Abstract
Cytochrome P450 (CYP) 3A is one of the most important subfamily of drug metabolising enzymes. Genetic factors such as breed and allelic variants might modify CYP3A expression and enzyme activity and lead to inter- and intra-individual variability in clinical response or toxicity. Cattle CYP3A gene cluster has been recently re-sequenced in 300 Piedmontese cattle by deep targeted sequencing. Thirteen missense single nucleotide variants (SNVs) were identified, and for five of them an impact on CYP3A activity was demonstrated in vitro. In the present work, we assessed the genetic frequency of these five SNVs in Limousine, a cattle breed widely raised for meat production in Veneto Region. A total of 215 cows were genotyped using specific melting curve assays. Only two missense SNVs out of five, both located on CYP3A28 coding sequence, were detected: rs384467435 and rs454167819. The former mutant allele was present in homozygosis in the 4% of tested cows, while in heterozygosis in the 24% of animals. The second SNV was detected only in heterozygosis in 11 cows out of 215 (i.e. 4.9%). These findings suggest the presence of missense SNVs, proved to halve CYP3A28 catalytic activity, in both Limousine and Piedmontese meat cattle breeds. As a consequence, these SNVs might impact on the kinetics of xenobiotics metabolised by CYP3A, including drugs and natural toxins like ivermectin and aflatoxins, thereby resulting in toxicity or accumulation of harmful residues in foodstuffs. This result paves the way for new considerations on the fate of xenobiotics in cattle farming.
Missense single nucleotide variants affecting CYP3A catalytic activity in vitro are present in Limousine and Piedmontese cattle breeds.
Rs384467435 is the only missense single nucleotide variant for which the mutant allele is recorded in homozygosis in approximately 4% of Limousine and Piedmontese cattle.
The polymorphisms here detected might affect the kinetics and the toxicity of xenobiotics metabolised by CYP3A in cattle.
HIGHLIGHTS
Introduction
In adult humans, the cytochrome P450 3A (CYP3A) is the most important subfamily of drug metabolising enzymes owing to its broad substrate specificity (70% of clinically used drugs, natural, anthropogenic and endogenous compounds). Several genetic, environmental and physiological factors may affect CYP3A expression and activity. In particular, human CYP3A4/5 genetic variants significantly contribute to ethnic, intra- and inter-individual variability in xenobiotic metabolism, leading to alterations in pharmacokinetics, clinical response and/or toxicity (Klein and Zanger Citation2013; Werk and Cascorbi Citation2014; Hohmann et al. Citation2016).
Cattle is one of the most important farm animal species, kept worldwide for milk and meat production. The bovine CYP3A (bCYP3A) subfamily consists of three genes, i.e. CYP3A28, CYP3A38, and CYP3A48 (Zancanella et al. Citation2010). They contribute to the oxidative metabolism of drugs commonly used in bovine practice, such as the macrocyclic lactone moxidectin, the pleuromutilin and macrolide antibiotics (e.g. tiamulin and tilmicosin, respectively) as well as the ionophore antibiotic monensin (Zweers-Zeilmaker et al. Citation1999; Dupuy et al. Citation2001; Nebbia et al. Citation2001). Furthermore, they are responsible for the bioactivation of natural toxins of current growing interest, i.e. aflatoxins and ergot alkaloids (Kuilman et al. Citation2000; Sales et al. Citation2012; Rosenkrans and Ezell Citation2015).
Breed-differences in bCYP3As expression and catalytic activity have been previously highlighted (Dacasto et al. Citation2005; Giantin et al. Citation2008; Ashwell et al. Citation2011; Zancanella et al. Citation2014). Conversely, little information is available on bCYP3A genetic polymorphisms and their possible consequences on the kinetics of xenobiotics, except for some effects on productivity traits (Sales et al. Citation2012, Citation2013). As an example, Brahman cattle show a greater tolerance to fescue toxicosis than Angus cattle (Brown et al. Citation1993), and this different sensitivity to ergot alkaloids has been associated to the presence of a polymorphism in CYP3A48 coding sequence (Sales et al. Citation2012). Specifically, cows with the mutant allele in homozygosis and grazing toxic tall fescue tended to reduce milk production and butterfat percentage, probably due to an altered CYP protein structure and function (Sales et al. Citation2012). These same authors identified further polymorphisms within the CYP3A48 regulatory region, and speculated that the reduction in calf growth was attributable to the consumption of ergot alkaloids (metabolised by CYP3A), either acquired directly by grazing than indirectly (as metabolites) through the milk (Sales et al. Citation2013).
Recently, thirteen bCYP3A missense single nucleotide variants (SNVs) were identified in Piedmontese beef cattle, thanks to the CYP3A gene cluster deep sequencing of 300 bulls. Subsequently, their functional impact was assessed by heterologous expression and measurement of CYP3A-dependent catalytic activity (Giantin et al. Citation2019). Five SNVs out of thirteen showed differences in the metabolism of CYP3A probe substrates: three CYP3A28 SNVs halved testosterone (TST) 6β-hydroxylation; one CYP3A38 variant triplicated TST 16β-hydroxylation, while a CYP3A48 SNV showed enhanced nifedipine oxidation. Interestingly, the reduction of TST 6β-hydroxylation obtained in vitro for rs384467435 was confirmed ex vivo in liver microsomes isolated from bulls homozygous for this same SNV (Giantin et al. Citation2019).
In food-producing species, inter-individual variability in xenobiotics metabolism might result in the accumulation of harmful residues in foodstuffs. Therefore, pharmacogenetic approaches are fundamental to either understand the fate of xenobiotics in the living animal than estimate the consumers’ risk of exposure to noxious chemicals (Hu et al. Citation2016; Jiang et al. Citation2018; Giantin et al. Citation2019). To improve our knowledge on this aspect, this study aims to measure the frequency of five harmful missense CYP3A SNVs in a population of Limousine cattle, a French meat cattle breed widely bred in Northern Italy (Mastrangelo et al. Citation2018). The five selected SNVs have been previously identified in Piedmontese breed and they were shown to affect CYP3A catalytic activity in vitro (Giantin et al. Citation2019).
Materials and methods
The genotyping of five CYP3A missense SNVs (Table ) was carried out on 215 Limousine cattle (female, 1–17 years old), bred in three different districts of Veneto Region (Italy): Rovigo (n = 129), Adria (n = 69), and Feltre (n = 17).
Table 1. Cattle cytochrome P450 3A missense single-nucleotide variants subject of investigation in the present study: IDs, functional impact on CYP3A activity and frequency in Limousine cattle breed.
Genomic DNA was isolated from archived whole blood samples collected within the framework of statutory surveillance activities for infectious diseases by district Official Veterinarians. The DNeasy Blood and Tissue Kit (Qiagen, Milan, Italy) was used and the nucleic acid extracts were then quantified by NanoDrop ND1000 Spectrophotometer (Thermoscientific, Waltham, Massachusetts, USA).
Genotyping was performed using FRET Hybprobe probes (Wittwer et al. Citation2001), as previously reported (Giantin et al. Citation2019). After PCR amplification, the melting profile of the heteroduplexes amplicon-probe, analysed using LightCycler 480 software release 1.5 (Roche, Basel, Switzerland), allowed the discrimination into wild-type (WT/WT), heterozygote (WT/MUT) and mutant (MUT/MUT) genotypes. For the correct interpretation of melting profiles, reference DNA samples corresponding to the different genotypes were used. They were selected after PCR end-point amplification, cloning, plasmid purification and confirmatory Sanger sequencing of at least ten colonies.
The genotype frequency of each variant was examined for deviations from Hardy-Weinberg Equilibrium (HWE) within the population using the Chi-square test implemented in the online calculator (https://www.coursehero.com/file/8442059/Court-lab-HW-calculator/). A p value < .05 indicated a deviation from HWE.
Results and discussion
The estimation of CYP3A pattern of allelic distribution in different ethnic groups is an approach widely used in human pharmacogenetics for its clinical implications and the actual need for personalised therapies (Hu et al. Citation2005; Szalai et al. Citation2015; Hu et al. Citation2017). Conversely, this study represents one of the few examples of the estimation of CYP3A genetic variants prevalence in bovine breeds (Sales et al. Citation2012, Citation2013; Giantin et al. Citation2019). The genotyping results are summed up in Table and described below.
Among the five target SNVs, only two of them (namely, NM_001099367.1:c.589G > A, and NM_001099367.1:c.1162A > G) were successfully identified and quantified in the tested population.
The NM_001099367.1:c.589G > A assay, distinguishing a SNV in exon 7 of CYP3A28, and causing the amino acid change Gly to Ser at residue 197, showed three different melting profiles (Figure ). The MUT/MUT genotype showed a single peak, with a melting temperature (Tm) of 60 °C; the WT/WT genotype, a peak at 66 °C; finally, the WT/MUT genotype showed both peaks, as expected. The mutant allele was detected both in homozygosis and heterozygosis in the 4% and 24.2% of animals, respectively. The frequency of MUT/MUT and WT/MUT genotypes was quite similar to that previously observed in Piedmontese cattle (3.4% and 17.7%, respectively: Giantin et al. Citation2019). However in Limousine population, in opposition to what observed in Piedmontese population, this SNV was consistent with HWE. This SNV is present in the official public repositories with the reference ID rs384467435, thus meaning that this variant is also present in Hereford cattle, whose DNA was used as template for the UMD3.1 bovine genome sequencing. Looking at the results as a whole, we might infer that this SNV is present in at least three different cattle breeds (Limousine, Piedmontese and Hereford). Therefore, we might suggest that rs384467435 is not a breed-specific SNV, and it could be subjected to selection only in Piedmontese population. Unfortunately the lack of data about the presence (and frequency) of such a SNV in other cattle breeds prevents us from inferring conclusive remarks. Anyway, this SNV is worth of mention, because in a recent study it was demonstrated to be the only missense variant (out of thirteen) enable to significantly reduce TST hydroxylation either in vitro, i.e. in V79 cells transiently transfected with CYP3A28 mutated sequence, than ex vivo, in liver microsomes isolated from 300 Piedmontese bulls (Giantin et al. Citation2019).
Figure 1. Output of NM_001099367.1:c.589G > A (A) and NM_001099367.1:c.1162A > G (B) melting curve genotyping assays (LightCycler 480 software release 1.5, Roche). WT/WT (wild type), WT/MUT (heterozygous), and MUT/MUT (mutant) genotypes are indicated with green, red and blue curves, respectively. The BLANK (no template control) is shown in purple.
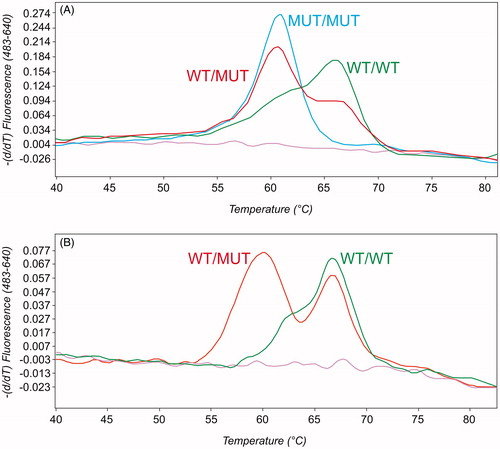
In the cohort of Limousine cattle, the genotyping of NM_001099367.1:c.1162A > G (identified in public databases as rs454167819) showed the alternative nucleobase only in heterozygosis (Table and Figure ). Indeed, only two different melting profiles were obtained. The WT/WT genotype was characterised by a single peak at 66 °C, while WT/MUT by 2 peaks at 60 °C and 66 °C (Figure ). All the animals showing the WT/MUT genotype (5.1%) were from the Rovigo district, presumably from the same progeny. The percentage of WT/MUT Limousine cows was lower when compared to that observed in Piedmontese cattle (i.e. 13%; Giantin et al. Citation2019), while the MUT/MUT genotype was completely absent in both breeds. This SNV has been proved to halve CYP3A28 catalytic activity (Giantin et al. Citation2019). Therefore, both WT/MUT and, even more dramatically, MUT/MUT genotypes might significantly affect veterinary drug kinetics, with consequences on their efficacy and residue formation. The low allele frequency might result from a man-driven selection (Bosse et al. Citation2019), yet the phenotype associated to this allele is still unknown. Nevertheless, further population studies encompassing a wider number of animals are required to confirm this hypothesis.
Concerning the remaining three target variants, two of them, i.e. NM_001099367.1:c.866C > T (rs433125080) and NM_001075888.2:c.1122G > T (a novel SNV recently identified in Piedmontese breed: Giantin et al. Citation2019), were completely absent in the Limousine population. The third one, i.e. NM_174513.3:c.1051G > A (rs137124349), was not detectable for technical limitations. Also in Piedmontese breed, NM_001099367.1:c.866C > T and NM_001075888.2:c.1122G > T were not detected or identified in only 1 bull out of 300, respectively (Giantin et al. Citation2019). This would confirm the extremely low frequency of these variants in both breeds. We would rule out a low efficiency of the assays because the probes allowed the identification of further missense SNVs in heterozygosis: rs481349500 and NM_001075888.2:c.1126T > C. Conversely, the FRET assay for NM_174513.3:c.1051G > A, previously set up and used for Piedmontese cattle genotyping (minor allele frequency 0.21: Giantin et al. Citation2019), was not efficient enough in Limousine samples because of the presence of an additional variant at residue 350. This synonymous variant (NM_174513.3:c.1050T > C or rs379152445), confirmed by Sanger sequencing, probably interfered with probes binding and therefore altered the melting curve profile. In this challenging situation, the amplicon cloning followed by Sanger sequencing is potentially the only viable strategy to assign the proper genotype.
Limousine and Piedmontese meat cattle breeds have been here considered because in a previous study they already showed differences in CYP3A catalytic activity (Dacasto et al. Citation2005). These two breeds share rs384467435 and rs454167819 missense variants with some slight differences in frequency, potentially attributable to the main features of the breed itself. Specifically, Limousine is a cosmopolitan breed imported in Italy, used worldwide for beef production in pure or cross-breeding systems, and showing a moderate level of genetic diversity compared to foremost Italian breeds (Mastrangelo et al. Citation2018). Conversely, Piedmontese is a local breed that has benefitted from proper breed management and has a sufficiently large effective population size and a low degree of recent consanguinity (Mastrangelo et al. Citation2018). In perspective, it could be of interest to extend the evaluation of these same SNVs in further local breeds (e.g. Rendena, Burlina), confined in small geographical areas and generally characterised by a low genetic diversity and a small population size.
The minor allele frequency (MAF) of the variants here identified resembles the one found in Nellore cattle in the xenobiotic sensor gene NR1I3 (Nuclear receptor 1 family I member 3, also known as Constitutive Androstane Receptor: Alexandre et al. Citation2014), and is lower than the one described in Braham and Angus and reciprocal crosses in CYP3A48 by Sales et al. (Citation2012, Citation2013). Even if low, the MAF here calculated should not be viewed as bad and might be regarded as a great opportunity to use these markers for rapidly increase the net effect of a chosen phenotype (Alexandre et al. Citation2014).
In the above mentioned studies (Sales et al. Citation2012, Citation2013; Alexandre et al. Citation2014), genotypes were associated with feed efficiency traits. However, feed efficiency in beef cattle is a phenotypic trait where several cellular processes and gene networks take part, such as cellular growth and proliferation, cell signalling, drug metabolism, protein synthesis, lipid metabolism and carbohydrate metabolism (Chen et al. Citation2011). All this makes feed efficiency phenotype a polygenic character, and it is difficult for a single gene to hardly affect it (Moore et al. Citation2009), unless this gene plays a fundamental role nor it is involved in many pathways (Alexandre et al. Citation2014).
Conversely, CYPs are a superfamily of genes basically involved in the biotransformation of xenobiotics and endogenous compounds (e.g. steroids, vitamin D); hence, a single genetic variant may potentially result in an altered phenotype. In human medicine, differences in CYP catalytic activity due to genetic polymorphisms may be of clinical significance with respect to drug efficacy, drug–drug interactions as well as overall xenobiotic toxicity (Werk and Cascorbi Citation2014; Szalai et al. Citation2015). In veterinary medicine, and particularly in food-producing species, these genetic alterations might impact also on the levels of harmful or unpleasant residues in foodstuffs. Notably, recent studies have described the effect of swine CYP3A29 and CYP2E1 SNVs on aflatoxin B1 and T-2 toxin bioactivation and skatole metabolism, respectively (Mörlein et al. Citation2012; Cheng et al. Citation2014; Wu et al. Citation2016; Zadinová et al. Citation2017; Jiang et al. Citation2018). Present results, and particularly those obtained for bCYP3A missense variant rs384467435 (MAF = 0.16), are therefore potentially relevant for the veterinary community, and should be considered in future pharmacogenetics studies. Anyway, the validation of genotype results with perspective phenotypic investigations, based on pharmacokinetics studies and residue analyses in genotyped cattle treated with drugs known to be metabolised by CYP3A (e.g. macrolides, macrocyclic lactones, …), is strongly recommended.
Finally, a further challenging issue in perspective could be to estimate the differential allelic-specific expression in cattle heterozygous for CYP3A SNVs, potentially resulting in changes in gene expression levels and, consequently, in further phenotypic variability (Hirota et al. Citation2004; Ghotbi et al. Citation2009).
Conclusions
In conclusion, this study confirm the presence of two (rs384467435 and rs454167819) out of five CYP3A missense variants affecting CYP3A activity in vitro in female Limousine cattle. Since the presence of these SNVs might result in differential clinical (e.g. a different drug mean residency time) and toxicological (e.g. increasing amounts of harmful residues) responses, further phenotypic insights are required in perspective.
Acknowledgements
This work was supported by grants from Regione del Veneto (DGR 2080/30.12.2015) to M.D. The authors thank Dr Manzan, Dr Randon and the student Giulia Scorrano for their technical assistance.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Alexandre PA, Gomes RC, Santana MHA, Silva SL, Leme PR, Mudadu MA, Regitano LCA, Meirelles FV, Ferraz JBS, Fukumasu H. 2014. Bovine NR1I3 gene polymorphisms and its association with feed efficiency traits in Nellore cattle. Meta Gene. 2:206–217.
- Ashwell MS, Fry RS, Spears JW, O'Nan AT, Maltecca C. 2011. Investigation of breed and sex effects on cytochrome P450 gene expression in cattle liver. Res Vet Sci. 90(2):235–237.
- Bosse M, Megens HJ, Derks MFL, de Cara ÁMR, Groenen MAM. 2019. Deleterious alleles in the context of domestication, inbreeding, and selection. Evol Appl. 12(1):6–17.
- Brown MA, Tharel LM, Brown AH, Jackson WG, Miesner JR. 1993. Milk production in Brahman and Angus cows on endophyte‐infected fescue and common bermudagrass. J Anim Sci. 71(5):1117–1122.
- Chen Y, Gondro C, Quinn K, Herd RM, Parnell PF, Vanselow B. 2011. Global gene expression profiling reveals genes expressed differentially in cattle with high and low residual feed intake. Anim Genet. 42(5):475–490.
- Cheng G, Liu C, Wang X, Ma H, Pan Y, Huang L, Hao H, Dai M, Yuan Z. 2014. Structure-function analysis of porcine cytochrome P450 3A29 in the hydroxylation of T-2 toxin as revealed by docking and mutagenesis studies. PLoS One. 9(9):e106769
- Dacasto M, Eeckhoutte C, Capolongoa F, Dupuy J, Carletti M, Calléja C, Nebbia C, Alvinerie M, Galtier P. 2005. Effect of breed and gender on bovine liver cytochrome P450 3A (CYP3A) expression and inter-species comparison with other domestic ruminants. Vet Res. 36(2):179–190.
- Dupuy J, Escudero E, Eeckhoutte C, Sutra JF, Galtier P, Alvinerie M. 2001. In vitro metabolism of 14C-moxidectin by hepatic microsomes from various species. Vet Res Comm. 25(5):345–354.
- Ghotbi R, Gomez A, Milani L, Tybring G, Syvänen AC, Bertilsson L, Ingelman-Sundberg M, Aklillu E. 2009. Allele-specific expression and gene methylation in the control of CYP1A2 mRNA level in human livers. Pharmacogenomics J. 9(3):208–217.
- Giantin M, Carletti M, Capolongo F, Pegolo S, Lopparelli RM, Gusson F, Nebbia C, Cantiello M, Martin P, Pineau T, et al. 2008. Effect of breed upon cytochrome P450 and phase II enzyme expression in cattle liver. Drug Metab Dispos. 36(5):885–893.
- Giantin M, Rahnasto-Rilla M, Tolosi R, Lucatello L, Pauletto M, Guerra G, Pezzato F, Lopparelli RM, Merlanti R, Carnier P, et al. 2019. Functional impact of cytochrome P450 3A (CYP3A) missense variants in cattle. Sci Rep. 9(1):19672.
- Hirota T, Ieiri I, Takane H, Maegawa S, Hosokawa M, Kobayashi K, Chiba K, Nanba E, Oshimura M, Sato T, et al. 2004. Allelic expression imbalance of the human CYP3A4 gene and individual phenotypic status. Hum Mol Genet. 13(23):2959–2969.
- Hohmann N, Haefeli WE, Mikus G. 2016. CYP3A activity: towards dose adaptation to the individual. Expert Opin Drug Metab Toxicol. 12(5):479–497.
- Hu GX, Dai DP, Wang H, Huang XX, Zhou XY, Cai J, Chen H, Cai JP. 2017. Systematic screening for CYP3A4 genetic polymorphisms in a Han Chinese population. Pharmacogenomics. 18(4):369–379.
- Hu S, Mazur CA, Feenstra KL, Lorenz JK, Merritt DA. 2016. Assessment of inhibition of porcine hepatic cytochrome P450 enzymes by 48 commercial drugs. Vet J. 211:26–31.
- Hu YF, He J, Chen GL, Wang D, Liu ZQ, Zhang C, Duan LF, Zhou HH. 2005. CYP3A5*3 and CYP3A4*18 single nucleotide polymorphisms in a Chinese population. Clin Chim Acta. 353(1–2):187–192.
- Jiang H, Wu J, Zhang F, Wen J, Jiang J, Deng Y. 2018. The critical role of porcine cytochrome P450 3A46 in the bioactivation of aflatoxin B1. Biochem Pharmacol. 156:177–185.
- Klein K, Zanger UM. 2013. Pharmacogenomics of cytochrome P450 3A4: recent progress toward the “missing heritability” Problem. Front Genet. 4:12.
- Kuilman ME, Maas RF, Fink-Gremmels J. 2000. Cytochrome P450-mediated metabolism and cytotoxicity of aflatoxin B(1) in bovine hepatocytes. Toxicol in vitro. 14(4):321–327.
- Mastrangelo S, Ciani E, Ajmone Marsan P, Bagnato A, Battaglini L, Bozzi R, Carta A, Catillo G, Cassandro M, Casu S, et al. 2018. Conservation status and historical relatedness of Italian cattle breeds. Genet Sel Evol. 50(1):35.
- Moore SS, Mujibi FD, Sherman EL. 2009. Molecular basis for residual feed intake in beef cattle. J Anim Sci. 87(14 Suppl):E41–E47.
- Mörlein D, Lungershausen M, Steinke K, Sharifi AR, Knorr C. 2012. A single nucleotide polymorphism in the CYP2E1 gene promoter affects skatole content in backfat of boars of two commercial Duroc-sired crossbred populations. Meat Sci. 92(4):739–744.
- Nebbia C, Ceppa L, Dacasto M, Nachtmann C, Carletti M. 2001. Oxidative monensin metabolism and cytochrome P450 3A content and functions in liver microsomes from horses, pigs, broiler chicks, cattle and rats. J Vet Pharmacol Ther. 24(6):399–403.
- Rosenkrans CF, Jr, Ezell NS. 2015. Relationships among ergot alkaloids, cytochrome P450 activity, and beef steer growth. Front Chem. 3:16.
- Sales MA, Larson MJ, Reiter ST, Brown AH, Jr, Brown MA, Looper ML, Coffey KP, Rosenkrans CF. Jr. 2012. Effects of bovine cytochrome P450 single-nucleotide polymorphism, forage type and body condition on production traits in cattle. J Anim Physiol Anim Nutr. 96(4):545–553.
- Sales MA, Murphy KY, Reiter ST, Brown AH, Jr, Brown MA, Looper ML, Rosenkrans CF. Jr. 2013. Effects of forage type, body condition and single-nucleotide polymorphisms in the bovine cytochrome P450 regulatory region on cow productivity. J Anim Physiol Anim Nutr. 97(1):91–96.
- Szalai R, Ganczer A, Magyari L, Matyas P, Bene J, Melegh B. 2015. Interethnic differences of cytochrome P450 gene polymorphisms may influence outcome of taxane therapy in Roma and Hungarian populations. Drug Metab Pharmacokinet. 30(6):453–456.
- Werk AN, Cascorbi I. 2014. Functional gene variants of CYP3A4. Clin Pharmacol Ther. 96(3):340–348.
- Wittwer CT, Herrman MG, Gundry CN, Elenitoba-Johnson KS. 2001. Real-time multiplex PCR assays. Methods. 25(4):430–442.
- Wu J, Chen R, Zhang C, Li K, Xu W, Wang L, Chen Q, Mu P, Jiang J, Wen J, et al. 2016. Bioactivation and regioselectivity of pig cytochrome P450 3A29 towards aflatoxin B1. Toxins. 8(9):267.
- Zadinová K, Stupka R, Stratil A, Čítek J, Vehovsky K, Lebedová N, Šprysl M, Okrouhlá M. 2017. Association analysis of SNPs in the porcine CYP2E1 gene with skatole, indole, and androstenedione levels in backfat of a crossbred pig population. Meat Sci. 131:68–73.
- Zancanella V, Giantin M, Dacasto M. 2014. Absolute quantification and modulation of cytochrome P450 3A isoforms in cattle liver. Vet J. 202(1):106–111.
- Zancanella V, Giantin M, Lopparelli RM, Patarnello T, Dacasto M, Negrisolo E. 2010. Proposed new nomenclature for Bos taurus cytochromes P450 involved in xenobiotic drug metabolism. J Vet Pharmacol Ther. 33(6):528–536.
- Zweers-Zeilmaker WM, Van Miert AS, Horbach GJ, Witkamp RF. 1999. In vitro complex formation and inhibition of hepatic cytochrome P450 activity by different macrolides and tiamulin in goats and cattle. Res Vet Sci. 66(1):51–55.
