their homology to other species was analysed using DNAMAN.
Isolation and culture of primary chicken hepatocytes
Chicken hepatocytes were isolated using an improved two-step collagenase method (Douaire et al. Citation1993). Hepatocytes were isolated from three 4-week-old Arbor Acres commercial chickens as previously described (Wang et al. Citation2013). Cell viability was verified by a trypan blue exclusion test. All procedures involving animals were approved by Changshu Institute of Technology Intramural Animal Use and Care Committee. The isolated hepatocytes were cultured at a density of 6 × 105 cells/ml in William’s E medium supplemented with 5% chicken serum, 100 U/ml penicillin-streptomycin, 10 µg/ml insulin and 30 mM NaCl at 37 °C with 5% CO2 in humidified incubator.
Transfection of hepatocytes with RNA oligonucleotides
The hepatocytes were cultured and plated as described above. After 24 h of culture, the cells were transfected with LNA-122 (5′-AAACACCATTGTCACACTCC-3′) (Exiqon, Vedbaek, Denmark) using X-tremeGENE HP DNA Transfection Reagent (Roche, Mannheim, Germany) as previously described (Wang et al. Citation2015). After 24 h of transfection, total RNA of the cells was isolated, and the expression levels of miR-122 and P4HA1 were detected by real-time qRT-PCR. Total proteins of the cells were isolated, and the expression of P4HA1 was detected by Western blot analysis.
RNA isolation and real-time qRT-PCR
Total RNA from hepatocytes cells and various tissues from 4-week-old chickens was isolated using TRIzol reagent (Invitrogen, Carlsbad, CA); RNA concentrations and quality were determined by a NanoDrop2000 spectrophotometer (Thermo Scientific, Wilmington, DE) and formaldehyde-agarose gel electrophoresis. The expression of miR-122 was detected by stem-loop real-time qRT-PCR as previously described (Wang et al. Citation2015). The expression levels of miR-122 were measured using the 2−ΔΔCT method and normalised to 18S rRNA (Applied Biosystems) (Livak and Schmittgen Citation2001). The mRNA expression of P4HA1 was detected according to the protocol of a PrimeScript RT reagent kit and SYBR Premix Ex Taq (Takara, Dalian, China), and was normalised to the mRNA level of β-actin. The sequences of primers used in this study are listed in Table S1.
Western blot analysis
The proteins of hepatocytes were isolated using the IP lysis buffer supplemented with 1 mM PMSF (Beyotime, Nantong, China) according to the manufacturer’s protocol. Proteins were separated on 10% SDS-PAGE gels and transferred to PVDF membranes (Roche). The membranes were incubated with primary antibodies to P4HA1 (Abcam, Hong Kong, China) for 2 h in TBST buffer and then incubated with HRP-conjugated secondary antibodies (Santa Cruz, Santa Cruz, CA) for 2 h. GAPDH was used as an internal control, and the GAPDH primary antibody was HRP-conjugated (Kangchen, Shanghai, China). Proteins were detected using an ECL kit (Beyotime) and visualised by a Biospectrum 410 (UVP, Upland, CA).
Plasmid construction
The sequence containing the precursor of miR-122 was PCR amplified from chicken genomic DNA. The PCR product was cloned into the pcDNA3.1 (+) vector (Invitrogen) using the HindIII and XhoI restriction sites to construct the miR-122 overexpression vector pcDNA3.1/miR-122. A negative control (NC) vector pcDNA3.1/NC-miRNA was constructed by inserting a sequence lacking the precursor of miR-122 into pcDNA3.1. The 3′ UTR fragment of P4HA1 containing the miR-122 binding site was amplified from chicken genomic DNA and inserted between SacI and HindIII sites within the pMIR-REPORT vector (Ambion, Carlsbad, CA) to construct the pMIR-P4HA1 vector. The binding site-mutated plasmid pMIR-mutP4HA1 was generated from pMIR-P4HA1 using overlap-extension PCR (). All constructed plasmids were verified by sequencing.
Cell culture, transfection and luciferase reporter assay
Chinese hamster ovary (CHO) cells were cultured as previously described (Wang et al. Citation2013). For a miRNA overexpression assay, the cells were seeded in 6-well plates for 24 h and transfected with pcDNA3.1/miR-122 using X-tremeGENE 9 DNA Transfection Reagent (Roche) as previously described (Wang et al. Citation2013). pcDNA3.1 was used as a control. After 48 h, total RNA from the cells was isolated and used to quantify the expression levels of miR-122 and P4HA1 by real-time qRT-PCR as described above. For a luciferase reporter assay, CHO cells were seeded in 24-well plates for 24 h, and transfection was performed as previously described (Wang et al. Citation2013). pcDNA3.1 and pcDNA3.1/NC-miRNA were used as controls. The cells were harvested and lysed in passive lysis buffer (PLB) 48 h post-transfection. Luciferase activities were measured using a Dual-Luciferase Reporter Assay System (Promega, Madison, WI) according to the manufacturer’s instructions. Firefly luciferase activity was measured in a Modulus single tube luminometer (Turner BioSystems, Sunnyvale, CA) and normalised to internal control Renilla luciferase activity.
Statistical analysis
All data are presented as the mean ± SEM of at least 3 independent experiments performed in duplicate or triplicate. Statistical analysis was performed using Student's t-test, one-way ANOVA or linear correlation test. P < 0.05 was considered statistically significant.
Results
Target prediction for miR-122
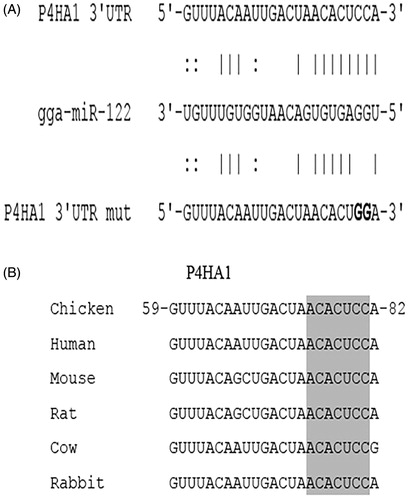
Three hundred sixty-four genes were predicted as targets of miR-122 in our previous study (Wang et al. Citation2015). Among these genes, P4HA1 has a miR-122 target site conserved across species. The 3′ UTR site of P4HA1 was predicted to stably interact with miR-122, and nucleotides 1–8 of miR-122 were completely complementary to the corresponding sequence (). The 3′ UTRs of P4HA1 in other species were also analysed using BLAST and DNAMAN. The chicken sequence of P4HA1 3′ UTR at the predicted miR-122 binding site is more than 80% identical to other species. The seed region of the target site is completely identical to those of other species ().
Figure 1. Complementarity between miR-122 and the predicted target site in the P4HA1 3′ UTR. (A) Schematic representation of the miR-122 binding site in the 3′ UTR of chicken P4HA1. The matched base pairs are connected by vertical lines, and the U:G wobble is connected by dots. The mutated nucleotides are in bold. (B) Sequence alignment of miR-122 target sites in P4HA1 in different species. The position of the target site in chicken P4HA1 is numbered, and the seed regions are highlighted in grey.
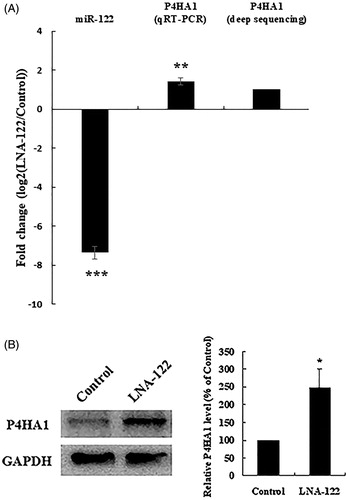
Association of increased P4HA1 mRNA expression with miR-122 knockdown in chicken hepatocytes
Among the differentially expressed genes (p < .05) identified by RNA-seq (Wang et al. Citation2015), P4HA1 is a known regulator of lipid metabolism and amino acid metabolism (Esau et al. Citation2006; Grimmer et al. Citation2006). The mRNA expression level of P4HA1 increased significantly in miR-122-knocked down chicken hepatocytes. We used real-time qRT-PCR to further determine the expression of P4HA1 mRNA. Compared with control hepatocytes, miR-122-inhibited hepatocytes showed increased mRNA expression of P4HA1 (). We used Western blotting to determine the expression of P4HA1 protein. Compared with control cells, miR-122-inhibited cells showed a significant increase in the protein level of P4HA1 (), indicating that miR-122 knockdown up-regulated not only P4HA1 mRNA but also its protein expression.
Figure 2. P4HA1 expression is up-regulated by miR-122 knockdown in chicken hepatocytes. (A) P4HA1 mRNA expression was increased by miR-122 knockdown. After transfection of control or LNA-122 in chicken hepatocytes, the expression of miR-122 was detected by real-time qRT-PCR and the expression of P4HA1 was detected by real-time qRT-PCR and RNA-seq . Sequencing data are from pooled samples of three 4-week-old chickens. Real-time qRT-PCR data are the means ± SEM of 3 independent experiments performed in duplicate and were analysed by student’s t-test. (B) P4HA1 protein expression was increased by miR-122 knockdown. After transfection of control or LNA-122 in chicken hepatocytes, the protein level of P4HA1 was detected by Western blotting and normalised to GAPDH. Left panel: Western blot analysis of P4HA1 in chicken hepatocytes transfected with LNA-122. Right panel: The protein level of P4HA1 was normalised to GAPDH, and the fold change relative to P4HA1 expression in controls is presented. Western blot data are the means ± SEM of 3 independent experiments and were analysed by student’s t-test. *p<.05; **p<.01; ***p<.001.
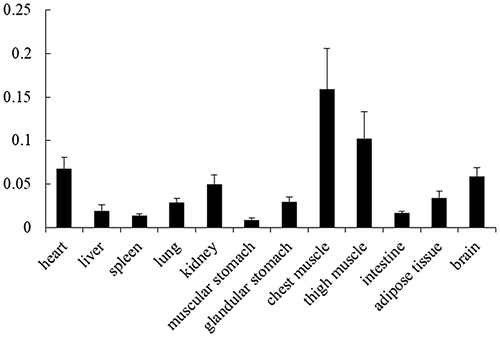
Expression pattern of P4HA1 mRNA
The expression of P4HA1 mRNA in 12 tissues from 4-week-old chickens was detected using real-time qRT-PCR. P4HA1 mRNA was expressed at high levels in skeletal muscle, at medial levels in the heart, kidney and brain, and at low levels in other tissues, such as the liver ().
miR-122 directly targets the 3′ UTR of P4HA1
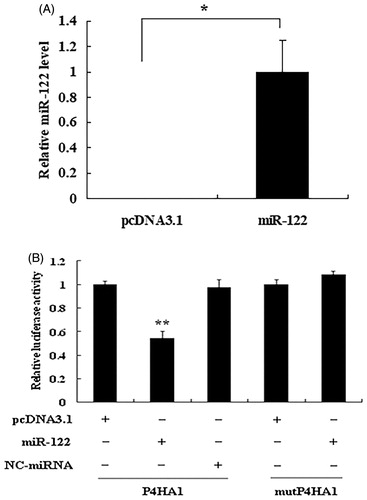
To validate the predicted target sites of miR-122 in the 3′ UTR of P4HA1, we cotransfected CHO cells with the pMIR-P4HA1 reporter vector and miR-122 overexpression vector pcDNA3.1/miR-122 or the control vector pcDNA3.1 or pcDNA3.1/NC-miRNA. pRL-CMV was also cotransfected to normalise the luciferase activity of pMIR-P4HA1. Compared with control samples transfected with pcDNA3.1, the samples transfected with pcDNA3.1/miR-122 showed a significant increase in miR-122 expression levels (). Compared with control cells, cells transfected with pcDNA3.1/miR-122 exhibited a decrease in the relative luciferase activity of pMIR-P4HA1 (). These data support direct targeting of the 3′ UTR of P4HA1 by miR-122. To further verify miR-122 targeting of P4HA1, we cotransfected CHO cells with the binding site-mutated plasmid pMIR-mutP4HA1 and pcDNA3.1/miR-122 or control pcDNA3.1. The resulting relative luciferase activity of pMIR-mutP4HA1 was not significantly different between the cells overexpressing miR-122 and the control cells (), indicating that the suppression of the relative luciferase activity by miR-122 was relieved by mutation of the target site.
Figure 4. miR-122 directly targets the 3′ UTR of chicken P4HA1 mRNA. (A) Overexpression of miR-122 in CHO cells. After CHO cells were transfected with control vector pcDNA3.1 or miR-122 overexpression vector pcDNA3.1/miR-122, the expression levels of miR-122 were detected by real-time qRT-PCR and normalised to 18S rRNA. (B) Target validation. CHO cells were cotransfected with pMIR-P4HA1 (Firefly luciferase) and pcDNA3.1 or pcDNA3.1/miR-122 or pcDNA3.1/NC-miRNA. The binding site-mutated vector pMIR-mutP4HA1 (Firefly luciferase) was cotransfected with pcDNA3.1 or pcDNA3.1/miR-122. pRL-CMV (Renilla luciferase) was used as an internal control. Relative luciferase activity was determined by Firefly luciferase activity normalised to Renilla luciferase activity. Data are the means ± SEM of at least 3 independent experiments performed in triplicate and analysed by student’s t-test or ANOVA. *p<.05; **p<.01.
Discussion
In our study, 364 genes were predicted as targets of miR-122. We verified P4HA1 targeting by miR-122 because it contains a nearly perfect target site for miR-122. P4HA1 is a key enzyme in collagen synthesis and folds procollagen polypeptide chains into stable triple helical molecules to regulate the maturation and secretion of collagen; inhibition of P4HA1 reduces collagen production (pig) (Kivirikko and Pihlajaniemi Citation1998; Rocnik et al. Citation1998). In the liver from morbidly obese women, P4HA1 is lower expressed than in the liver of women who have massive weight loss, implying it is involved in lipid metabolism in liver (Elam et al. Citation2009). P4HA1 is also involved in proliferation of and collagen maturation in the hepatic stellate cells (HSCs) from mouse, and miR-122 suppresses the proliferation of and collagen maturation in HSCs by reducing the expression of P4HA1 and thereby improves the therapeutic efficacy of adipose tissue-derived MSCs in the treatment of liver fibrosis (Lou et al. Citation2017).
LNA-122-treated chicken hepatocytes showed significant inhibition of miR-122. This result is consistent with the results in mammals (African green monkey, mouse) (Elmen, Lindow, Schutz, et al. Citation2008; Elmen, Lindow, Silahtaroglu, et al. Citation2008). Both real-time PCR and RNA-seq showed increased expression of P4HA1 mRNA in LNA-122-treated chicken hepatocytes, demonstrating that it was regulated by miR-122 and suggesting that it may be a target of miR-122. Esau and Su used anti-miR-122 to treat mice and found a 2-fold increase in the mRNA level of P4HA1, which is very similar to our results (Esau et al. Citation2006; Su et al. Citation2011). We also showed that P4HA1 protein levels were increased by miR-122 knockdown, further demonstrating that P4HA1 is regulated by miR-122.
P4HA1 has high expression levels in skeletal muscle and low expression levels in the liver. This expression pattern is opposite to that of miR-122, which is expressed at high levels in the liver but low levels in skeletal muscle (chicken) (Wang et al. Citation2015). This is consistent with the result in previous report in which the expression of P4HA1 is inversely correlated with miR-122 in human and rat HSCs and in the mouse liver. miR-122 and P4HA1 have opposite functions on collagen production (Li et al. Citation2013). This result again supports that P4HA1 mRNA expression in chicken skeletal muscle and liver is regulated by miR-122.
A homologous target fragment of mouse P4HA1 was cloned to construct the 3′ UTR reporter vectors pMIR-mmuP4HA1 and cotransfected with miR-122 overexpression or control vectors into CHO cells. Compared with control cells, the cells transfected with pcDNA3.1/miR-122 showed a decrease in the relative luciferase activity of pMIR-mmuP4HA1 (Figure S1). This result indicates that mouse P4HA1 was also a target of miR-122. In the target mutation assay, we mutated the nucleotides corresponding to 2nd and 3rd nucleotides of the seed region according to a previous report (human) (Stern-Ginossar et al. Citation2008). The results not only further validated the target site of P4HA1 but also indicated the importance of complementarity between the seed region and target site. Human, rat and mouse P4HA1 was also investigated in previous reports in which this gene was validated as a target of miR-122 by a dual-luciferase reporter assay, target mutation assay and inhibition assay. As shown in these reports, miR-122 regulates P4HA1 by repressing the expression of its mRNA (Esau et al. Citation2006; Li et al. Citation2013).
Conclusions
In summary, the results of the present study support P4HA1 as a target gene of miR-122 in chicken liver.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bartel DP. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 116:281–297. Epub 2004/01/28.
- Chang J, Nicolas E, Marks D, Sander C, Lerro A, Buendia MA, Xu C, Mason WS, Moloshok T, Bort R, et al. 2004. miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biol. 1:106–113. Epub 2006/12/21.
- Chang J, Provost P, Taylor JM. 2003. Resistance of human hepatitis delta virus RNAs to dicer activity. J Virol. 77:11910–11917.
- Cirera S, Birck M, Busk PK, Fredholm M. 2010. Expression profiles of miRNA-122 and its target CAT1 in minipigs (Sus scrofa) fed a high-cholesterol diet. Comparat Med. 60:136–141. AprEpub 2010/04/24.
- Douaire M, Belloir B, Guillemot JC, Fraslin JM, Langlois P, Mallard J. 1993. Lipogenic enzyme and apoprotein messenger RNAs in long-term primary culture of chicken hepatocytes. J Cell Sci. 104:713–718.
- Elam MB, Cowan GS, Jr., Rooney RJ, Hiler ML, Yellaturu CR, Deng X, Howell GE, Park EA, Gerling IC, Patel D, et al. 2009. Hepatic gene expression in morbidly obese women: implications for disease susceptibility. Obesity. 17:1563–1573.
- Elmen J, Lindow M, Schutz S, Lawrence M, Petri A, Obad S, Lindholm M, Hedtjarn M, Hansen HF, Berger U, et al. 2008. LNA-mediated microRNA silencing in non-human primates. Nature. 452:896–899. Epub 2008/03/28.
- Elmen J, Lindow M, Silahtaroglu A, Bak M, Christensen M, Lind-Thomsen A, Hedtjarn M, Hansen JB, Hansen HF, Straarup EM, et al. 2008. Antagonism of microRNA-122 in mice by systemically administered LNA-antimiR leads to up-regulation of a large set of predicted target mRNAs in the liver. Nucleic Acids Res. 36:1153–1162.
- Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, Watts L, Booten SL, Graham M, McKay R, et al. 2006. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metabol. 3:87–98.
- Gatfield D, Le Martelot G, Vejnar CE, Gerlach D, Schaad O, Fleury-Olela F, Ruskeepaa AL, Oresic M, Esau CC, Zdobnov EM, et al. 2009. Integration of microRNA miR-122 in hepatic circadian gene expression. Genes Dev. 23:1313–1326.
- Grimmer C, Balbus N, Lang U, Aigner T, Cramer T, Muller L, Swoboda B, Pfander D. 2006. Regulation of type II collagen synthesis during osteoarthritis by prolyl-4-hydroxylases: possible influence of low oxygen levels. Am J Pathol. 169:491–502.
- Kivirikko KI, Pihlajaniemi T. 1998. Collagen hydroxylases and the protein disulfide isomerase subunit of prolyl 4-hydroxylases. Adv Enzymol Relat Areas Mol Biol. 72:325–398.
- Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. 2005. Silencing of microRNAs in vivo with 'antagomirs'. Nature. 438:685–689. Epub 2005/11/01.
- Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. 2002. Identification of tissue-specific microRNAs from mouse. Curr Biol. 12:735–739. Epub 2002/05/15.
- Laudadio I, Manfroid I, Achouri Y, Schmidt D, Wilson MD, Cordi S, Thorrez L, Knoops L, Jacquemin P, Schuit F, et al. 2012. A feedback loop between the liver-enriched transcription factor network and miR-122 controls hepatocyte differentiation. Gastroenterology. 142:119–129. Epub 2011/09/17.
- Lee RC, Feinbaum RL, Ambros V. 1993. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 75:843–854. Dec 3Epub 1993/12/03.
- Li J, Ghazwani M, Zhang Y, Lu J, Li J, Fan J, Gandhi CR, Li S. 2013. miR-122 regulates collagen production via targeting hepatic stellate cells and suppressing P4HA1 expression. J Hepatol. 58:522–528. Epub 2012/11/28.
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. Epub 2002/02/16.
- Lou G, Yang Y, Liu F, Ye B, Chen Z, Zheng M, Liu Y. 2017. MiR-122 modification enhances the therapeutic efficacy of adipose tissue-derived mesenchymal stem cells against liver fibrosis. J Cell Mol Med. 21:2963–2973.
- Mignone F, Grillo G, Licciulli F, Iacono M, Liuni S, Kersey PJ, Duarte J, Saccone C, Pesole G. 2004. UTRdb and UTRsite: a collection of sequences and regulatory motifs of the untranslated regions of eukaryotic mRNAs. Nucleic Acids Res. 33:D141–D146. Epub 2004/12/21.
- Rocnik EF, Chan BM, Pickering JG. 1998. Evidence for a role of collagen synthesis in arterial smooth muscle cell migration. J Clin Invest. 101:1889–1898. Epub 1998/06/13.
- Stern-Ginossar N, Gur C, Biton M, Horwitz E, Elboim M, Stanietsky N, Mandelboim M, Mandelboim O. 2008. Human microRNAs regulate stress-induced immune responses mediated by the receptor NKG2D. Nat Immunol. 9:1065–1073. Epub 2008/08/05.
- Su J, Baigude H, McCarroll J, Rana TM. 2011. Silencing microRNA by interfering nanoparticles in mice. Nucleic Acids Res. 39:e38. Epub 2011/01/08.
- Wang X, Shao F, Yu J, Jiang H, Gong D, Gu Z. 2015. MicroRNA-122 targets genes related to liver metabolism in chickens. Comp Biochem Physiol B Biochem Mol Biol. 184:29–35.
- Wang XG, Shao F, Wang HJ, Yang L, Yu JF, Gong DQ, Gu ZL. 2013. MicroRNA-126 expression is decreased in cultured primary chicken hepatocytes and targets the sprouty-related EVH1 domain containing 1 mRNA. Poult Sci. 92:1888–1896. Epub 2013/06/19.
- Wienholds E, Plasterk RH. 2005. MicroRNA function in animal development. FEBS Letters. 579:5911–5922. Epub 2005/08/23.
- Xu H, He JH, Xiao ZD, Zhang QQ, Chen YQ, Zhou H, Qu LH. 2010. Liver-enriched transcription factors regulate microRNA-122 that targets CUTL1 during liver development. Hepatology. 52:1431–1442. Epub 2010/09/16.
- Yekta S, Shih IH, Bartel DP. 2004. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 304:594–596. Epub 2004/04/24.