 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
By regulating the expression of low-density lipoprotein receptors (LDLR), the SEC24A protein modulates the clearance of plasma cholesterol in mice. The whole-genome sequencing of five Duroc boars that sired a Duroc purebred experimental population (LIPGEN population, 350 individuals) revealed the segregation of one putative nonsense mutation located at the SEC24A (rs336401826, c.44G>A) gene. Genotyping of this polymorphic site in the LIPGEN population highlighted the existence of a significant departure from the Hardy Weinberg equilibrium (χ2 = 5.05, p-value = 0.024527). Despite the fact that SEC24A inactivation should affect serum lipid concentrations, we did not observe an association between SEC24A genotype and such traits. A thorough revision of the annotation of the SEC24A rs336401826 SNP revealed that this polymorphism is probably located in the 5’UTR of the SEC24A gene, rather than at the beginning of the first exon, hence not introducing the predicted premature stop codon that could truncate the SEC24A protein. Given that the annotation of loss-of-function mutations in pigs is far from perfect, careful manual curation of such mutations is recommended before undertaking the analysis of their effects at the experimental level.
One mutation (c.44 G>A) predicted to disrupt the SEC24A protein segregates in a Duroc pig population.
This polymorphism is not associated with serum lipid levels despite its expected effect on such traits.
Manual curation of the annotation of this putative nonsense polymorphism has revealed that it maps to the 5’UTR of the SEC24A gene.
HIGHLIGHTS
Introduction
An important fraction of proteins synthesised in the endoplasmic reticulum need to enter the secretory pathway to reach other intracellular organelles, the cell surface or the extracellular space (Bonifacino and Glick Citation2004). The transport of these proteins to the Golgi complex, where they can undergo post-translational modifications, is mediated by vesicles coated with coat protein complex II (COPII), which are assembled through the recruitment of SEC24 and other proteins (Lee et al. Citation2004). Indeed, the SEC24 subunit of the COPII complex contributes to cargo selection by recognising specific molecular signals (Lee et al. Citation2004). In mammals, there are multiple forms of COPII coat structures specialised in different types of cargos and, in this context, four paralogous SEC24 genes (SEC24A to SEC24D) have been reported so far (Zanetti et al. Citation2011). In mice, the inactivation of the SEC24A gene is not lethal but it causes a reduction in plasma cholesterol because the secretion of proprotein convertase subtilisin/kexin type 9 (PCSK9), which is a key mediator in the lysosomal degradation of low-density lipoprotein (LDL) receptors (Lagace Citation2014), is shut off, thus leading to an hepatic upregulation of LDL receptors and, in consequence, to an increased cholesterol clearance (Chen et al. Citation2013).
In a previous experiment (Mármol-Sánchez et al. Citation2020, Citation2021), we sequenced the whole genomes of the five Duroc boars that sired a purebred population of 350 Duroc barrows (LIPGEN population), as reported by Gallardo et al. (Citation2008, Citation2009). The analysis of these five whole-genome sequences (WGS) revealed the segregation of a putative nonsense mutation located in the SEC24 Homolog A, COPII Coat Complex Component (SEC24A) gene (our unpublished data). This mutation was recorded at Ensembl repositories (https://www.ensembl.org/index.html) with the rs336401826 (c.44 G>A, SEC24A) identifier. The goal of the current work was to investigate the segregation of this putative nonsense SNP in the LIPGEN population, as well as to determine its association with lipid-related traits in order to verify whether SEC24A inactivation in pigs has the same consequences on fat metabolism than those previously reported in mice.
Materials and methods
Sequencing the genomes of five Duroc boars
In brief, genomic DNA was purified from blood samples belonging to the five boars that sired a Duroc population of 350 offspring (LIPGEN population), as reported by Gallardo et al. (Citation2008, Citation2009). Subsequently, these genomic DNA samples were sequenced at the Centre Nacional d’Anàlisi Genòmica (CNAG, Barcelona, Spain). Paired-end multiplex libraries were prepared according to the instructions of the manufacturer with the KAPA PE Library Preparation kit (Kapa Biosystems, Wilmington, MA) and processed to generate 100 bp paired-end reads on a HiSeq2000 Illumina instrument. Base calling and quality control analyses were performed with the Illumina RTA sequence analysis pipeline. Quality-checked filtered reads were then processed and any remaining sequence adapters were trimmed by making use of the Trimmomatic software (v.0.36) with default parameters (Bolger et al. Citation2014). Trimmed paired-end sequences were aligned against the pig Sscrofa11.1 reference genome (Warr et al. Citation2020) with the BWA-MEM algorithm (Li Citation2013) and default settings. Sequence alignment map (SAM) formatted files were then sorted and transformed into binary (BAM) formatted files and PCR duplicates were subsequently identified with the MarkDuplicates package from Picard tool (https://broadinstitute.github.io/picard/) and removed to perform INDEL realignment with the IndelRealigner package from the Genome Analysis Toolkit (GATK v.3.8, McKenna et al. Citation2010). Base quality score recalibration (BQSR) was implemented with BaseRecalibrator from GATK v.3.8. Variant calling was performed using the HaplotypeCaller tool according to GATK best practices recommendations. Finally, SNP variants were retained with SelectVariants package and quality filtered using the FilterVariants tool from GATK v.3.8, according to the following parameters: QD < 2.0, FS > 60.0, MQ < 40.0, MQRankSum < −12.5, ReadPosRankSum < −8.0, SOR > 3.0.
Retrieval of the whole-genome sequences from 120 pigs available at public repositories
A total of 120 whole-genome sequences from wild boars and domestic pigs (Sus scrofa) were retrieved from the NCBI Sequence Read Archive (SRA) database (https://www.ncbi.nlm.nih.gov/sra). The 120 selected genome sequences (Supplementary Table 1) were classified into Asian domestic pigs (ADM, N = 40), Asian wild boars (AWB, N = 20), European domestic pigs (EDM, N = 40) and European wild boars (EWB, N = 20). Further details about breeds and sample origins can be found in Mármol-Sánchez et al. (Citation2020, Citation2021). All SRA files from selected porcine individuals were downloaded from SRA public repositories and converted into fastq format by using the fastq-dump 2.8.2 tool available in the SRA-toolkit package (ncbi.github.io/sra-tools/). Subsequent processing, SNP calling and filtering were performed analogously to what has been reported in the previous section for the 5 whole-genome sequenced Duroc sires.
Isolation of genomic DNA and genotyping
The manipulation of Duroc pigs followed Spanish national guidelines and it was approved by the Ethical Committee of Institut de Recerca i Tecnologia Agroalimentàries (IRTA). Genomic DNA was purified from blood samples collected from the offspring of the five sequenced Duroc boars by using a standard phenol-chloroform extraction protocol (Gallardo et al. Citation2008, Citation2009). All Duroc pigs from the LIPGEN population were typed with the Porcine SNP60 BeadChip (Illumina) in a previous experiment (González-Prendes et al. Citation2017). Moreover, a dedicated TaqMan assay was used by the Veterinary Molecular Genetics Service from the Universitat Autònoma de Barcelona (http://svgm.es/es/Home) to genotype the rs336401826 G > A SEC24A (N = 320) mutation by employing a QuantStudio 12 K flex Real Time PCR System. The HWChisq module from the HardyWeinberg R package (https://CRAN.R-project.org/package=HardyWeinberg) was used to compute a Chi square test of independence to infer whether genotype frequencies deviate from the Hardy Weinberg equilibrium (Table ).
Table 1. Absolute genotypic frequencies of the rs336401826 (c.44G>A) mutation of the porcine SEC24A gene in a Duroc pig population (N = 350) and their departure from the Hardy-Weinberg equilibrium (HWE).
Association analyses with lipid traits
A summary of the lipid-related phenotypes considered in our experiment is provided in Supplementary Table 2. The measurement of lipid traits has been previously described in detail (Gallardo et al. Citation2008, Citation2009; Quintanilla et al. Citation2011). Serum lipid traits were measured after weaning and before starting the fattening period (∼45 days of age), as well as prior to slaughter (∼190 days of age), according to specific experimental protocols for determining total serum cholesterol (Richmond Citation1992), low density lipoprotein (LDL, Friedewald et al. Citation1972), high density lipoprotein (HDL, Rafai and Warnick Citation1994) and triglyceride (Fossati and Prencipe Citation1982) concentrations. Intramuscular fat (IMF) was measured by Near Infra-red Transmittance (NIT, Infratec 1625, Tecator Hoganas, Sweden), while muscle cholesterol content was assessed following Cayuela et al. (Citation2003). A chromatography analysis of methyl esters (Mach et al. Citation2006) was used to determine the fatty acid composition in the C:12 to C:22 range and they were subsequently expressed as a percentage of the IMF for the gluteus medius (GM) and longissimus dorsi (LD) skeletal muscles. Association analyses were performed by applying a univariate linear mixed model framework with the Genome-Wide Efficient Mixed-Model Association (GEMMA) software (Zhou and Stephens Citation2012) and the following statistical model:
where y is the vector of phenotypic observations for every individual; α is a vector containing the intercept plus the included fixed effects, i.e. batch effect with four categories (all traits), farm origin effect with three categories (all traits) and data of extraction with two categories within batch (serum lipids traits). The α vector also contains the regression coefficients of the following covariates: (1) IMF content in the LD or GM skeletal muscles for LD and GM fatty acid composition, (2) backfat thickness (measured between 3rd and 4th ribs) and live weight at slaughterhouse for serum lipids measured at 45 and 190 days of age, and (3) backfat thickness (measured between 3rd and 4th ribs) for IMF content; W is the incidence matrix relating phenotypes with the corresponding effects; x is the vector of the genotypes corresponding to the set of selected polymorphisms; δ is the allele substitution effect of the analysed polymorphism; u is a vector of random individual genetic effects with a n-dimensional multivariate normal distribution MVNn (0, λ τ − 1 K), where τ − 1 is the variance of the residual errors, λ is the ratio between the two variance components and K is a known relatedness matrix derived from the SNPs; and ε is the vector of residual errors. A false discovery rate (FDR) approach (Benjamini and Hochberg Citation1995) was used for multiple testing correction.
Chromosome-wide association analyses
Genotyping data for the SEC24A rs336401826 G>A nonsense mutation were merged with the genotypes of 3684 SNPs mapping to chromosome 2 (SSC2) and generated with the Porcine SNP60 BeadChip (Illumina, San Diego, CA) as reported by González-Prendes et al. (Citation2017). The GEMMA software (Zhou and Stephens Citation2012) was used to perform this association analysis by considering the methods reported previously and chromosome-wide significance was assessed at the nominal level (p-value <0.05) as well as after correction for multiple testing using the Bonferroni approach.
Multiple sequence alignment of SEC24A transcripts from different species
We used the Clustal Omega tool of the EMBL-EBI sequence analysis toolkit (Madeira et al. Citation2019) to generate a multiple sequence alignment of the annotated coding sequence (CDS) of the 1st exon of the SEC24A transcripts retrieved from eight available porcine assemblies (Sscrofa11.1, USMARC, Bamei, Meishan, Rongchan, Pietrain, Landrace and Large White), as well as from the human assembly (GCRh38.p13). Moreover, the MultAlin software (Corpet Citation1988) was employed to generate a multiple alignment of the porcine SEC24A protein with other homologous SEC24A proteins annotated in human (GCRh38.p13), cattle (ARS-UCD1.2), mouse (GRCm38.p6) and dog (CanFam3.1) assemblies.
Results and discussion
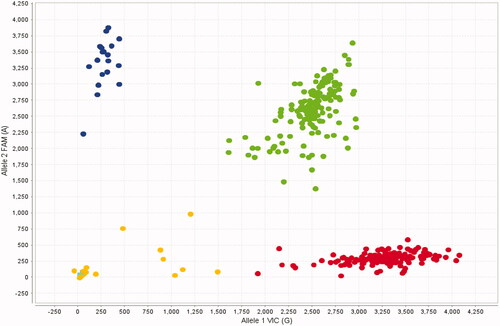
A significant departure from the Hardy Weinberg equilibrium was found (χ2 = 5.05, p-value = 0.024527, Table ) for the rs336401826 G>A SEC24A variant, mainly because there was a slight deficit of the two homozygous genotypes. Genotyping errors are one of the most common causes of observing Hardy-Weinberg departures, but that does not seem to be the case because all three AA, GA and GG genotypes are clearly discernible (Figure ). Another potential reason for the Hardy-Weinberg departure observed by us would be that the frequencies of the SNP might be altered by selection or stochastic factors directly acting on it or on a linked nearby mutation. Nevertheless, the statistical significance of the departure was so weak that no biological inferences should be made. More importantly, no missing genotypes were detected, thus evidencing that this putative nonsense mutation is not lethal. As previously said, this result was anticipated because SEC24A inactivation is expected to just reduce plasma cholesterol concentration via up-regulating LDL receptor membrane concentration due to a specific block in the secretion of PCSK9 (Chen et al. Citation2013). The rs336401826 (c.44G>A, Trp15*) variant is located at codon 15 of a gene encoding a protein of 1133 amino acid residues, so the premature stop codon should provoke the full inactivation of the protein and marked changes in blood lipid levels.
Figure 1. Allelic discrimination plot obtained for the rs336401826 SNP using a TaqMan assay. The x- and y-axes indicate the fluorescence values of the VIC and FAM dyes, respectively, while the dots are individual sample points. The following pig SEC24A genotypes are depicted in this plot: AA (upper left side, N = 19), GA (middle, N = 149) and GG (down right side, N = 152). Samples that were not successfully genotyped are represented by dots in the lower left side. It can be seen that the three SEC24A genotypes are clearly discernible.
Contrary to our expectations, statistical analyses did not show any significant association between SEC24A rs336401826 genotypes (GG, N = 152; GA, N = 149 and AA, N = 19) and serum cholesterol, LDL, HDL or triglyceride concentrations at 45 and 190 days (Supplementary Table 3). In contrast, significant associations were found between the nonsense rs336401826 mutation and C14:0, C16:0, C16:1 and C20:2 fatty acid contents in the GM skeletal muscle (Table ), although only at the nominal level (p-value < 0.05). Data about average GM C14:0, C16:0, C16:1 and C20:2 contents in pigs with different rs336401826 genotypes are displayed in Table .
Table 2. Nominally significant associations (p-value < 0.05) between fatty acid composition traits and c.44G>A (rs336401826, SEC24A) genotypes (GG, N = 152; GA, N = 149 and AA, N = 19)a.
Table 3. Average C20:2, C16:1, C14:0 and C16:0 fatty acids contents in the gluteus medius (GM) skeletal muscle of pigs with GG and AA genotypes for the rs336401826 (c.44G>A) mutation in the porcine SEC24A gene.
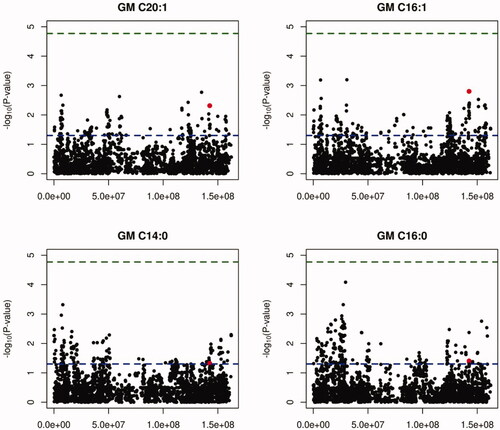
A total of 2951 SNPs mapping to the SSC2 porcine chromosome were successfully analysed in chromosome-wide association analyses, which revealed that the rs336401826 SEC24A polymorphism was among the SNPs displaying the most significant associations with GM C16:1 (nominal p-value = 1.577E-03) and GM C20:2 (nominal p-value = 4.841E-03) phenotypes (Figure ). It should be noted that C14:0, C16:1 and C20:2 fatty acids are found in very low amounts in the intramuscular fat of the GM muscle, so it is challenging to measure them with accuracy. On account of this, the results obtained in the current association analysis should be interpreted with caution until replicated in other porcine populations.
Figure 2. Manhattan plot depicting the association of the SEC24A rs336401826 variant plus 2950 additional SNPs mapping to SSC2 with C20:2, C16:1, C14:0 and C16:0 fatty acids contents in the gluteus medius (GM) muscle. The blue line represents the chromosome-wide nominal p-value of significance (α = 0.05), while the green line represents the p-value after correction for multiple testing applying the Bonferroni approach. The chromosomal position of the rs336401826 G>A SNP is highlighted in red.
When we assessed the segregation patterns of the SEC24A mutation in the 120 WGS from European and Asian pigs and wild boars, the frequency of the A allele was higher than that of the G allele, despite its potential deleterious effect on gene function, and this was observed in European and Asian domestic breeds as well as in European and Asian wild boars (Supplementary Table 1). In the EDM group, the G allele was predominantly found in Duroc pigs (N = 7; 2 GG, 4 GA and 1 AA). Aside the Duroc breed, only one GA Yucatan pig and three GG Yucatan, Yorkshire and Large White pigs carried the G-allele, while all the remaining EDM pigs harboured AA genotypes (Supplementary Table 1). Despite the high frequency of the A-allele in most porcine breeds, the G-allele is the reference allele in the Sscrofa11.1 porcine assembly (Warr et al. Citation2020). The most likely reason of this outcome is that this assembly was built using PacBio RS II deep long-read sequencing from one single female pig belonging to the Duroc breed (Warr et al. Citation2020), in which the G variant is frequent.
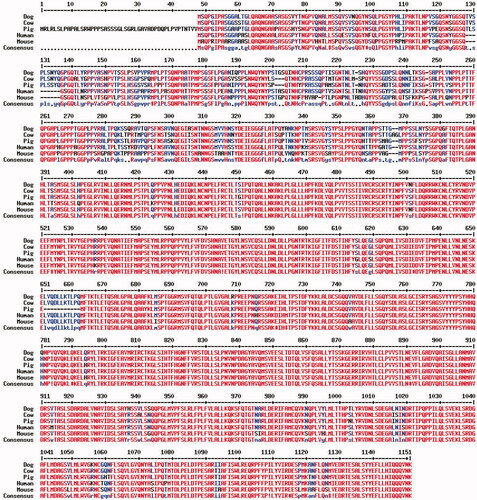
In the light of these results, that were not consistent with a deleterious effect of the nonsense rs336401826 G>A SEC24A mutation, we decided to further investigate the genomic context of this SNP. By doing so, we determined that the common predicted ATG translation start codon is located 141 nucleotides downstream from the predicted start codon of SEC24A exon 1 in the Sscrofa11.1 assembly (Supplementary Figure 1). Interestingly, in the Rongchang, Large White and Landrace assemblies the SEC24A exon 1 coding sequence was correctly annotated, whereas other available porcine assemblies like USMARC, Meishan, Bamei and Pietrain contained a wrong annotation, analogously to what was found for the Sscrofa11.1 assembly (Supplementary Figure 1). The alignment of the mature SEC24A protein sequences from human, mouse, pigs, cattle and dogs also evidenced that the porcine sequence contains an initial segment that is not present in the remaining species (Figure ), a result that once again suggests that the starting codon of the porcine SEC24A sequence is incorrectly annotated. Altogether, these findings would imply that the rs336401826 G>A SEC24A polymorphism is located within the 5’UTR of the SEC24A transcripts, rather than at codon 15 of the coding region.
Figure 3. Multiple sequence alignment showing the sequence of the protein encoded by the SEC24A gene in different species (human, mouse, cattle, dog and pig). The portion of the pig SEC24A protein that is not homologous to the SEC24A protein sequences from the remaining species is clearly visible, evidencing that the translation start codon of the porcine SEC24A protein sequence is incorrectly annotated in the Sscrofa11.1 assembly.
Although the current Sscrofa11.1 assembly (Warr et al. Citation2020) represents a considerable improvement with regard to the previous one (Sscrofa10.2, Groenen et al. Citation2012), there are still annotation problems that need to be corrected. The annotation of nonsense mutations can be particularly error-prone when gene and/or transcript structures are not well known or when the existence of nearby compensatory polymorphisms is ignored. Another important source of error can be pseudogenes, which are expected to contain a much higher rate of deleterious mutations than that observed in functional genes. As a whole, these findings reinforce the need of manually curating the annotation of loss-of-function mutations in pigs in order to be able to predict their functional consequences.
Conclusions
Rather than a nonsense mutation, the rs336401826 G>A SEC24A variant is a benign 5’UTR polymorphism with no harmful consequences. In general, the annotation of loss-of-function mutations is challenging and this is why manual curation becomes essential to prevent wrongful annotations. At this point, it is difficult to assess how many putative loss-of-function mutations are incorrectly annotated in the current pig genome assembly, but caution is advised. Careful inspection and verification of the annotation of loss-of-function mutations is recommended before undertaking the analysis of their effects at the experimental level.
Supplemental Material
Download MS Word (332.4 KB)Supplemental Material
Download MS Excel (14.3 KB)Supplemental Material
Download MS Excel (13 KB)Supplemental Material
Download MS Excel (13.2 KB)Acknowledgments
The authors are indebted to Selecció Batallé S.A. for providing the animal material used in this study. We gratefully acknowledge J. Reixach and J. Soler (IRTA) for their collaboration in the experimental protocols.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The 3,684 SNPs included in this study were obtained from published porcine SNP60 BeadChip data reported by González-Prendes et al. (Citation2017). Lipid-related traits and genotype data can be accessed at the Figshare public repository through the following link: https://figshare.com/projects/SEC24A/92765. Whole-genome sequencing data from the 5 Duroc boars are available at the Sequence Read Archive (SRA) database (BioProject: PRJNA626370).
Additional information
Funding
References
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B. 57(1):289–300.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Bonifacino JS, Glick BS. 2004. The mechanisms of vesicle budding and fusion. Cell. 116(2):153–166.
- Cayuela JM, Garrido MD, Bañón SJ, Ros JM. 2003. Simultaneous HPLC analysis of alpha-tocopherol and cholesterol in fresh pig meat. J Agric Food Chem. 51(5):1120–1124.
- Chen XW, Wang H, Bajaj K, Zhang P, Meng ZX, Ma D, Bai Y, Liu HH, Adams E, Baines A, et al. 2013. SEC24A deficiency lowers plasma cholesterol through reduced PCSK9 secretion. Elife. 2013(2):444.
- Corpet F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16(22):10881–10890.
- Fossati P, Prencipe L. 1982. Serum triglycerides determined colorimetrically with an enzyme that produces hydrogen peroxide. Clin Chem. 28(10):2077–2080.
- Friedewald WT, Levy RI, Fredrickson DS. 1972. Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin Chem. 18(6):499–502.
- Gallardo D, Quintanilla R, Varona L, Díaz I, Ramírez O, Pena RN, Amills M. 2009. Polymorphism of the pig acetyl-coenzyme A carboxylase alpha gene is associated with fatty acid composition in a Duroc commercial line. Anim Genet. 40(4):410–417.
- Gallardo D, Pena RN, Amills M, Varona L, Ramírez O, Reixach J, Díaz I, Tibau J, Soler J, Prat-Cuffi JM, et al. 2008. Mapping of quantitative trait loci for cholesterol, LDL, HDL, and triglyceride serum concentrations in pigs. Physiol Genomics. 35(3):199–209.
- González-Prendes R, Quintanilla R, Cánovas A, Manunza A, Figueiredo Cardoso T, Jordana J, Noguera JL, Pena RN, Amills M. 2017. Joint QTL mapping and gene expression analysis identify positional candidate genes influencing pork quality traits. Sci Rep. 7:39830.
- Groenen MAM, Archibald AL, Uenishi H, Tuggle CK, Takeuchi Y, Rothschild MF, Rogel-Gaillard C, Park C, Milan D, Megens H-J, et al. 2012. Analyses of pig genomes provide insight into porcine demography and evolution. Nature. 491(7424):393–398.
- Lagace TA. 2014. PCSK9 and LDLR degradation: regulatory mechanisms in circulation and in cells. Curr Opin Lipidol. 25(5):387–393.
- Lee MCS, Miller EA, Goldberg J, Orci L, Schekman R. 2004. Bi-directional protein transport between the ER and Golgi. Annu Rev Cell Dev Biol. 20:87–123.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv. 3:13033997. http://arxiv.org/abs/1303.3997.
- Mach N, Devant M, Díaz I, Font-Furnols M, Oliver MA, García JA, Bach A, 2006. Increasing the amount of n-3 fatty acid in meat from young Holstein bulls through nutrition. J Anim Sci. 84(11):3039–3048.
- Madeira F, Park YM, Lee J, Buso N, Gur T, Madhusoodanan N, Basutkar P, Tivey ARN, Potter SC, Finn RD, et al. 2019. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 47(W1):W636–W641.
- Mármol-Sánchez E, Luigi-Sierra MG, Castelló A, Guan D, Quintanilla R, Tonda R, Amills M. 2021. Variability in porcine microRNA genes and its association with mRNA expression and lipid phenotypes. Genet Sel Evol. 53(1):43.
- Mármol-Sánchez E, Luigi-Sierra MG, Quintanilla R, Amills M. 2020. Detection of homozygous genotypes for a putatively lethal recessive mutation in the porcine argininosuccinate synthase 1 (ASS1) gene. Anim Genet. 51(1):106–110.
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. 2010. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20(9):1297–1303.
- Quintanilla R, Pena RN, Gallardo D, Cánovas A, Ramírez O, Díaz I, Noguera JL, Amills M. 2011. Porcine intramuscular fat content and composition are regulated by quantitative trait loci with muscle-specific effects. J Anim Sci. 89(10):2963–2971.
- Rafai N, Warnick GR. 1994. Lipoproteins and apolipoproteins. Laboratory measurement of lipids. Washington (DC): AACC.
- Richmond W. 1992. Analytical reviews in clinical biochemistry: the quantitative analysis of cholesterol. Ann Clin Biochem. 29(6):577–597.
- Warr A, Affara N, Aken B, Beiki H, Bickhart DM, Billis K, Chow W, Eory L, Finlayson HA, Flicek P, et al. 2020. An improved pig reference genome sequence to enable pig genetics and genomics research. Gigascience. 9(6):1–14.
- Zanetti G, Pahuja KB, Studer S, Shim S, Schekman R. 2011. COPII and the regulation of protein sorting in mammals. Nat Cell Biol. 14(1):20–28.
- Zhou X, Stephens M. 2012. Genome-wide efficient mixed-model analysis for association studies. Nat Genet. 44(7):821–824.
