Abstract
Milk protein genes are known to be highly polymorphic. Several studies have shown the influence of milk proteins genetic variants and casein haplotypes on milk nutritional and technological properties. From 1990, the analysis of caseins polymorphism gained new attention due to the concern about possible negative effects of CSN2*A1 on human health. As a consequence, the CSN2*A2 variant gained interest and milk produced by with A2A2 β-casein cows is now available in different countries. Aim of the present paper was to analyse how casein variants’ and haplotypes’ frequencies changed in the Italian Holstein breed, due to the health ‘claims’ about CSN2*A2, and the possible effects on milk technological properties. Data were compared also with Italian Jersey cattle. A total of 223,655 Holstein and of 622 Jersey were genotyped using Illumina beadchips and data of 62 SNP in the casein cluster were analysed to reconstruct casein genotypes and haplotypes. The results demonstrate that although the selection towards β-casein A2 is not so effective in Italy, an increase of the frequency of this allele to the disadvantage of A1 and B alleles, included in the most favourable genotypes and haplotypes for cheese making, is occurring and should not be forced. Indeed, although a selection for the favourable κ-casein B allele, and against the unfavourable E allele, is occurring and is limiting the general loss of haplotypes associated with good technological properties, the sharp decrease of the favourable B-B and I-B β- κ-haplotypes, is an alert of the risk of losing useful biodiversity.
After the concern about the β-caseinA1 variant on human health, a selection favoring the A2 allele was carried out in different countries.
In Italy milk is mostly destined to cheese-making and even without a direct selection for theβ-casein A2 caseins allelic frequencies are changing.
Genotyping data should be used to monitor and maybe contrast the reduction of variants associated with milk favorable technological properties.
Highlights
Introduction
Milk protein genes are known to be highly polymorphic in ruminants and the effects of this variability on milk nutritional and technological properties is well documented (Caroli et al. Citation2009). Nevertheless, casein variants were rarely taken into consideration in selection schemes. Starting from the 1990s, after some concerns in New Zealand for potential negative effects of β-casein A1 milk consumption on human health (Elliott Citation1992; McLachlan Citation1996), a company, A2 Corporation, started marketing milk produced only by β-casein A2A2 individuals. The bioactive peptide with opioid properties β-casomorphin-7 (BCM7), one of the several peptides released during milk protein digestion, was suspected to be the risk factor in human disease as Type 1 diabetes, coronary heart disease, infant death syndrome, neurological disorders (e.g. autism and schizophrenia), and milk allergy (Elliott Citation1992; McLachlan Citation1996, Citation2001; Laugesen and Elliott Citation2003; Woodford Citation2007). CSN2*A2 and CSN2*A3 variants differ from CSN2*A1, CSN2*B, and CSN2*C for the amino acid substitution of a histidine (His) with a proline (Pro) at position 67 in the mature protein. The presence of His67 determines the enzymatic cleavage which releases BCM7 in the last three variants. Besides the epidemiological studies finding a correlation between milk consumption and the aforementioned diseases, other studies failed to demonstrate a relationship (Hunter et al. Citation2003; Truswell Citation2005; Chin-Dusting et al. Citation2006; Venn et al. Citation2006; Cass et al. Citation2008), and no clinical trials on human beings were carried out to verify it. Therefore, in 2009 a European Food Safety Authority report (EFSA Citation2009) stated that a cause–effect relationship between BCM7 or related peptides and the aetiology of the diseases could not be established.
More recently, new researches pointed at possible intolerances and gastrointestinal effects of BCM7, associating CSN2*A1A1 milk consumption to delayed intestinal transit, looser stool consistency, and intestinal inflammation, and found an overall evidence in animal trials and in vitro studies (Brooke-Taylor et al. Citation2017). Anyway, as many of these studies were sponsored by A2 Corporation, which had the interest to demonstrate that A2 milk is better than other commercial milks, were carried out on small cohorts and in people intolerant to milk and usually without controlling the rest of the diet, or did not result in significant differences that can directly be imputed to the CSN2*A1 variant, further validations underlying BCM7 mechanism role and assessing whether negative effects are general or limited to sensitive individuals are needed to prove its the effects on human population (Hedge Citation2019; Summer et al. Citation2020). Nevertheless, CSN2*A2A2 milk is now marketed in various countries and CSN2*A2 is considered the allele to be selected for. Aim of the present study was to analyse how the frequencies of the main casein variants and haplotypes changed in the Italian Holstein breed due to the unhealthy ‘claims’ about CSN2*A1, and the possible effects on milk technological properties. Data were compared also with Italian Jersey cattle.
Materials and methods
A total of 223,655 Holstein and of 622 Italian Jersey (IJ) individual samples were genotyped using different Illumina Beadchips. Of the Holstein samples, 213,124 can be considered as older individuals (oHF), whose genotyping data were already stored by the National Association of Italian Holstein and Jersey Breeds (ANAFIJ), whereas the 10,531 remaining samples, together with the 622 IJ, were Italian Holstein recently analysed thanks to the PSRN project (rIH). oHF included animals born from 1952 to 2017 and coming from different countries, with the prevalence of USA individuals (50.86%), but with the Italian samples being the second most represented (23.85%), Canada the third (9.82%), and other countries accounting for a small percentage of the total dataset.
The IJ and rIH were genotyped using the GeneSeek Genomic Profiler 3 (26 K, 53 samples), GeneSeek Genomic Profiler 4 (30 K, 13,027 samples), GeneSeek MD (48 K, 6,621 samples) and Labogena MD (42 K, 3,678 samples). DNA was extracted from blood using a commercial kit and subsequently analysed by LGS (Cremona, Italy), Geneseek (Scotland, UK) and IFN (Berlin, Germany). Blood sampling was performed in accordance with Italian Animal Protection Law (Legislative Decree No. 26/2014, https://www.gazzettaufficiale.it).
For the Holstein breed the possibility to impute missing SNPs was also tested and the oHF population was used to verify which SNP could successfully be imputed. Imputation was performed using pedimpute (Nicolazzi et al., Citation2013).
Casein genotyping data were pruned from total genotyping data and analysed using various R software packages (http://cran.r-project.org), and the intragenic haplotypes for each casein gene and the casein cluster haplotypes were analysed using Phase 2.1 software (Stephens et al. Citation2001; Stephens and Scheet Citation2005).
Results
SNP in the casein cluster
Considering upstream, intergenic and downstream variants a total of 62 SNP found on the chips used were considered as belonging to the casein cluster. Since 6 SNP corresponded to other SNP called with different names (BCN_8356 = CSN2_1; BCNAB = BCN_8267 = CSN2_3; BCN_8178 = CSN2_4; BCN_8163 = CSN2_5; KappaCasein12951_1 = KappaCasein12951_2 = KappaCasein12951_3; GNSC355 = BovineHD4100005327), we finally considered 54 unique SNP (Supplementary Table 1). For some SNP a low number of individuals were actually genotyped and were imputed in the remained oHF population: only 11 SNP were genotyped in at least the 30% of the oHF, 30 SNP were genotyped in at least about the 20%, 3 further SNP were genotyped in at least the 8.40% and the remaining 20 SNP were genotyped in about the 1%. Genotypes frequencies for 6 of these last 20 SNP were significantly different from the ones of the genotyped individuals. As this was indicative of imputation errors, we excluded all the SNP imputed with less than the 10% of the individuals genotyped in the oHF population from the subsequent analyses on the rIH and IJ population. One SNP (HM27109-BTC-060711) whose genotype frequencies in imputed oHF were significantly different from the ones of the genotyped individuals, although genotyped in more than the 35% of the oHF, was maintained since in the imputed rIH the difference in imputed versus genotyped individuals was not statistically different.
Table 1. SNP with at least 10% of genotyped individuals in the older Holstein Friesian archived genotyped individuals (oHF), and with a minor allele frequency of at least 1% in the genotyped oHF, recently analysed Italian Holstein (rIH), and Italian Jersey (IJ).
Of the 31 SNP remaining, 5 more were excluded from subsequent analyses as they were not polymorphic in the genotyped oHF, rIH and IJ. Therefore, 26 SNP could be used to reconstruct or impute the casein genotypes of each individual (Table ). Of these, 12 had to be imputed in rIH: 7 SNP were successfully genotyped in less than the 2.2% of the rIH, whereas the remaining 5 had to be imputed in about half of the population, since a number of individual ranging from 48.10 to 51.30% were successfully genotyped. For these 12 SNP the imputed genotype frequencies were not significantly different from the ones of the genotyped individuals. In the IJ, 7 SNP in the non-coding regions of the genes had only missing genotypes, therefore for the reconstruction of intragenic and casein cluster haplotypes only missense mutations were taken into consideration. Since BCN_849, although listed as missense mutation, was reported only in genomic projects, but never confirmed at the protein level, and was found only in oHF with a minor allele frequency of 0.002, also this SNP was not included in the intragenic and casein cluster haplotype reconstruction.
Allele frequencies of the β- and k-casein alleles
Table shows the results of the β- and k-casein allele frequencies obtained from the intragenic haplotype reconstruction in the three populations, and these are reported as a function of time in the oHF considered both in its total (toHF) and only in the Italian sample (IoHF). At the CSN2 gene, the A2 allele resulted the most frequent in all three populations, and the difference in frequency to the A1 allele increased sensibly after 1990. It appeared that before 1990 the frequency of the A1 allele was even higher than that of A2, but this could be due to the limited number of samples analysed (166). For CSN3, the A allele is still the most frequent in the Holstein, but the frequency of the B allele is constantly increasing (from 18.6% before 1990 to more than 30% nowadays), whereas in the Jersey B is the predominant variant (90%).
Table 2. Changes in the frequency (%) of the β- and k-casein alleles obtained by intragenic haplotype reconstruction in the total worldwide and Italian Holstein archived genotyped individuals (toHF and IoHF, respectively), recently analysed Italian Holstein (rIH), and Italian Jersey (IJ) from before 1990 to nowadays.
Haplotypes frequencies
Table shows the results of the β-k casein haplotype frequencies obtained in rIH and IJ, compared with previously reported haplotypes frequencies in the Italian Holstein breed (Boettcher et al. Citation2004; Chessa et al. Citation2014). As it can be noted the predominant haplotype in the Holstein is the A2-A haplotype (about 48%), followed by the A1-A (22–28%), which clearly depends on the high frequency of the k-casein A variant.
Table 3. Frequency (%) of the β-k-casein haplotype found in the recently analysed Italian Holstein (rIH), and Italian Jersey (IJ) populations. Haplotypes data were compared with previously published papers (Boettcher et al. Citation2004; Chessa et al. Citation2014).
Discussion
SNP in the casein cluster
The Illumina Beadchip used are mainly focused on β- and k-casein. As a matter of fact the SNP related to CSN1S1 gene that are necessary to distinguish alleles of potential interest, as C, E, F and G are not in the genotyping chips and therefore no useful intragenic haplotypes could be analysed at the CSN1S1. Moreover, at the CSN1S2 gene only the A allele was described in the Holstein and Jersey breeds and the D allele, which can be genotyped using the chips, was absent in the Jersey and had a frequency lower than 0.01 and 0.04 in the oHF and rIH, respectively. Thus, no intragenic haplotype analysis was necessary. A further molecular analysis could help confirm the presence of the D allele in the analysed samples and exclude the possibility of genotyping errors. As for the CSN2 gene considered only in the genotyped populations, the SNP responsible for the C allele was found only in oHF at a frequency lower than 0.01, whereas the one responsible for F allele was found both in oHF and in IJ with a frequency lower than 0.01. Again, a further molecular analysis could help confirm the presence of the alleles in the analysed samples and exclude the possibility of genotyping errors.
Allele frequencies of the β- and k-casein alleles
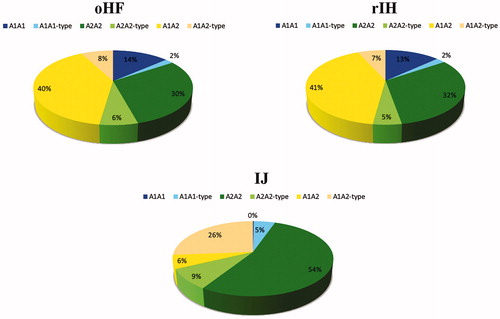
Considering the debate about the negative effects of CSN2*A1, although the responsibility of BCM7 on the lower digestibility of milk cannot be considered verified in human, milk produced by A2 homozygous cows is commercialised and sold at higher prices than other milks. Actually, it should also be considered that A2, A3, D, E, H2 and I variants all behave in the same way, not releasing the BCM7 and we can define them as A2 type. Variants A1, B, C, F, G, H1, instead, all possess the His67 and potentially release BCM7, and can be defined as A1 type. Thus, to maintain genetic variability, all the variants should be considered. To produce the so called A2 milk, cows homozygous for the A2 allele, together with the homozygous and the heterozygous for the A2-type alleles could be used, since no BCM7 will be released. If we consider the genotypes distribution, the total A2A2 (real A2A2 and A2A2-type) accounted for 36% in the oHF, 37% in the rIH, and 63% in the IJ, the total A1A1 (real A1A1 and A1A1-type) for the 16% in oHF, 15% in rIH, and 5% in IJ, and the total A1A2 (real A1A2 and A1A2-type) for the 48% in oHF and rIH and the 32% in IJ (Figure ).
Figure 1. Frequency (%) of real A1A1, A1A1-type, real A2A2, A2A2-type, real A1A2 and A1A2-type genotypes in the older Holstein Friesian individuals (oHF), recently analysed Italian Holstein (rIH), and Italian Jersey (IJ) populations.
Because of the marketing success of the A2A2 milk in the Anglo-Saxon countries, the semen of A2A2 bulls seems to be used more often in Italy too, although, if we look at the data, an increase of the frequency of A2 was already going on before 2007, when the paper of Kamiński et al. (Citation2007), increased the European community interest about the A1/A2 effects and lead to the EFSA report of 2009. As a matter of fact, the frequency of A2 allele increased from 45.6% in toHF, and 39.2% in IoHF, to 56.4% in the recently analysed (rIH) samples (Table ), but this increase was higher in the first decades after 1990: +5% in cows born in 1990–2000, +3.2% in 2000–2010, +0.4% in 2010–2017 with respect to toHF; +9.8% in 1990–2000, +4.1% in 2000–2010, +2.7 in 2010–2017 with respect to IoHF. rIH A2 frequency increased of about the 2.2% and 0.6%, respectively, with respect to the last toHF and IoHF data. Thus, it seems that other selection strategies, instead of direct selection for increasing A2 allele frequency, are shifting casein variants frequency. Anyway, since its increment in the last years with respect to toHF is higher than in 2010–2017, maybe some selection towards A2 allele is taking place and should be monitored. Simultaneously the frequencies of A1 and B decreased from about the 41% to 36.5% and from 4.2% (toHF) and 7.5% (IoHF) to 2.8%, respectively. In the Jersey breed the high frequency of A2 was confirmed (73.0%), with the B variant being the second most frequent variant (17.3%). It has to be noted that the β-casein B variant is considered positively associated with milk technological properties, although it belongs to the A1-type variants. Therefore, selecting against this variant could determine a worsening of milk technological properties. In the Holstein populations a selection for the B allele of k-casein, known for its positive association with cheese-making properties, seems to be carried out, determining an increase of its frequency (from 18.6% in toHF and 24.9% in IoHF to 32.2% in rIH) and a decrease of A (from 67.4% in toHF and 61.8% in IoHF to 57.2% in rIH) allele (Table ).
Recently De Noni et al. (Citation2015) analysing different types of cheese (Supplementary Table 2), found that not all cheeses, even if obtained by cows carrying the A1 protein variant, contained BCM7 before digestion. After in vitro digestion of some cheeses containing BCM7, it resulted that the amounts of BCM7 formed at the intestinal step seemed insufficient to exert a physiological activity. Researches are going on to analyse if this is also true in vivo, but it has to be considered that different peptides released by cheeses could exert an effect, and BCM7 alone could not be the sole responsible of negative effects. The casein cluster is a complex system able to exert a global effect both on nutritional and technological properties of milk, due to the high number of bioactive peptides released and to the arrangement of the casein genes in specific haplotypes. Therefore, to analyse the effect of a single variant of a single casein instead of considering the casein cluster as a whole, could lead to misleading results, since other peptides could exert synergic or opposing activity to that of BCM7. This strengthens the importance of considering the casein cluster as a whole and to verify the effects of different haplotypes on milk nutritional and technological properties.
Previous researches have shown that B-A1-B haplotype (Chessa et al. Citation2014), and the combination of BB-BB β-k genotypes (Comin et al. Citation2008) are associated with the best cheese-making attitude traits, thus selection against A1 and B could reduce the cheese-making properties of milk. The β-κ-casein haplotypes found in the rIH actually demonstrate that the increase of the B allele and the decrease of the E allele at the k-casein are compensating the increase of the allele A2 at the β-casein, generally maintaining a balanced proportion of haplotypes associated with good cheese-making properties (A1-B haplotype increased its frequency to 8.1% from less than 4–6%), with respect to the haplotypes associated with poor cheese-making properties. Nevertheless, the frequency of the B-B (from 6% to 1.2%) and of the I-B (from 3% to less than 1%) haplotypes, associated with good cheese-making properties, has decreased sharply and this should be monitored. In the Jersey, the predominant haplotype was obviously the A2-B, but the frequency of the B-B haplotype was also high, representing 17.1% of the total haplotypes.
Conclusions
After the concern about the β-casein A1 variant on human health, a selection favouring the A2 allele was carried out in different countries. The present work shows that in Italy different forces than a direct selection for the β-casein A2 allele seem to be the main responsible for a change in caseins allelic frequency. We are assisting to the drastic reduction of the frequency of some alleles and haplotypes, such as the β-casein B allele, included in genotypes and haplotypes associated with favourable technological properties. Considering that milk in Italy is mostly used for cheese-making, the selection of other alleles at other casein loci, such as the B variant of κ-casein, is somehow compensating for the loss of genetic and haplotype variability, although some haplotypes associated with good cheese-making properties have now a very low frequency. Researches are ongoing to analyse if the β-casein A1 variant is really responsible of a lower digestibility of milk, but most studies are focusing on the single variant, instead of considering the milk as the complex mixture of nutrients that are all together responsible for its properties. Moreover, it seems that the quantity of BCM7 found in cheeses is insufficient to exert a biological effect in vivo. Therefore, unless new solid scientific reports on the negative effects of BCM7 and on its presence in cheeses are published, the increasing availability of genotyping data should be used for a better understanding of the real effects of casein genotypes and haplotypes on both nutritional and technological properties of milk, and could help addressing selection schemes better suited for the Italian dairy market.
Ethical Approval
The experimental method was performed in accordance with Italian Animal Protection Law (Legislative Decree No. 26/2014, https://www.gazzettaufficiale.it).
Supplemental Material
Download MS Word (33.9 KB)Acknowledgment
This study was supported by ‘Latteco project’, sottomisura 10.2 of the PSRN Biodiversity 2014–2020.
References
- Boettcher PJ, Caroli A, Stella A, Chessa S, Budelli E, Canavesi F, Ghiroldi S, Pagnacco G. 2004. Effects of casein haplotypes on production traits in Italian Holstein and Brown cattle. J Dairy Sci. 87(12):4311–4317.
- Brooke-Taylor S, Dwyer K, Woodford K, Kost N. 2017. Systematic review of the gastrointestinal effects of A1 compared with A2 β-casein. Adv Nutr. 8(5):739–748.
- Caroli AM, Chessa S, Erhardt GJ. 2009. Invited review: milk protein polymorphisms in cattle: effect on animal breeding and human nutrition. J Dairy Sci. 92(11):5335–5352.
- Cass H, Gringras P, March J, McKendrick I, O’Hare AE, Owen L, Pollin C. 2008. Absence of urinary opioid peptides in children with autism. Arch Dis Child. 93(9):745–750.
- Chessa S, Bulgari O, Rizzi R, Calamari L, Bani P, Biffani S, Caroli AM. 2014. Selection for milk coagulation properties predicted by Fourier transform infrared spectroscopy in the Italian Holstein-Friesian breed. J Dairy Sci. 97(7):4512–4521.
- Chin-Dusting J, Shennan J, Jones E, Williams C, Kingwell B, Dart A. 2006. Effect of dietary supplementation with beta-casein A1 or A2 on markers of disease development in individuals at high risk of cardiovascular disease. Br J Nutr. 95(1):136–144.
- Comin A, Cassandro M, Chessa S, Ojala M, Dal Zotto R, De Marchi M, Carnier P, Gallo L, Pagnacco G, Bittante G. 2008. Effects of composite beta- and kappa-casein genotypes on milk coagulation, quality, and yield traits in Italian Holstein cows . J Dairy Sci. 91(10):4022–4027.
- De Noni I, Stuknytė M, Cattaneo S. 2015. Identification of β-casomorphins 3 to 7 in cheeses and in their in vitro gastrointestinal digestates. LWT – Food Sci Technol. 63(1):550–555.
- EFSA. 2009. Scientific Report of EFSA prepared by a DATEX Working Group on the potential health impact of β-casomorphins and related peptides. European Food Safety Authority Science Report. 1–107.
- Elliott RB. 1992. Epidemiology of diabetes in Polynesia & New Zealand. Epidemiology and etiology of Insulin-Dependent Diabetes in the Young. (Eds) Levy-Marchal C and Czernichow p. 66–71.
- Hedge N. 2019. Research on A1 and A2 milk: A1 milk is not a matter of health concern. Indian J Anim Sci. 89(7):707–711.
- Hunter LC, O'Hare A, Herron WJ, Fisher LA, Jones GE. 2003. Opioid peptides and dipeptidyl peptidase in autism. Dev Med Child Neurol. 45(2):121–128.
- Kamiński S, Cieslińska A, Kostyra E. 2007. Polymorphism of bovine beta-casein and its potential effect on human health. J Appl Genet. 48(3):189–198.
- Laugesen M, Elliott R. 2003. Ischaemic heart disease, type 1 diabetes, and cow milk A1 beta-casein. N Z Med J. 116(1168):U295.
- McLachlan CN. 1996. New Zealand Patent Application 306584.
- McLachlan CN. 2001. Beta-casein A1, ischaemic heart disease mortality, and other illnesses. Med Hypotheses. 56(2):262–272.
- Nicolazzi E L, Biffani S, Jansen G. 2013. Short communication: imputing genotypes using PedImpute fast algorithm combining pedigree and population information. Journal of Dairy Science. J Dairy Sci. 96(4):2649–2653. doi:10.3168/jds.2012-6062. 23462161
- Stephens M, Scheet P. 2005. Accounting for decay of linkage disequilibrium in haplotype inference and missing-data imputation. Am J Hum Genet. 76(3):449–462.
- Stephens M, Smith N, Donnelly P. 2001. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 68(4):978–989.
- Summer A, Di Frangia F, Ajmone Marsan P, De Noni I, Malacarne M. 2020. Occurrence, biological properties and potential effects on human health of β-casomorphin 7: current knowledge and concerns. Crit Rev Food Sci. 1–19.DOI:10.1080/10408398.2019.1707157
- Truswell AS. 2005. The A2 milk case: a critical review. Eur J Clin Nutr. 59(5):623–631.
- Venn BJ, Skeaff CM, Brown R, Mann JI, Green TJ. 2006. A comparison of the effects of A1 and A2 beta-casein protein variants on blood cholesterol concentrations in New Zealand adults. Atherosclerosis. 188(1):175–178.
- Woodford K. 2007. Devil in the milk: Illness, health and politics: A1 and A2 milk. Wellington, New Zealand: Craig Potton Publishing.
