?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
Data on the prevalence of major contagious pathogens in bulk tank milk (BTM) in Italy are generally not available. The availability of Real-Time PCR procedures (qPCR) to perform BTM analysis by represents an important step to define herd health status. Therefore, a cross-sectional epidemiological study was designed to assess the prevalence of contagious pathogens and Prototheca spp in BTM samples. The study was performed on 581 herds from four districts in the west Lombardy region of Italy. Additionally, the relationship between pathogens in BTM and SCC or milk yield; the presence of an association between four risk factors (district, herd size, average milk yield and SCC) with pathogens in BTM were assessed. The overall data showed that S. aureus was recovered in 42% of the herds, Str. agalactiae in 10%, Prototheca spp in 11% and M. bovis in 1.5% of the herds. The GLM model applied showed a significant influence of BTM results, district, herd size and their interactions on SCC and on milk yield variance. Particularly, S. aureus or Str. agalactiae have a significant effect on milk yield variability and, in a lesser extent, on SCC. The very high prevalence of contagious pathogens significantly affects milk characteristics and yield, thus affecting economic sustainability of the herds, and suggests the need to implement control programmes to decrease the prevalence of contagious pathogens, This will also allow to decrease the use of antimicrobials and to improve cow welfare.
First study on a large sample of Italian dairy herds on the prevalence of contagious pathogens in bulk tank milk samples. The prevalence value observed exceeded 50%.
First study estimating the prevalence of M. bovis in bulk tank milk in a large sample of Italian dairy herds, and the prevalence observed was 1.5%.
Prevalence of contagious pathogens has a significant influence on milk yield and SCC.
Bulk tank milk SCC confirmed to have a low accuracy to identify infected herds.
Highlights
Introduction
The need to know the health status of dairy herds is essential to increase herd efficiency and sustainability (Pulina et al. Citation2017). Among the diseases affecting dairy cows, mastitis is still the most important one significantly affecting the profits of dairy farmers (Summer et al. Citation2015; Goncalves et al. Citation2018; Heikkilä et al. Citation2018). Furthermore, this aspect is important when protocols based on a prudent use of antibiotics should be applied on such as selective dry cow therapy (Trevisi et al. Citation2014; Zecconi, Sesana, et al. Citation2018). Indeed, the presence of contagious bacteria (Mycoplasma bovis, Staphylococcus aureus, Streptococcus agalactiae) requires a larger use of antimicrobials in affected herds, compared to contagious-free herds (Zecconi et al. Citation2004; Zecconi Citation2007).
Despite the large importance of dairy production in Italy, data on the major contagious pathogens prevalence in bulk tank milk is not available either at national or regional level, except for Str. agalactiae data in few regions. Whereas, these data are available for other countries or regions (Piepers et al. Citation2007; Riekerink et al. Citation2008; Katholm et al. Citation2012).
Bulk tank milk (BTM) sampling is considered the procedure with the best cost/benefit ratio to assess the presence of contagious pathogens in a herd, being simple to perform and representative of the whole herd milk production (Jayarao and Wolfgang Citation2003). However, it has two major critical points: it is affected by the dilution factor, the chances to have a positive result are influenced by the number of infected cows and by their level of milk production; there are relatively high chances of contamination both during milking procedures and sampling. Indeed, the recovery of pathogens with reservoirs outside mammary gland in BTM (i.e. environmental pathogens) cannot be considered as a sure sign of the presence of infected cows, because it can be mainly due to environmental contamination. The level of BTM contaminations could also impair the accuracy of conventional microbiological analysis (Jayarao and Wolfgang Citation2003). For these reasons, BTM sampling should be restricted to the diagnosis of contagious pathogens, having their reservoir in the udder, and a positive result means the presence of at least one positive cow in the herd. Unfortunately, a negative result will not assure that the herd is free from contagious pathogens, even if it is unlikely, due to the dilution problem previously described.
The availability of molecular methods to perform microbiological analysis of BTM by the development of Real-Time PCR procedures (qPCR) represents an important positive step in applying BTM sampling to detect the presence of contagious pathogens. These procedures are targeting specific bacteria, therefore, the effects of contamination can be minimised, since bacteria species other than the ones considered in the specific analysis will be not amplified. The sample can be frozen, or a preserving agent can be added, thus facilitating the delivery and storage of the samples. Furthermore, these techniques are semi-quantitative, therefore it can be estimated the bacteria count for a specific pathogen (see Supplementary Table S1). The qPCR is a tool largely applied in Nordic countries (Koskinen et al. Citation2010; Katholm et al. Citation2012), but there aren’t any epidemiological study applying this technique on BTM reported for Italy at a regional or national level. This technique allows to identify three well-known contagious pathogens (Str. agalactiae, S. aureus and M. bovis) and an environmental one (Prototheca spp), an algae characterised by a large diffusion and high cell count, once an outbreak is established (Pieper et al. Citation2012).
The availability of qPCR technology allowed to plan a cross-sectional epidemiological study having the aims to measure the prevalence of contagious pathogens and Prototheca spp in BTM samples in Lombardy; to assess the relationship between pathogens in BTM and SCC or milk yield; to identify the presence of an association between four risk factors (district, herd size, average milk yield and SCC) with pathogens in BTM. The relationship between bacteria load in BTM and qPCR results were also investigated to gain useful information for the application of this technique in practice.
Material and methods
Study herds
Bulk tank milk (BTM) samples were collected in all the 581 dairy farms associated to Regional Breeders Association (ARAL) and performing monthly milk tests, within four districts of ARAL in the western area of Lombardy region. The four districts include Como and Lecco provinces (district 1), Milano and Lodi provinces (district 2), Pavia province (district 3) and Varese province (district 4). The four districts include all the three main geographical areas of the region (alpine, sub-alpine and Po valley).
ARAL provided also a database with the following information: herd size, herd location, yearly individual average milk yield and yearly individual average SCC of milk test records.
Sampling
Three BTM samples were collected per dairy farm over a period 4 months, from September 2016 to January 2017. For each dairy farm, the samples were collected three times within a 10 days period by ARAL technical service personnel. Samples were taken from the top of the tank using a clean, sanitised dipper after the milk was agitated for 5–10 min as suggested (Hogan et al. Citation1999). The samples were immediately frozen at −20 °C, delivered in a refrigerated truck within 4 h to ARAL laboratories and maintained at −20 °C until processed.
Molecular analysis
Bulk tank milk samples were analysed using qPCR. This technique showed to have a sensitivity of Se and Sp of qPCR respectively ≥0.95 and ≥0.99 for the contagious pathogens (Paradis et al. Citation2012; Hiitiö et al. Citation2016; Timonen et al. Citation2017).
A commercial diagnostic kit was used (Mastitis 4E kit; DNA Diagnostic A/S), following producer’s instruction. This kit allows bacterial DNA extraction, identification and quantification of S. aureus, Str. agalactiae M. bovis and Prototheca spp using qPCR. The reaction conditions of qPCR were as follows: 95 °C for 1 min, 40 amplification cycles at 95 °C for 5 s and 60 °C for 25 s. Cycle threshold (Ct) values were considered positive when the value were ≤37, as suggested by the manufacturer. The qPCR reactions were performed on Stratagene Mx3005P (Agilent Technologies Inc., Santa Clara, CA).
The producer of the kit supplies also an interpretative scheme correlating Ct obtained with bacteria counts as reported in Supplementary Table S1.
Statistical analyses
Data description and Armitage-Cochrane trend test were calculated by XLSTAT 2019 1.1 (Addinsoft, Boston, USA). Data were also analysed by ANOVA by using a generalised linear model applying GLM procedure of SAS 9.4 (SAS Institute Cary NC, USA). The model was:
where Y = dependent variables (SCC, average milk yield); µ = general mean; Di = effect of district (i = 1–4); Bj = effect of BTM results (j = negative, Str.agalactiae +ve; S.aureus +ve; M.bovis +ve; Prototheca spp + ve), Hk = effect of herd size (k = <45; 46–80; 81–120; 121–180; >180 lactating cows), Bj(Di) = effect of BTM nested in district, Bj(Hk) = effect of BTM nested in herd size, eijk = residual error.
A binary logistic analysis was performed by Procedure Logistic of SAS 9.4 (SAS Institute Cary NC, USA) in order to identify the risk factors associated with herd health status (presence/absence of one of the four udder pathogens considered) by calculating odds ratio, an estimate of relative risk (Thrusfield Citation2005). The risk factors considered were district, herd size, average milk yield (yearly average of milk records) and SCC (yearly average of milk records). The final models were described in terms of odds ratios and 95% confidence intervals.
Results
Herd characteristics
The study considered 581 dairy herds in four districts of Lombardy region, and herd size include a wide range of lactating cows (median 96.6, range 5–781). These districts represented all the different geographical areas of Lombardy (alpine, sub-alpine and Po valley). The herd characteristics considered in this study were reported in Supplementary Table S2.
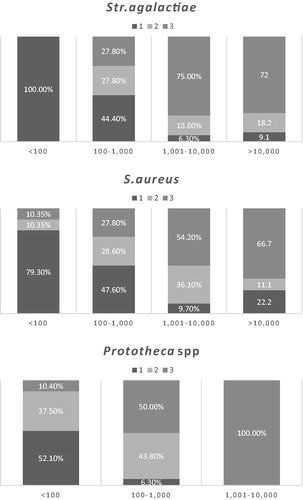
qPCR ct values interpreted as suggested by the producer (Supplementary Table 1) and frequency of BTM positive results suggest that a higher bacteria count (estimated by the number of Ct of the sample resulted positive, the lower the cycle the higher the bacteria count) will result in a higher number of consecutive positive samples. We analysed this aspect comparing the frequency of positive results with the estimated bacteria counts by Cohrane-Armitage test. Figure showed a significant trend (p < .0001) for the increase in frequency of positive BTM as bacteria count increases for S. aureus, Str. agalactiae and Prototheca spp. The analysis for Mycoplasma bovis was not performed due to the low number of positive samples.
Pathogen prevalence among provinces
The analysis in triplicate of bulk milk samples (BTM) showed the results reported in Table . In relatively few cases a combination of pathogens were diagnosed in BTM and more precisely: S. aureus and Str. agalactiae in 10 herds, Prototheca spp and S. aureus in 9 herds, Prototheca spp and Str. agalactiae in 3 herds and combinations of Str. agalactiae, S. aureus, Prototheca spp. and M. bovis or Str. agalactiae and S. aureus was found in one herd each.
Table 1. Prevalence of positive bulk tank milk samples for the four bacteria species considered.
The overall data showed as S. aureus was recovered in 42% of the herds with significant differences in their frequencies among the districts considered. Indeed, district 4 had a significant higher prevalence of S. aureus positive herds, when compared with all the other districts. Differences among district’s prevalence for all the other pathogens were not statistically significant, with the single exception for Prototheca spp.
Factors affecting milk production and SCC variability
The GLM model applied showed as both SCC and milk yield variability were significantly influenced by the factors considered, even if R2 values were largely different (Table ). Indeed, R2 for SCC has a value of 15%, while it is 40% for milk yield. The influence of district on SCC was the only factor resulted as not significant in the statistical analyses. BTM results, alone or combined with the other factors (district, herd size), were always statistically significant.
Table 2. Analysis of variance of the GLM model applied to assess the role of the factors considered on SCC and milk yield.
The analyses of the results for each single factor (Table ) showed that district 4 had the highest SCC mean values among the four districts considered (differences significant at α = 0.05). The same effect was observed for the two smaller herd size classes when compared with the other classes. Herd with BTM positive for Str. agalactiae showed the highest SCC values, when compared with the other bacteria.
Table 3. Results of contrast analysis related to the three main factors considered for SCC and milk yield.
Milk yield showed to be statistically higher in districts 2–4 when compared to all the other districts, while district 1 showed lowest values (differences significant at α = 0.05). Smaller herds, as expected, were significantly associated to the lowest production levels, and average milk yield increases as herd sizes increases. As expected, the lowest mean productions were observed in S. aureus and Str. agalactiae positive herds, while in all the other classes the differences were not statistically significant.
The interaction between BTM results and herd size was furthermore analysed (Table ). The analysis of the influence of this interaction showed as lowest SCC values were always observed for herds with negative BTM for all herd sizes, out of smaller ones. On the other side, the higher SCC values were always observed in contagious pathogen (Str. agalactiae, S. aureus) positive herds.
Table 4. Results of contrast analysis related to the interaction of BTM results and herd size and for SCC and milk yield.
When milk yield was considered, the differences between Str. agalactiae and S. aureus positive BTM herds and negative ones were significantly different in three out of five hers size classes (1–45, 81–120, >180).
Risk factors analysis
The analysis of the association between four factors (district, herd size, average milk yield and SCC) and the presence of the four udder pathogens considered were performed by binary logistic regression analysis, and it gave interesting results (Table ).
Table 5. Odds ratio (confidence limits 95%) for risk factors associated to the presence of the four pathogens considered in BTM.
When M. bovis was considered, SCC showed a consistent significant negative association with positive BTM in all SCC classes considered, out of >500,000 cells/ml class. In other words, BTM with an average SCC counts >200,000 cells/ml have a significant lower risk to be M. bovis positive when compared with BTM with SCC ≤200,000 cells/ml.
Herd size showed to be consistently and significantly associated to the presence of BTM positive for Str. agalactiae, Prototheca spp and M. bovis. Indeed, odds ratios for all herd size classes were significantly and positively associated with positive BTM when compared with reference herd size (<45 cows). Interestingly, herd size was not associated to S. aureus positive BTM, with the exception of herds in 81-120 class.
The analysis of the association between average milk yield and positive BTM gave opposite results. Only in the case of S. aureus a production <34 kg/d showed to be consistently and significantly associated to an increased risk of infection. These risks decrease as milk production increases, still staying significant.
Finally, when compared with district 1, all the other districts showed a higher risk for a positive BTM when Str. agalactiae, Prototheca spp and M. bovis were considered. Only for S. aureus the results were not consistent with a significant negative association for district 3 and a significant positive association for district 4.
Discussion
In our knowledge this is the first study performed in Italian dairy herds, either at regional or national levels, to estimate contemporarily the prevalence of three contagious pathogens (Str. agalactiae, S. aureus and M. bovis) and an environmental one (Prototheca spp). The study was made possible for the availability of the qPCR technology, which allows to analyse BTM samples targeting specific bacteria and the capability of Regional Breeder Association (ARAL) to collect and deliver BTM samples from herds across a very large territory.
There are some limitation to consider and they are related to the sampling that involved only the registered dairy herds (64.5% of the dairy herds in the same area), and that the district considered did not cover the entire Region. Despite these limitations, the number of herds sampled and their geographical distribution can be considered a good estimate of the situation at regional level.
The diagnostic tool applied confirmed to be a feasible approach to assess the prevalence of pathogens in BTM in our Region as shown in studies performed in other Countries (Francoz et al. Citation2012; Katholm et al. Citation2012).
The data confirmed also that bacteria count (estimated by Ct values) is positively correlated to frequency of BTM positive results (Francoz et al. Citation2012). Therefore, herds with a high prevalence of contagious mastitis or Prototheca spp have a higher likelihood to be identified by qPCR even with a single sampling.
The results of this epidemiological study showed as contagious pathogens have been detected in more than 50% of the herds considered. This number is much larger than expected. Indeed, a previous investigation performed in 2002 with conventional bacteriological methods showed a prevalence of about 31%, even though Mycoplasma were not considered in this study (Piccinini et al. Citation2003). The presence of this very high prevalence of contagious pathogens is of high concern because their effects are associated to a decrease in milk yield and quality. Moreover, these infections require very often the use of antimicrobials, particularly at drying-off (Zecconi et al. Citation2003; Zecconi Citation2007). Therefore, herds with contagious pathogens need to maintain blanket dry-cow therapy to control the infections, thus impairing the collective efforts to reduce the use of antimicrobials and the application of selective dry-cow therapy.
This study reports, for the first time, the prevalence of M. bovis in BTM from a large sample of Italian dairy herds. The prevalence observed is low, when compared to values observed in other countries (Lysnyansky et al. Citation2016; Timonen et al. Citation2017), but not negligible, and this suggests the importance to investigate furthermore the epidemiology of these bacteria in Italian dairy herds. As observed in previous studies Mycoplasma infections are associated to large herds (Fox Citation2012).
The frequency of Prototheca spp. is comparable to Str. agalactiae. However Prototheca spp. have their reservoir in animal gut and in the environment and their presence in BTM may be related to a contamination during milking. Therefore, the recovery of Prototheca spp. in BTM should be considered with caution.
The prevalence of BTM positive samples within districts showed to be statistically different in most of the cases when S. aureus was considered, and in few cases for the other pathogens, confirming that the epidemiology of S. aureus is different from the other contagious bacteria and involves both internal (virulence) and external factors (Zecconi et al. Citation2003; Zucali et al. Citation2009; Mazzilli et al. Citation2015; Zecconi et al. Citation2019).
As expected, the presence of contagious pathogens or Prototheca spp has a significant effect on milk yield variability and, in a lesser extent, on SCC (Gallo et al. Citation2002). Indeed, a significant influence of BTM status and its interactions with district and herd size on SCC and milk yield variance was observed. A depth analysis of these effects is beyond the aims of this paper. However, previous studies showed different bacteria affect differently both SCC and milk yield (Heikkilä et al. Citation2018) as well as herd size and management affect bacteria prevalence (Halasa et al. Citation2007).
When milk yield is considered in relation to the pathogen recovered in BTM some differences can be observed. Indeed, the highest mean differences in milk yield were observed in Str. agalactiae positive smaller herds, when compared with BTM negative ones. S. aureus showed a similar pattern with a smaller difference in comparison with negative herds, while M. bovis did not showed to have any influence on milk yield or SCC. This data confirm the negative influence of S. aureus or Str. agalactiae on milk production impairing economic sustainability of dairy herds.
The different epidemiological patterns and the effects of the pathogens on the variability of SCC and milk yield suggested to investigate the risks associated to the herd characteristics and to BTM results to identify and prioritise herd risks.
This analysis confirmed previous considerations. Indeed, district was always associated to an increased risk for a positive BTM for all the four pathogens considered. Only in the case of district 3, a significant preventive effect was observed, when compared to reference district (1). Herd size showed very high risks associated to all the pathogens out of S. aureus. However, this bacteria is the only one consistently associated to the lower level of production when compared with milk yield >34 kg/d. SCC showed to have a very poor predictive value for all the pathogens considered, out of M. bovis. Indeed, in this latter case odds ratio were significantly <1 and, therefore, suggest a protective effect. This result can be explained by the presence of M. bovis only in BTM of large herds. In these herds, a relative low frequency of infected cows can be expected, otherwise clinical cases should be observed. Therefore, we hypothesise that the negative effects on SCC due to infection are compensated by high production and low SCC of healthy cows, as confirmed by the data analysing herd-size classes (Table ).
Overall, these data support previous observation on the need to implement new diagnostic approaches based on epidemiological assessment of herd health status (Zecconi et al. Citation2019). Furthermore, SCC confirmed to have a low accuracy at the levels currently observed in Italian dairy herds and the need to improve somatic cells accuracy by implementing new methods such as differential cell counts (Zecconi, Vairani, et al. Citation2018).
Conclusions
This paper reports for the first time the results of a large-scale epidemiological study on the prevalence of the three major contagious pathogens and Prototheca spp in Italian dairy herds. The high prevalence of BTM positive for S. aureus and, in a lesser extent for the other two contagious pathogens considered are of particular concern, being higher than expected. The results of the study for the four risk factors considered (district, herd size, milk yield and SCC) confirm that S. aureus have a different epidemiological pattern than the other pathogens considered. This information should be considered to define intervention priorities and protocols. The results of this study suggest the need to implement control programmes, aiming to decrease the prevalence of contagious pathogens to increase the sustainability of the herds and to improve milk quality and safety.
Supplemental Material
Download MS Word (14.7 KB)Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Fox LK. 2012. Mycoplasma mastitis causes, transmission, and control. Vet Clin N Am-Food Anim Pract. 28:225–237.
- Francoz D, Bergeron L, Nadeau M, Beauchamp G. 2012. Prevalence of contagious mastitis pathogens in bulk tank milk in Québec. Can Vet J-Rev Vet Can. 53:1071–1078.
- Gallo L, Carnier P, Cassandro M, Cesarini F. 2002. Health disorders and their association with production and functional traits in Holstein Friesian cows. Italian J Anim Sci. 1:197–210.
- Goncalves JL, Cue RI, Botaro BG, Horst JA, Valloto AA, Santos MV. 2018. Milk losses associated with somatic cell counts by parity and stage of lactation. J Dairy Sci. 101:4357–4366.
- Halasa T, Huijps K, Osteras O, Hogeveen H. 2007. Economic effects of bovine mastitis and mastitis management: a review. Vet Q. 29:18–31.
- Heikkilä AM, Liski E, Pyörälä S, Taponen S. 2018. Pathogen-specific production losses in bovine mastitis. J Dairy Sci. 101:9493–9504.
- Hiitiö H, Simojoki H, Kalmus P, Holopainen J, Pyörälä S, Taponen S. 2016. The effect of sampling technique on PCR-based bacteriological results of bovine milk samples. J Dairy Sci. 99:6532–6541.
- Hogan JS, Gonzales RN, Harmon RJ, Nickerson SC, Oliver SP, Pankey JW, Smith KL. 1999. Laboratory handbook on bovine mastitis. Revised ed. Madison WI: National Mastitis Council Inc.
- Jayarao BM, Wolfgang DR. 2003. Bulk-tank milk analysis – A useful tool for improving milk quality and herd udder health. Vet Clin N Am-Food Anim Pract. 19:75–92.
- Katholm J, Bennedsgaard TW, Koskinen MT, Rattenborg E. 2012. Quality of bulk tank milk samples from Danish dairy herds based on real-time polymerase chain reaction identification of mastitis pathogens. J Dairy Sci. 95:5702–5708.
- Koskinen MT, Wellenberg GJ, Sampimon OC, Holopainen J, Rothkamp A, Salmikivi L, van Haeringen WA, Lam T, Pyörälä S. 2010. Field comparison of real-time polymerase chain reaction and bacterial culture for identification of bovine mastitis bacteria. J Dairy Sci. 93:5707–5715.
- Lysnyansky I, Freed M, Rosales RS, Mikula I, Khateb N, Gerchman I, van Straten M, Levisohn S. 2016. An overview of Mycoplasma bovis mastitis in Israel (2004-2014). Vet J. 207:180–183.
- Mazzilli M, Piccinini R, Scali F, Zecconi A. 2015. Pattern characterization of genes involved in non-specific immune response in Staphylococcus aureus isolates from intramammary infections. Res Veter Sci. 103:54–59.
- Olde Riekerink RGM, Barkema HW, Kelton DF, Scholl DT. 2008. Incidence rate of clinical mastitis on canadian dairy farms. J Dairy Sci. 91:1366–1377.
- Paradis ME, Haine D, Gillespie B, Oliver SP, Messier S, Comeau J, Scholl DT. 2012. Bayesian estimation of the diagnostic accuracy of a multiplex real-time PCR assay and bacteriological culture for 4 common bovine intramammary pathogens. J Dairy Sci. 95:6436–6448.
- Pieper L, Godkin A, Roesler U, Polleichtner A, Slavic D, Leslie KE, Kelton DF. 2012. Herd characteristics and cow-level factors associated with Prototheca mastitis on dairy farms in Ontario, Canada. J Dairy Sci. 95:5635–5644.
- Piepers S, De Meulemeester L, de Kruif A, Opsomer G, Barkema HW, De Vliegher S. 2007. Prevalence and distribution of mastitis pathogens in subclinically infected dairy cows in Flanders, Belgium. J Dairy Res. 74:478–483.
- Piccinini R, Binda E, Zecconi A, Zanini E. 2003. Prevalence study on bulk milk tank cultures in 1000 dairy herds in Lombardia (Italy). 42nd NMC Annual Meeting, Fort Worth; Jan 26–29. Madison WI: National Mastitis Council Inc.
- Pulina G, Francesconi AHD, Stefanon B, Sevi A, Calamari L, Lacetera N, Dell’Orto V, Pilla F, Marsan PA, Mele M, et al. 2017. Sustainable ruminant production to help feed the planet. Italian J Anim Sci. 16:140–171.
- Summer A, Franceschi P, Formaggioni P, Malacarne M. 2015. Influence of milk somatic cell content on Parmigiano-Reggiano cheese yield. J Dairy Res. 82:222–227.
- Thrusfield M. 2005. Veterinary Epidemiology. III ed. Oxford: Blackwell Science Ltd.
- Timonen AAE, Katholm J, Petersen A, Motus K, Kalmus P. 2017. Within-herd prevalence of intramammary infection caused by Mycoplasma bovis and associations between cow udder health, milk yield, and composition. J Dairy Sci. 100:6554–6561.
- Trevisi E, Zecconi A, Cogrossi S, Razzuoli E, Grossi P, Amadori M. 2014. Strategies for reduced antibiotic usage in dairy cattle farms. Res Veter Sci. 96:229–233.
- Zecconi A, Piccinini R, Fox KL. 2003. Epidemiologic study of intramammary infections with Staphylococcus aureus during a control program in nine commercial dairy herds. JAVMA. 223:684–688.
- Zecconi A, Piccinini R, Fox KL. 2004. Epidemiological study of non-contagious intramammary infections in nine commercial dairy herds following a Staphylococcus aureus control programme. JVetMed B. 51:333–336.
- Zecconi A, Scali F, Bonizzi L, Ferrari N, Ferrero F, Grillo G, Lanfranchi P, Mortarino M, Sala V, Taloni D, et al. 2019. Risk prioritization as a tool to guide veterinary public health activities at regional level. Veterinaria Italiana. 55:113–121.
- Zecconi A, Sesana G, Vairani D, Cipolla M, Rizzi N, Zanini L. 2018. Somatic cell count as a decision tool for selective dry cow therapy in Italy. Ital J Anim Sci. 17:1–6.
- Zecconi A, Vairani D, Cipolla M, Rizzi N, Zanini L. 2018. Assessment of subclinical mastitis diagnostic accuracy by differential cell count in individual cow milk. Ital J Anim Sci. 18:435–440.
- Zecconi A. 2007. Contagious mastitis control. FIL-IDF Bull. 416:34–40.
- Zucali M, Bava L, Sandrucci A, Tamburini A, Piccinini R, Dapra V, Tonni M, Zecconi A. 2009. Milk flow pattern, somatic cell count and teat apex score in primiparous dairy cows at the beginning of lactation. Ital J Anim Sci. 8:103–111.