Abstract
The establishment and maintenance of a balanced gut microbiota in early life play a pivotal role in pigs. This study aims to evaluate the effect of the administration of a single early-life probiotic on piglet faecal microbiota and growth performance until weaning. Forty-eight hours after birth (d0), 820 piglets were allocated into 4 groups (205 piglets/16 litters/group) and orally inoculated as follows: 1) Control (CO: 4 mL of pure water); 2) Saccharomyces (SA: 4 mL containing a total of 1 × 1010 CFU of Saccharomyces cerevisiae var. boulardii CNCM-1079; 3) Enterococcus (EF: 4 mL containing a total of 1 × 1010 CFU of Enterococcus faecium lactiferm WS200); 4) a mix of the two probiotics at the same doses (SAEF). At d7 and d18, the piglets were weighed, and faeces from the piglets (18 piglets/group from 6 sows/group) and their mothers were analysed for a microbial profile by sequencing the v3-v4 regions of the 16S rRNA gene. Data were arranged in a 2x2 factorial design. The probiotic supplement improved piglet ADG in the periods d7-d18 (p < .0001) and d0-d18 (p < .05). From d7 to d18, the SA group tended to have lower mortality than the CO group (p = .08). The probiotic supplement significantly affected the microbial beta diversity at d7 (p < .05). The SA probiotic favoured the colonisation of Erysipelatoclostridium and Christensenella, and the EF probiotic the colonisation of Lachnospiraceae. These results highlighted that the administration of a single early-life probiotic supplement could improve piglet performance and shape the faecal microbial profile.
A single dose of E. faecium or S. cerevisiae improved piglet performance in the pre-weaning period.
The early administration of probiotics shaped the faecal microbial profile of the piglets and contributed to improved growth performance.
Highlights
Introduction
Pre-weaning mortality is currently considered one of the most important causes of reduced production efficiency and is a welfare issue in swine herds (Heuß et al. Citation2019). According to the literature, there are several factors associated with piglet perinatal death, such as low body weight at birth, litter size and homogeneity, colostrum intake in the first hours of life and the appropriate development of the piglet immune system which plays a fundamental role in their robustness and survivability (Heuß et al. Citation2019; Revilla et al. Citation2019). In fact, it is generally recognised that young piglets have an immature mucosal immune system; it develops in a programmed process (Stokes et al. Citation2004) in which the intestinal microbiota is strongly involved (Stokes Citation2017) ).
Knowledge regarding the succession of microbial colonisation in the pig gut has intensively increased in recent years (Bian et al. Citation2016; Han et al. Citation2018). As for other mammals, gut colonisation in pigs begins at birth at the moment of initial contact with the bacteria present in the sow vaginal tract and, subsequently, with sow faeces (Tannock et al. Citation1990). More recently, the maternal transfer of bacteria via the umbilical cord blood already during gestation has been recognised (Leblois et al. Citation2017).
A number of studies have reported that the early commensal colonisation of the piglet intestinal microbiota can influence piglet robustness under experimental conditions. An appropriate early commensal colonisation can have beneficial effects on the development of the intestinal and systemic immune system and nutrient absorption, resulting in better growth performance and in a reduction of the negative effects related to the post-weaning diarrhoeal syndrome (Hooper et al. Citation2001; Taras et al. Citation2006; Kenny et al. Citation2011; Stokes Citation2017; Trevisi et al. Citation2018; Luise et al. Citation2019). In fact, it has been shown that management and dietary interventions in the post-natal period affect the establishment of intestinal microbiota and the normal architecture of the mucosal immune system by promoting interaction between lymphoid cells which favours the maturation of the intestinal and systemic immune systems (Everaert et al. Citation2017). In this context, early colonisation with beneficial microbes, such as probiotics, in the first days of life has provided interesting evidence in promoting the intestinal eubiosis of piglets (Wang et al. Citation2016; Kiros et al. Citation2019; Wang et al. Citation2019). However, the solution tested in previous studies suffered from a lack of practicality as the probiotics in those studies were administered (manually) for several days while there were no data regarding the effect of a single probiotic intervention in early life which could result in a more applicable solution in husbandry. Furthermore, relatively little is known regarding the short- and mid-term effects of an early single oral administration of probiotics on the gut microbial profile of nursery piglets under field conditions. Therefore, in the present study, the hypothesis that the administration of a single early-life probiotic supplement to suckling piglets would affect their gut microbiota, driving it to a more mature microbiota represented by one of the sows was developed. The aim of the present study was to evaluate the effect of the administration of a single early-life probiotic supplement on pre-weaning piglet growth, health and the faecal microbial profile.
Materials and methods
Animals and experimental design
A total of 820 piglets from 64 litters (average 13 piglets per litter) were included in the study. At 48 h after birth (d0), the piglets were allocated into 4 groups (205 piglets/16 litters/group) and orally supplied as follows: 1) Control group, CO: 4 mL of pure water; 2) Saccharomyces group, SA: solution containing a total of 1 × 1010 CFU of Saccharomyces cerevisiae var. boulardii CNCM-1079 (Lallemand Animal Nutrition Italia, Castel d’Azzano, Italy); 3) Enterococcus group, EF: solution containing a total of 1 × 1010 colony-forming unit (CFU) of Enterococcus faecium lactiferm WS200 (Chr Hansen Italia, Ciano d'Enza, Italy) and 4) a mix of the two probiotics at the same doses;,SAEF: solution containing a total of 1 × 1010 CFU of Saccharomyces cerevisiae var. boulardii CNCM-1079 and 1 × 1010 CFU di Enterococcus faecium lactiferm WS200) (Supplementary Table 1). The solutions were prepared by mixing probiotic powder in pure water (4 mL) and were administered orally to the piglets using a 10 mL syringe.
Table 1. The effect of the early-life oral administration of probiotics on the mortality and exclusion rate of piglets in the experiment.
During the lactation period, the sows had free access to feed and water. The piglets were raised with their mothers in farrowing crates of 4.5 m2 with a slatted floor and had free access to water. The farrowing room temperature was maintained at 23˚C, and heat lamps were used in the first week after birth. After the first week, the heat lamps were used only at night.
The mortality of the piglets was recorded during the entire study. The piglets were individually weighed at 7 (d7) and 18 (d18) days after the start of the trial. The piglets were then weaned at 21 days of age.
At d7 and d18, a faecal swab from 18 piglets/group (from 6 litters balanced for mother parity and representative of their litter body weight) and their respective mothers (6 sows/group) were collected in sterile tubes, snap-frozen in liquid nitrogen and then preserved at −80 °C until analysis.
Microbiota analysis
Total bacterial DNA was extracted from the faecal samples using a FastDNATM Spin Kit for Soil (MP Biomedicals, Europe, LLC) following the manufacturer’s instructions. The quantity and purity of the DNA isolated were checked using a NanoDrop spectrophotometer (Fisher Scientific, 13 Schwerte, Germany) and agarose gel electrophoresis. Thus, the analysis of the microbial profile was carried out using sequencing of the v3-v4 regions of the 16S rRNA gene. Briefly, amplicons were produced using the primers: Pro341F:50-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGGGNBGCASCAG-30 and Pro805R: 50GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGACT ACNVGGGTATCTAATCC-30 and PlatinumTM Taq DNA Polymerase High Fidelity (Termo Fisher Scientific, Italy). The libraries were prepared using the standard protocol for MiSeq Reagent Kit v3 and were sequenced on the MiSeq platform (Illumina Inc., San Diego, Ca, USA). The raw data (fastq format) obtained are publicly available at the European Nucleotide Archive (ENA) under accession number PRJEB40203.
Statistical analysis
The probiotic effect on faecal microbiota and performance was analysed using a 2 × 2 factorial model (two levels of probiotic Saccharomyces cerevisiae var. boulardii and two levels of Enterococcus faecium lactiferm WS200). In detail, to study the effect of the probiotic Saccharomyces cerevisiae var. boulardii CNCM-1079 (Prob SA), the SA and SAEF groups were considered while, to study the effect of Enterococcus faecium lactiferm WS200 (Prob EF), the EF and SAEF groups were considered; the CO group was considered to be a negative control for both probiotics.
Data regarding performance were analysed using an analysis of variance (ANOVA) model including Prob SA, Prob EF and their interaction, the litter of origin, the parity of the sow (1-2; 3-4-5; ≥6) as fixed factors, and the initial body weight of the piglets as a covariate. Piglet mortality and exclusion from the trial due to severe health impairment were analysed using a GENMOD procedure (SAS software version 9.3) with a binomial distribution and the litter of origin as the experimental unit. The model included Prob SA, Prob EF and their interaction and the parity of the sows (1-2; 3-4-5; ≥6), as fixed factors and the initial body weight (BW) of the litter and the litter size as covariates. In the case of a statistically significant effect of Prob SA and Prob EF interaction, the effect of the probiotic supplementation as compared to the CO group was tested with the following contrasts: SA vs. CO; EF vs. CO and SAEF vs. CO. The effects were considered to be significant when p was <.05 whereas when p was >.05 but <.10, the differences were considered to indicate a trend towards a significant effect.
Bioinformatic and statistical analysis of microbiota
The sequences generated (approximately ∼460 bp) were analysed using the DADA2 package (version 1.5.0) and workflow (Callahan et al. Citation2016) in R version 3.6 (http://www.R-project.org). Taxonomy was assigned using the Silva Database (release 132) (Quast et al. Citation2013).
Alpha (Chao, Shannon and InvSimpson indices) and beta (the Bray–Curtis dissimilarity) diversity, and the abundance of taxa at the different taxonomy levels were calculated and analysed with R software (version 3.6) using PhyloSeq (McMurdie and Holmes Citation2013), Vegan (Dixon Citation2003), lme4 (Bates et al. Citation2015), DESeq2 (Love et al. Citation2014) and MixOmics (Lê Cao et al. Citation2011) packages.
The statistical analysis was initially carried out using the ANOVA (alpha indices) and PERMANOVA (beta diversity) models which included the effect of time (d7 and d18) and of the sample (piglet vs. sow). Data regarding the piglets and the sows were subsequently analysed individually. For alpha diversity, a mixed model including Prob SA, Prob EF and their interaction, the effect of time (d7 and d18), the litter, and the repetition in each piglet per time as factors was carried out; for beta diversity, a PERMANOVA model (Adonis test) including Prob SA, Prob EF and their interaction, the litter and the effect of time (d7 and d18) was carried out.
The data at each time point (d7 and d18) were then analysed separately in order to point out possible substructures in the data. The ANOVA and PERMNOVA models, including Prob SA, Prob EF, their interaction and the litter as factors were carried out for the alpha and beta diversity indices, respectively. Before performing the Adonis test, the homogeneity of dispersion was tested (betadisper function) and was not significant for any factor.
The difference in the taxonomic composition of Prob SA and Prob EF as compared with the CO group at each time point was analysed using the DESeq2 package, based on negative binomial generalised linear models and applying the Benjamini-Hochberg method for multiple testing correction (Love et al. Citation2014). Furthermore, in order to identify the discriminant taxa of each group, the multivariate sparse Partial Least Squares Discriminant Analysis (sPLS-DA) supervised approach was carried out on the microbial data at d7 and d18 separately (Lê Cao et al. Citation2011). The microbial data had previously been normalised using total sum scaling normalisation coupled with the centred log‐ratio (CLR) transformation. The optimal number of components and the optimal number of variables selected for each component included in the sPLS‐DA model were selected based on the average balanced classification error rate with centroid distance over 100 repeats of the 5‐fold cross‐validation of a sPLS‐DA model. To validate the results, the stability of frequency scores of the selected amplicon sequence variants (ASVs) was calculated (“perf” function) with 5‐fold cross‐validation and 100 repetitions; the ASVs showing a correlation with the treatment of >0.5 and a stability of ≥65% were considered discriminative. The effects were considered significant when p was <.05 whereas when p was >.05 but <0.10, the differences were considered to indicate a trend towards a significant effect.
Results
A total of 88 piglets (10.7%) were excluded from the trial due to severe impairment of health conditions (piglet behaviour was characterised by limited movement and attention, no milk intake and visible growth stoppage or loss of body weight) or death. During the periods between d0-d7 and d0-d18, no significant effect of the administration of the probiotics on mortality and exclusion percentage rate was observed while, in the period between d7-d18, a trend towards interaction between the two probiotics was observed (p = .059) (). The comparisons carried out showed that the SA group tended to have a reduced percentage of mortality and exclusion percentage as compared with the CO group (p = .08).
The average initial piglet body weight per litter had a negative effect on the mortality and the exclusion percentages at d0-d7 (p < .001), at d7-d18 (p = .008) and at d0-d18 (p < .001) (Table ).
Piglet performance
Table reports the effect of the oral administration of probiotics on piglet BW and average daily gain (ADG). The probiotic administrations did not affect BW at d7. At d18, a significant effect of the interaction of the two probiotics (p < .0001) was found. The contrast with the CO group showed an increase in BW at d18 in both the SA (p <.005) and the EF (p < .0001) groups while no difference was observed between the CO and SAEF groups.
Table 2. The effect of the oral early-life administration of probiotics on the body weight and growth performance of piglets in the pre-weaning phase.
The probiotic supplementation did not affect the piglet ADG in the period between d0-d7. The interaction between Prob EF and Prob SA regarding piglet ADG at d7-d18 and at d0-d18 was statistically significant (p < .0001 and p < .001, respectively). Comparison with the CO group showed an increase in ADG for the period d7-d18 and d0-d18 in both the SA (p < .0001 and p < .005 for d7-d18 and d0-d18, respectively) and EF (p < .0001 and p < .05 for d7-d18 and d0-d18, respectively) groups while no difference was observed when compared to the SAEF group.
In addition to the effect of the probiotic administration, the BWs and the ADGs at all the time points were influenced by the initial body weight of the piglets and the mothers’ parity (p < .05; except for the ADG at d7-d18 for which the mother’s parity had a p = .086). Litter and litter size did not influence BW, but significantly influenced the ADG at d0-d7 (p < .001), the ADG at d7-d18 (only the litter, p = .036) and the ADG at d0-d18 (only a trend for litter size, p = 0.057)
Faecal microbiota profile
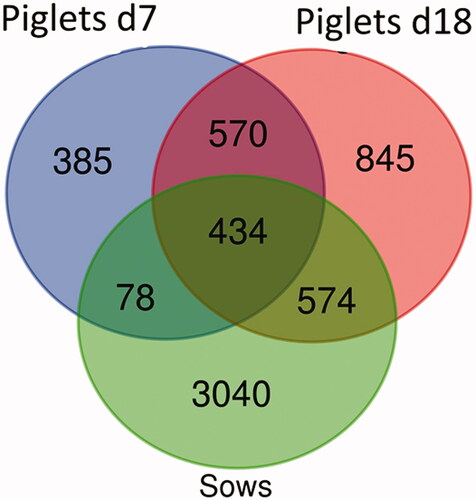
Three samples were excluded from the analysis due to the poor quality of the DNA extracted. After sequencing, a total of 49,936.28 reads distributed among the samples were obtained (). The relative rarefaction curves showed a tendency to a plateau for all the samples, suggesting that the sequencing depth was sufficient to describe the variability within the microbial communities analysed (Supplementary Figure 1). Taxonomic assignment allowed obtaining 18 phyla, 23 classes, 51 families and 150 genera. At the phylum level, Firmicutes (53%), Bacteroidetes (32%) and Proteobacteria (8%) were the most abundant phyla; at the family level, Ruminococcaceae (16%), Lactobacillaceae (15%) and Prevotellaceae (9%) were the most abundant families. Figure shows the Venn plot reporting the number of common and unique ASVs of the piglets at the two time points and of the sows. Sows showed the largest number of ASVs. The sows had 3040 unique ASVs and 434 ASVs in common with the piglets at both time points; 17 ASVs were in common with the piglets at d7 and 574 ASVs were in common with the piglets at d18. The piglets had 570 common ASVs in their faecal microbiome between d7 and d18 while 385 were unique at d7 and 845 were unique at d18.
Figure 1. Venn diagram: number of unique and common amplicon sequence variants (ASVs) of sows and piglets at d7 and d18.
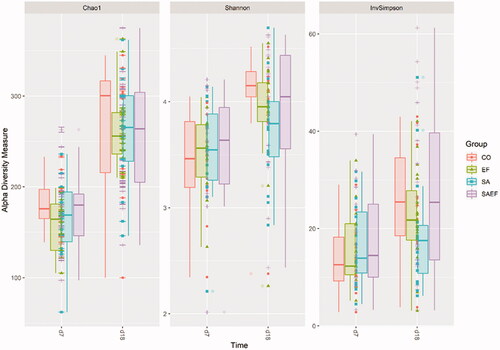
Supplementary Figure 2 shows the alpha diversity indices Chao1, Shannon and InvSimpson for piglets and sows at d7 and d18 which increased as a function of time (age) and sample (piglets vs. sows) (p < .001). No effect of Prob SA and Prob EF and no statistical difference between the four groups (CO, SA, EF, SAEF) on the alpha diversity was observed at either time point (Figure ).
Figure 2. Chao1, Shannon and InvSimpson alpha indices of piglets divided into the four experimental groups and the two time points. Groups: CO = 4 ml of pure water; SA= 4 mL of solution containing a total of 1 × 1010 CFU of Saccharomyces cerevisiae var. boulardii CNCM-1079 (Lallemand Animal Nutrition); EF = 4 mL of solution containing a total of 1 × 1010 UFC di Enterococcus faecium lactiferm WS200 (Chr Hansen) and SAEF 4 mL of solution containing the mix of two probiotics (1 × 1010 CFU of Saccharomyces cerevisiae var. boulardii CNCM-1079 and 1 × 1010 CFU of Enterococcus faecium lactiferm WS200).
The Adonis test showed a significant effect of time (R = 0.03; p = .001) and of sample (piglets vs. sows) (R = 0.18; p = .001) on the Bray-Curtis distance matrix. The effect of time and sample type was visualised by the non-metric multidimensional scaling (NMDS) plot (Supplementary Figure 3) in which two clusters belonging to the sows and the piglets, respectively, were clearly separated. The dispersion of the samples taken from sows was less than the dispersion of the samples taken from the piglets.
Considering only the data of the piglet microbiota, a significant effect of time (R = 0.072; p = .001) and litter (R = 0.007; p = .001) was observed on the faecal microbial composition (beta-diversity); furthermore, a tendency towards the interaction between time and Prob SA (R = 0.009; p = .07) was observed (Table ). The NMDS plot with Bray-Curtis distance matrix for piglet microbiota is reported in Supplementary Figure 3, and a clear separation of the samples by time can be observed.
Table 3. The results of the Adonis test for time and oral early-life administration of probiotics in the faecal microbiota of suckling piglets.
Considering the two time points independently, at d7 both the probiotic supplementations significantly affected the beta diversity (Prob SA: R = 0.02; p = .04; Prob EF: R = 0.02; p = .04) while, at d18, a tendency for Prob SA (R = 0.18; p = .07) was observed. Supplementary Figures 4 and 5 show the NMDS plots with the Bray-Curtis distance matrix for the piglet microbiota at d7 and d18, respectively, and an overlap of the samples among the groups can be observed. Litter significantly affected the faecal microbial structure at both time points (d7: R = 0.45; p = .001; d18: R = 0.35; p = .001).
The results for the taxonomic differences between the probiotic groups and the CO group are reported in Tables and for d7 and d18, respectively. At d7, the groups supplied with the Prob SA (SA and SAEF groups) had a higher abundance of bacteria belonging to the genus Erysipelatoclostridium (adj p = .009) while the groups supplied with the Prob EF (EF and SAEF groups) showed a higher abundance of Lachnospiraceae_UCG_004, Sanguibacteroides, Alistipes genera (adj p < .05) and a lower abundance of the genus Erysipelatoclostridium (adj p = .002) (Table ). At d18, the groups supplied with the Prob SA had a higher abundance of bacteria belonging to the family Enterobacteriaceae (adj p < .0001) and genus Escherichia_Shigella (adj p < .0001), a lower abundance of the family Saccharimonadaceae (adj p = .007) and the genus Hydrogenoanaerobacterium (adj p = .050). The groups supplied with the Prob EF showed a lower abundance of bacteria belonging to the families Clostridiales_vadinBB60_group and Lachnospiraceae (adj p = .026) (Table ).
Table 4. The effect of the oral early-life administration of probiotics on the faecal microbial taxa of suckling piglets at day 7.
Table 5. The effect of the oral early-life administration of probiotics on the faecal microbial taxa of suckling piglet at day 18.
The PLS-DA was carried out on the data aggregated at the genus level for the two time points to identify the discriminant taxa for each specific group (Figure ).
Figure 3. The results of the PLS-DA analysis on the faecal microbiota of piglets at d7. (A) Individual score plot of the samples along the first two components. (B) Table reporting the most discriminant genera per group; Value.var expresses the variance explained by the single genera; Freq, expresses the frequencies by which the genera were chosen among the 100 repetitions of the cross validation; PC stands for the principal component which discriminates the genera; Direction expresses the group for which the genera were discriminated. (C and D) Contribution plot represented the contribution of each genus on the first and second component, respectively. Genus contribution ranked from bottom (most important) to top. Groups: CO = 4 ml of pure water; SA= 4 mL of solution containing a total of 1 × 1010 CFU of Saccharomyces cerevisiae var. boulardii CNCM-1079 (Lallemand Animal Nutrition); EF = 4 mL of solution containing a total of 1 × 1010 UFC di Enterococcus faecium lactiferm WS200 (Chr Hansen) and SAEF 4 mL of solution containing the mix of two probiotics (1 × 1010 CFU of Saccharomyces cerevisiae var. boulardii CNCM -1079 and 1 × 1010 CFU of Enterococcus faecium lactiferm WS200).
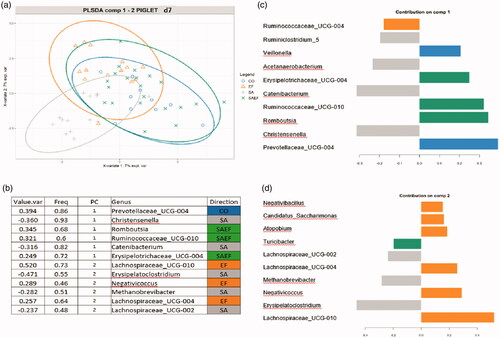
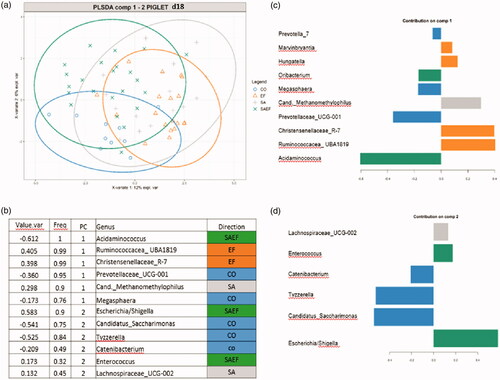
Figure 4. The results of the PLS-DA analysis on the faecal microbiota of the piglets at d18. (A) Individual score plot of the samples along the first two components. (B) Table reporting the most discriminant genera per group; Value.var expresses the variance explained by the single genera; Freq, expresses the frequencies by which the genera were chosen among the 100 repetitions of the cross validation; PC stands for the principal component which discriminates the genera; Direction expresses the group for which the genera were discriminated. (C and D) Contribution plot represented the contribution of each genus on the first and second component, respectively. Genus contribution ranked from bottom (most important) to top. Groups: CO = 4 ml of pure water; SA= 4 mL of solution containing a total of 1 × 1010 CFU of Saccharomyces cerevisiae var. boulardii CNCM-1079 (Lallemand Animal Nutrition); EF = 4 mL of solution containing a total of 1 × 1010 UFC di Enterococcus faecium lactiferm WS200 (Chr Hansen) and SAEF 4 mL of solution containing the mix of two probiotics (1 × 1010 CFU of Saccharomyces cerevisiae var. boulardii CNCM -1079 and 1 × 1010 CFU of Enterococcus faecium lactiferm WS200).
At d7, the CO group was discriminated by bacteria belonging to the genera Prevotellaceae_UCG-004 and Veillonella; the SA group was discriminated by bacteria belonging to the genera Christensenella, Erysipelatoclostridium and Methanobrevibacter; the EF group was discriminated by bacteria belonging to the genera Lachnospiraceae_UCG-010, Lachnospiraceae_UCG-004 and Negativicoccus and finally, the SAEF group was discriminated by bacteria belonging to the genera Ruminococcaceae_UCG-010, Romboutsia and Erysipelotrichaceae_UCG-004 (Figure ).
At d18, the CO group was discriminated by bacteria belonging to the genera Prevotellaceae_UCG-001, Candidatus_Saccharimonas and Tyzzerella; the SA group was discriminated by bacteria belonging to the genera Methanomethylophilus and Lachnospiraceae_UCG-002; the EF group was discriminated by bacteria belonging to the genera Christensenellaceae_R-7 and Ruminococcacea_ UBA1819 and the SAEF group was discriminated by bacteria belonging to the genera Acidaminococcus, Escherichia/Shigella and Enterococcus (Figure ).
Discussion
In this study, the oral administration of S. cerevisiae var. boulardii CNCM-1079 or E. faecium lactiferm WS200 within forty-eight hours after birth led to an improvement in piglet growth performance during the suckling period, with a greater effect after the first week post supplementation, and influenced the piglet faecal microbiota.
These probiotics differ with respect to their natural habitat; S. cerevisiae var. boulardii is not naturally harboured in the pig gut while E. faecium naturally colonises the gastrointestinal tract of mammals (Banla et al. Citation2019). Although both probiotics have already been authorised for use as a feed additive in piglets in the EU, there are no studies in which they have been used as a single oral supplement or as a probiotic mixture in suckling pigs. Some attempts at using the combination of the latter two have been observed in other species, with controversial effects (Maia et al. Citation2001; Beauchemin et al. Citation2003; Emmanuel et al. Citation2007; Chiquette et al. Citation2015).
In the present study, S. cerevisiae var. boulardii showed a positive effect on productive performance in the second part of the suckling period and a tendency to decrease piglet mortality between the first and the third weeks of life. In agreement with Hancox et al. (Citation2015), no significant effect of a single oral administration of S. cerevisiae on piglet ADG in the first week of life (d0-d7) was observed; on the other hand, improvement of the ADG for the period between d7-d18 and for the overall suckling period observed in the present study was in line with the results of Kiros et al. (Citation2019). In fact, in the study of Kiros et al. (Citation2019) the piglets supplied with another strain of S. cerevisiae by oral gavage every two days starting from birth to weaning (28 of age) began to improve BW 10 days post-administration and showed a significant increase in the ADG when considering the total sucking period.
As far as the administration of the probiotic E. faecium was concerned, the present results added to and agreed with the literature (Zeyner and Boldt Citation2006; Wang et al. Citation2016), demonstrating improved growth performance in piglets supplemented with this probiotic. Although the experimental designs are not strictly comparable since both Wang et al. (Citation2016) and Zeyner and Boldt (Citation2006) used different strains of E. faecium at different doses and for longer periods than in the present study, their results support the present results, emphasising that a single administration of this bacterial species was able to promote piglet growth until weaning.
Although no data regarding post-weaning performance were available in the present experiment, a higher BW at weaning is generally associated with higher robustness of the piglets, and with higher lifetime growth performance and long-term weight (Dunshea et al. Citation2003; Collins et al. Citation2017; Revilla et al. Citation2019); thus, the positive effect of a single E. faecium and S. cerevisiae early administration in piglets in the post-weaning phase should not be excluded.
The combined administration of both the probiotics tested had a negative effect on piglet growth performance. The negative effect of the probiotic mixture on piglet performance in the present study may have been due to a competition of the probiotics for binding sites and nutrients in the gut (Chapman et al. Citation2011). The mucus-binding ability of several strains of E. faecium has already been shown in pigs (Jin et al. Citation2000; Laukova et al. Citation2004). This specific capacity is apparently not documented for S. cerevisiae in swine; however, it possesses a set of cell wall-associated proteins which confer adhesion to diverse biotic and abiotic surfaces (Brückner and Mösch Citation2012). Thus, the interference of S. cerevisiae on E. faecium development, by its aggregative properties, not necessarily associated with mucus adhesion cannot be excluded. Otherwise, it may be possible that the administration of the probiotic mixture increased the energy expenditure more by stimulating local immune activity at the expense of growth performance (Martin et al. Citation2003; Trevisi et al. Citation2010). Supplementation with the same S. cerevisiae in milk given to calves stimulated the innate immune response and raised markers of acute-phase reaction in blood serum (Fomenky et al. Citation2018). A strain of E. faecium (NCIMB 11181) showed the stimulatory effects of both cell-mediated and humoral immunity in broilers when supplemented at doses of 1 or 2 × 108 CFU E. faecium/kg in the diet (Wu et al. Citation2019). However, no information is available regarding the effect of the combination of both probiotics in piglets regarding the activation of innate or acquired immunity.
Given the pivotal role of beneficial microbes in modulating the gut microbial ecosystem, and the robust interplay between microbiota and the host, the microbial profiles of sows and piglets at different time points were evaluated. In young animals, time (age) is one of the main factors affecting the gut microbiota as a result of the progressive settlement of a stable and diverse microbiota. This is linked either to interaction with the environment, including exposure to maternal microbiota and to dietary change, and physiological changes, such as the development of the immune system and of gut maturation (Thompson et al. Citation2008; Starke et al. Citation2013; Chen et al. Citation2017S). In line with the literature, the results obtained in the present study clearly confirmed the dynamics of the modification in the faecal microbiota during the suckling period. This was characterised by an increase in alpha diversity values and a tendency to resemble the microbial structure of the adult pig microbiota represented by the sows with which the piglets shared increasing numbers of ASVs with ageing (Chen et al. Citation2017; Grześkowiak et al. Citation2019; Motta et al. Citation2019).
In addition to age, the microbial profile, in terms of beta diversity and taxa abundance, was modulated by the probiotic supplementation. Considering the alpha indices, the effect of probiotics is controversial (Guevarra et al. Citation2019). The present results showed that alpha diversity was affected by time but not by the administration of the probiotics. The present results are in contrast with a previous study of Kiros et al. (Citation2018) in which the oral administration of S. cerevisiae showed a reduction in piglet alpha diversity, supporting the hypothesis that early supplementation of suckling piglets with a yeast probiotic may anticipate the stability of the gut microbiota before weaning (Chen et al. Citation2017; Kiros et al. Citation2018). On the other hand, as far as what concerns the probiotic E. faecium, the present results are consistent with those reported by Wang et al. (Citation2016) in whose study no significant difference in intestinal microbial diversity of new-born piglets was found.
Regarding beta diversity, the effect of the probiotics supplementation was significant at d7 while it displayed a trend at d18 (only for the S. cerevisiae supplementation). These results suggested that a single oral administration of probiotics within the first forty-eight hours of life may have a time-limited effect on the piglet intestinal microbiota. However, soon after the first sampling time, creep feed was introduced to the piglets, and that might have affected the intestinal microbiota and lessened the effect of the probiotic administration.
The different response to the probiotic supplementation is noteworthy in terms of alpha and beta diversity. The absence of the effect of the probiotics regarding alpha diversity indicated that, inside each individual pig, the variability, in terms of quantity of different taxa, was not changed by the supplementation. Conversely, the significant effect obtained by the Adonis test suggested that the supplementation with probiotics was able to increase the microbial similarity among the samples in the same group since the differences in the microbial structures among the groups were higher than the differences within each group. This could be important for the microbiota exchanges among piglets over a long-term perspective and, for instance, when the piglets are mixed at weaning. Moreover, increased microbial similarity among the pigs, thanks to the probiotic supplementation, may increase the stability of the intestinal microbial profile and, thus, reduce the introduction of potentially pathogenic bacteria.
The supplementation of S. cerevisiae particularly favoured the colonisation of Erysipelatoclostridium and Christensenella genera which was significantly different in comparing taxa abundance and also as discriminant features of the SA group using PLS-DA analysis. This could be an effect of the anaerobic environment promoted by the live yeast which increased the concentration of the strictly anaerobic bacteria ( El Hassan et al. Citation1996; Marden et al. Citation2008; Chaucheyras-Durand and Durand, Citation2010) ), such as Erysipelatoclostridium and Christensenella, which are generally considered common taxa of the suckling piglet intestinal microbiota (Chen et al. Citation2017; Hasan et al. Citation2018). In this context, previous studies have reported that, in ruminants and horses, S. cerevisiae supplementation significantly improved the digestibility of dietary cellulose as a result of an increase in specific activities of the fibrolytic enzymes (Jouany et al. Citation1998; Chaucheyras-Durand and Fonty Citation2001, Jouany et al. Citation2009). Accordingly, Stanley et al. (Citation2016) showed a significant favourable correlation between the relative abundance of Erysipelotrichaceae and the feed conversion rate (FCR) performance in broilers.
The supplementation of E. faecium influenced the abundance of genera belonging to the Lachnospiraceae family, which is one of the dominant genera of the intestinal microbiota of neonatal pigs (Sagheddu et al. Citation2016; Chen et al. Citation2017). This family is composed of more than 24 genera, including Ruminococcus, Blautia and Dorea, known for their production of volatile fatty acids (VFAs: butyrate, propionate and acetate) considered positive for intestinal health and for providing energy to the host (Biddle et al. Citation2013). These assumptions could partially explain the increased growth performance of piglets in the EF group as compared with the control group, as has already been suggested by Yang et al. (Citation2017) in whose study the Lachnospiraceae family was associated with improved feed efficiency in pigs. However, in the present study, the VFAs were not analysed; thus, additional studies are needed to verify this hypothesis.
The SAEF group was discriminated by some families in common with the SA (for instance, Erysipelatoclostridiaceae at d7) and the EF groups (for instance the Ruminococcaceae which were discriminant for EF and SAEF at d7 at d18, respectively); however, it was also discriminated at d18 by genera belonging to the groups of Escherichia/Shigella. The discrimination of SAEF by Escherichia/Shigella can, in some way, explain the poorer growth performance observed in these groups; in fact, bacteria belonging to the groups of Escherichia/Shigella which are known to be present in the intestinal tract of young pigs (Konstantinov et al. Citation2006), if present in high concentrations, they can disturb the gut microbial ecosystem and increase the risk of diarrhoea; thus, they are not considered beneficial to the health of piglets (Gresse et al. Citation2017). Considering other species, such as ruminants, the effect of an E. faecium and S. cerevisiae mixture is controversial; a mixture of E. faecium EF212 and S. cerevisiae was associated with increased concentrations of acute phase proteins in the plasma of feedlot steers and, to explain the results, the authors hypothesised an increase in bacterial translocation (Emmanuel et al. Citation2007) while, in the studies of Chiquette et al. (Citation2015), a decrease in acute phase proteins in the plasma of dairy cows both during adaptation and subacute ruminal acidosis challenge was found. In the present study, the negative effect of the interaction between the probiotics was seen with respect to piglet growth, but was not seen on the microbial profile at the two time points. This could indicate that the negative effect of the probiotic mixture on the gut microbiota was active before the sampling times. In addition, as previously mentioned, the hypothesis that the piglets in the SAEF group had more energy for local immune activity, thus having the poorer performance, should not be excluded.
Conclusion
In conclusion, the present results showed that the administration of a single, early-life probiotic could improve piglet performance and shape the piglet faecal microbial profile during the suckling period, supporting the key importance of early microbial colonisation in the mid-term period. Erysipelotrichaceae and Lachnospiraceae families were favoured by the S. cerevisiae and E. feaecium supplementation, respectively, and could be candidate bacterial families for the promotion of piglet growth performance. This suggested that there was not a univocal stimulation of the microbiota for growth promotion. Additional studies are needed to clarify and more fully explain the biological mechanisms (e.g., the effect on the intestinal mucosa) of early-life probiotics on the growth and survival performance of piglets. Furthermore, future studies aimed at evaluating the effect of early-life probiotic supplementation in the post-weaning phase would be desirable to support probiosis as a robust strategy for improving piglet robustness and reducing antibiotic use.
Ethical approval
The procedures were approved by the Ethic-Scientific Committee for Experiments on Animals of the University of Bologna with approval number 104993/15th May 2019. The animals involved in the present study were reared on a conventional farm following Council Directive 2008/120/EC.
Disclosure statement
The Authors declare that there is no conflict of interest.
Additional information
Funding
References
- Banla LI, Salzman NH, Kristich CJ. 2019. Colonization of the mammalian intestinal tract by enterococci. Curr Opin Microbiol. 47:26–31.
- Bates D, Maechler M, Bolker B, Walker S. 2015. lme4: Linear mixed-effects models using Eigen and S4. R package version 1.1-8. http://CRAN.R-project.org/package=lme4.
- Beauchemin KA, Yang WZ, Morgavi DP, Ghorbani GR, Kautz W, Leedle JA. 2003. Effects of bacterial direct-fed microbials and yeast on site and extent of digestion, blood chemistry, and subclinical ruminal acidosis in feedlot cattle. J Anim Sci. 81(6):1628–1640.
- Bian G, Ma S, Zhu Z, Su Y, Zoetendal EG, Mackie R, Liu J, Mu C, Huang R, Smidt H, et al. 2016. Age, introduction of solid feed and weaning are more important determinants of gut bacterial succession in piglets than breed and nursing mother as revealed by a reciprocal cross-fostering model. Environ Microbiol. 18(5):1566–1577.
- Biddle A, Stewart L, Blanchard J, Leschine S. 2013. Untangling the genetic basis of fibrolytic specialization by Lachnospiraceae and Ruminococcaceae in diverse gut communities. Diversity. 5(3):627–640.
- Brückner S, Mösch HU. 2012. Choosing the right lifestyle: adhesion and development in Saccharomyces cerevisiae. FEMS Microbiol Rev. 36(1):25–58.
- Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. 2016. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. 13(7):581–583.
- Chapman CMC, Gibson GR, Rowland I. 2011. Health benefits of probiotics: Are mixtures more effective than single strains? Eur J Nutr. 50(1):1–17.
- Chaucheyras-Durand F, Durand H.Beneficial Microbes 2010. 11p. 3–9. Wageningen Academic PublishersProbiotics in animal nutrition and health
- Chaucheyras-Durand F, Fonty G. 2001. Establishment of cellulolytic bacteria and development of fermentative activities in the rumen of gnotobiotically-reared lambs receiving the microbial additive Saccharomyces cerevisiae CNCM I-1077. Reprod Nutr Dev. 41(1):57–68.
- Chen L, Xu Y, Chen X, Fang C, Zhao L, Chen F. 2017. The maturing development of gut microbiota in commercial piglets during the weaning transition. Front Microbiol. 8:1688.
- Chiquette J, Lagrost J, Girard CL, Talbot G, Li S, Plaizier JC, Hindrichsen IK. 2015. Efficacy of the direct-fed microbial Enterococcus faecium alone or in combination with Saccharomyces cerevisiae or Lactococcus lactis during induced subacute ruminal acidosis. J Dairy Sci. 98(1):190–203.
- Collins CL, Pluske JR, Morrison RS, McDonald TN, Smits RJ, Henman DJ, Stensland I, Dunshea FR. 2017. Post-weaning and whole-of-life performance of pigs is determined by live weight at weaning and the complexity of the diet fed after weaning. Animal Nutrition. 3(4):372–379.
- Dixon P. 2003. VEGAN, a package of R functions for community ecology. J Veg Sci Title. 14(6):927–930.
- Dunshea FR, Kerton DK, Cranwell PD, Campbell RG, Mullan BP, King RH, Power GN, Pluske JR. 2003. Lifetime and post-weaning determinants of performance indices of pigs. Aust J Agric Res. 54(4):363–370.
- El Hassan SM, Newbold CJ, Edwards IE, Topps JH, Wallace RJ. 1996. Effect of yeast culture on rumen fermentation, microbial protein flow from the rumen and live-weight gain in bulls given high cereal diets. Anim Sci. 62(1):43–48.
- Emmanuel DG, Jafari A, Beauchemin KA, Leedle JA, Ametaj BN. 2007. Feeding live cultures of Enterococcus faecium and Saccharomyces cerevisiae induces an inflammatory response in feedlot steers. J Anim Sci. 85(1):233–239.
- Everaert N, Van Cruchten S, Weström B, Bailey M, Van Ginneken C, Thymann T, Pieper R. 2017. A review on early gut maturation and colonization in pigs, including biological and dietary factors affecting gut homeostasis. Anim Feed Sci Tech. 233:89–103.
- Fomenky BE, Chiquette J, Lessard M, Bissonnette N, Talbot G, Chouinard YP, Ibeagha-Awemu EM. 2018. Saccharomyces cerevisiae var. boulardii CNCM I-1079 and Lactobacillus acidophilus BT1386 influence innate immune response and serum levels of acute-phase proteins during weaning in Holstein calves. Can J Anim Sci. 98(3):576–588.
- Gresse R, Chaucheyras-Durand F, Fleury MA, Van De Wiele T, Forano E, Blanquet-Diot S. 2017. Gut microbiota dysbiosis in postweaning piglets: understanding the keys to health. Trends Microbiol. 25(10):851–873.
- Grześkowiak Ł, Dadi TH, Zentek J, Vahjen W. 2019. Developing gut microbiota exerts colonisation resistance to Clostridium (syn. Clostridioides) difficile in piglets. Microorganisms. 7:218.
- Guevarra RB, Lee JH, Lee SH, Seok MJ, Kim DW, Kang BN, Johnson TJ, Isaacson RE, Kim HB. 2019. Piglet gut microbial shifts early in life: causes and effects. J Anim Sci Biotechnol. 10:1–0.
- Han GG, Lee JY, Jin GD, Park J, Choi YH, Kang SK, Chae BJ, Kim EB, Choi YJ. 2018. Tracing of the fecal microbiota of commercial pigs at five growth stages from birth to shipment. Sci Rep. 8(1):1–9.
- Hancox LR, Le Bon M, Richards PJ, Guillou D, Dodd CE, Mellits KH. 2015. Effect of a single dose of Saccharomyces cerevisiae var. boulardii on the occurrence of porcine neonatal diarrhoea. Animal. 9(11):1756–1759.
- Hasan S, Junnikkala S, Peltoniemi O, Paulin L, Lyyski A, Vuorenmaa J, Oliviero C. 2018. Dietary supplementation with yeast hydrolysate in pregnancy influences colostrum yield and gut microbiota of sows and piglets after birth. PloS One. 13(5):e0197586.
- Heuß EM, Pröll-Cornelissen MJ, Neuhoff C, Tholen E, Große-Brinkhaus C. 2019. Invited review: Piglet survival: benefits of the immunocompetence. Animal. 13(10):2114–2124.
- Hooper LV, Wong MH, Thelin A, Hansson L, Falk PG, Gordon JI. 2001. Molecular analysis of commensal host-microbial relationships in the intestine. Science. 291(5505):881–884.
- Jin LZ, Marquardt RR, Zhao X. 2000. A strain of Enterococcus faecium (18C23) inhibits adhesion of enterotoxigenic Escherichia coli K88 to porcine small intestine mucus. Appl Environ Microbiol. 66(10):4200–4204.
- Jouany JP, Mathieu F, Senaud J, Bohatier J, Bertin G, Mercier M. 1998. Effect of Saccharomyces cerevisiae and Aspergillus oryzae on the digestion of nitrogen in the rumen of defaunated and refaunated sheep. Anim Feed Sci Technol. 75(1):1–3.
- Jouany JP, Medina B, Bertin G, Julliand V. 2009. Effect of live yeast culture supplementation on hindgut microbial communities and their polysaccharidase and glycoside hydrolase activities in horses fed a high-fiber or high-starch diet. J Anim Sci. 87(9):2844–2852.
- Kenny M, Smidt H, Mengheri E, Miller B. 2011. Probiotics - do they have a role in the pig industry? Animal. 5(3):462–470.
- Kiros TG, Derakhshani H, Pinloche E, D’Inca R, Marshall J, Auclair E, Khafipour E, Van Kessel A. 2018. Effect of live yeast Saccharomyces cerevisiae (Actisaf Sc 47) supplementation on the performance and hindgut microbiota composition of weanling pigs. Sci Rep. 8(1):3.
- Kiros TG, Luise D, Derakhshani H, Petri R, Trevisi P, D’Inca R, Auclair E, van Kessel AG. 2019. Effect of live yeast Saccharomyces cerevisiae supplementation on the performance and cecum microbial profile of suckling piglets. PLoS One. 14(7):e0219557.
- Konstantinov SR, Awati AA, Williams BA, Miller BG, Jones P, Stokes CR, Akkermans AD, Smidt H, De Vos WM. 2006. Post‐natal development of the porcine microbiota composition and activities. Environ Microbiol. 8(7):1191–1199.
- Laukova A, Strompfova V, Ouwehand A. 2004. Adhesion properties of enterococci to intestinal mucus of different hosts. Vet Res Commun. 28(8):647–655.
- Lê Cao KA, Boitard S, Besse P. 2011. Sparse PLS discriminant analysis: biologically relevant feature selection and graphical displays for multiclass problems. BMC Bioinformatics. 12:253.
- Leblois J, Massart S, Li B, Wavreille J, Bindelle J, Everaert N. 2017. Modulation of piglets’ microbiota: differential effects by a high wheat bran maternal diet during gestation and lactation. Sci Rep. 7(1):1–1.
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15(12):550.
- Luise D, Bertocchi M, Motta V, Salvarani C, Bosi P, Luppi A, Fanelli F, Mazzoni M, Archetti I, Maiorano G, et al. 2019. Bacillus sp. probiotic supplementation diminish the Escherichia coli F4ac infection in susceptible weaned pigs by influencing the intestinal immune response, intestinal microbiota and blood metabolomics. J. Anim. Sci. Biotechnol. 10:74.
- Maia OB, Duarte R, Silva AM, Cara DC, Nicoli JR. 2001. Evaluation of the components of a commercial probiotic in gnotobiotic mice experimentally challenged with Salmonella enterica subsp. enterica ser. Typhimurium. Vet Microbiol. 79(2):183–189.
- Marden JP, Julien C, Monteils V, Auclair E, Moncoulon R, Bayourthe C. 2008. How does live yeast differ from sodium bicarbonate to stabilize ruminal pH in high-yielding dairy cows? J Dairy Sci. 91(9):3528–3535.
- Martin LB, Scheuerlein A, Wikelski M. 2003. Immune activity elevates energy expenditure of house sparrows: a link between direct and indirect costs? Proc Biol Sci. 270(1511):153–158.
- McMurdie PJ, Holmes S. 2013. Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PloS One. 8(4):e61217.
- Motta V, Luise D, Bosi P, Trevisi P. 2019. Faecal microbiota shift during weaning transition in piglets and evaluation of AO blood types as shaping factor for the bacterial community profile. PloS One. 1614(5):e0217001.
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41(Database issue):D590–6.
- Revilla M, Friggens NC, Broudiscou LP, Lemonnier G, Blanc F, Ravon L, Mercat MJ, Billon Y, Rogel-Gaillard C, Le Floch N, et al. 2019. Towards the quantitative characterisation of piglets' robustness to weaning: a modelling approach. Animal. 13(11):2536–2546.
- Sagheddu V, Patrone V, Miragoli F, Puglisi E, Morelli L. 2016. Infant early gut colonization by Lachnospiraceae: high frequency of Ruminococcus gnavus. Front Pediatr. 4:57.
- Stanley D, Hughes RJ, Geier MS, Moore RJ. 2016. Bacteria within the gastrointestinal tract microbiota correlated with improved growth and feed conversion: challenges presented for the identification of performance enhancing probiotic bacteria. Front Microbiol. 7:187.
- Starke IC, Pieper R, Neumann K, Zentek J, Vahjen W. 2013. Individual responses of mother sows to a probiotic Enterococcus faecium strain lead to different microbiota composition in their offspring. Benef Microbes. 4(4):345–356.
- Stokes CR. 2017. The development and role of microbial-host interactions in gut mucosal immune development. J Anim Sci Biotech. 8:1–0.
- Stokes CR, Bailey M, Haverson K, Harris C, Jones P, Inman C, Pié S, Oswald IP, Williams BA, Akkermans ADL, et al. 2004. Postnatal development of intestinal immune system in piglets: implications for the process of weaning. Anim Res. 53(4):325–334.
- Tannock GW, Fuller R, Pedersen K. 1990. Lactobacillus succession in the piglet digestive tract demonstrated by plasmid profiling. Appl Environ Microbiol. 56(5):1310–1316.
- Taras D, Vahjen W, Macha M, Simon O. 2006. Performance, diarrhea incidence, and occurrence of Escherichia coli virulence genes during long-term administration of a probiotic Enterococcus faecium strain to sows and piglets. Sci J Anim Sci. 84(3):608–617.
- Thompson CL, Wang B, Holmes AJ. 2008. The immediate environment during postnatal development has long-term impact on gut community structure in pigs. Isme J. 2(7):739–748.
- Trevisi P, Corrent E, Messori S, Casini L, Bosi P. 2010. Healthy newly weaned pigs require more tryptophan to maximize feed intake if they are susceptible to Escherichia coli K88. Livest Sci. 134(1-3):236–238.
- Trevisi P, Priori D, Jansman AJM, Luise D, Koopmans SJ, Hynönen U, Palva A, van der Meulen J, Bosi P. 2018. Molecular networks affected by neonatal microbial colonization in porcine jejunum, luminally perfused with enterotoxigenic Escherichia coli, F4ac fimbria or Lactobacillus amylovorus. PLoS One. 13(8):e0202160.
- Wang YB, Du W, Fu AK, Zhang XP, Huang Y, Lee KH, Yu K, Li WF, Li YL. 2016. Intestinal microbiota and oral administration of Enterococcus faecium associated with the growth performance of new-born piglets. Benef Microbes. 7(4):529–538.
- Wang Y, Gong L, Wu YP, Cui ZW, Wang YQ, Huang Y, Zhang XP, Li WF. 2019. Oral administration of Lactobacillus rhamnosus GG to newborn piglets augments gut barrier function in pre-weaning piglets. J Zhejiang Univ Sci B. 20(2):180–192.
- Wu Y, Zhen W, Geng Y, Wang Z, Guo Y. 2019. Effects of dietary Enterococcus faecium NCIMB 11181 supplementation on growth performance and cellular and humoral immune responses in broiler chickens. Poultry Sc. 98(1):150–163.
- Yang H, Huang X, Fang S, He M, Zhao Y, Wu Z, Yang M, Zhang Z, Chen C, Huang L. 2017. Unraveling the fecal microbiota and metagenomic functional capacity associated with feed efficiency in pigs. Front Microbiol. 8:1555.
- Zeyner A, Boldt E. 2006. Effects of a probiotic Enterococcus faecium strain supplemented from birth to weaning on diarrhoea patterns and performance of piglets. J Anim Physiol Anim Nutr. 90(1-2):25–31.
