Abstract
Protein chaperones help proteins adopt and maintain native conformations and play vital roles in cellular processes where proteins are partially folded. They comprise a major part of the cellular protein quality control system that protects the integrity of the proteome. Many disorders are caused when proteins misfold despite this protection. Yeast prions are fibrous amyloid aggregates of misfolded proteins. The normal action of chaperones on yeast prions breaks the fibers into pieces, which results in prion replication. Because this process is necessary for propagation of yeast prions, even small differences in activity of many chaperones noticeably affect prion phenotypes. Several other factors involved in protein processing also influence formation, propagation or elimination of prions in yeast. Thus, in much the same way that the dependency of viruses on cellular functions has allowed us to learn much about cell biology, the dependency of yeast prions on chaperones presents a unique and sensitive way to monitor the functions and interactions of many components of the cell's protein quality control system. Our recent work illustrates the utility of this system for identifying and defining chaperone machinery interactions.
Abbreviations
| NEF | = | nucleotide exchange factor |
| CTD | = | C-terminal domain |
| NBD | = | nucleotide-binding domain |
Introduction
Amyloid-based yeast prions propagate as self-assembling fibers of single types of protein. The commonly studied prions [URE3] and [PSI+] are composed of the cellular proteins Ure2 and Sup35, respectively.Citation1 They propagate in the cytosol and are transmitted between cells with cytoplasm during cell division or cell fusion (i.e. mating). To persist in an expanding yeast population, prions must reproduce to keep pace with cell division. To do so, they must continually grow and be fragmented into more numerous self-propagating pieces. Growth can occur spontaneously by addition of soluble protein to fiber ends.Citation2,3 Replication by fragmentation of fibers requires action of the Hsp104-driven protein disaggregation machinery, which includes the Hsp70 system comprised of Hsp70 and its co-chaperones of the Hsp40 (J-protein) and nucleotide exchange factor (NEF) families.Citation4–10
A major role of this disaggregation machinery is to preserve viability of cells exposed to stresses that cause protein denaturation by restoring solubility of aggregated proteins.Citation11 Hsp104 works with the Hsp70 system to extract proteins from aggregates, extruding them one at a time through the central channel of the Hsp104 hexamer.Citation12–15 This machinery acts in a similar way to extract protein monomers from prion fibers, which causes the fibers to break in a way that allows the resulting pieces to continue self assembly ().Citation9,16
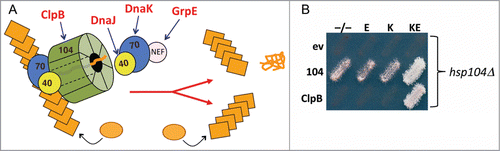
Figure 1. E. coli disaggregation machinery propagates prions in yeast. (A) Yeast prions propagate as amyloid fibers (stacked orange rectangles) that grow when soluble protein (orange circles) is added to fiber ends. Replication of fibers requires cooperation of the Hsp70 system (Hsp70, Hsp40 and NEF) with Hsp104 (green), which extracts proteins from the fiber thereby causing the fiber to break into 2 self-assembling prion fibers. E. coli counterparts of machinery components are indicated in red. (B) Cells lacking chromosomal Hsp104 (hsp104Δ) express Hsp104 (104) or E. coli ClpB from a plasmid, or carry the corresponding empty vector (ev), as indicated on the left. As indicated above, cells also carry plasmids for expressing E. coli Hsp70 DnaK (K), the DnaK NEF GrpE (E), both (KE) or the empty vectors (−/−). Medium shown allows growth only if cells propagate prions. The combination of ClpB, DnaK and GrpE (BKE) is both necessary and sufficient for prion propagation when Hsp104 is absent.
Hsp70 binds and releases exposed hydrophobic peptides on unfolded proteins in an ATP-regulated reaction cycle controlled primarily by Hsp40 and NEF family co-chaperones.Citation17 Hsp40s bind similar hydrophobic substrates and have a J-domain that interacts with and activates the ATPase domain of Hsp70. These activities help coordinate Hsp70 ATP hydrolysis with substrate binding. NEFs then facilitate release of ADP, which allows rebinding of ATP and release of substrate to reinitiate the cycle.
Because yeast prions depend on the disaggregation machinery, they cannot propagate in cells lacking Hsp104 or in which activity of this machinery is compromised.Citation4,18,19 There is extensive functional redundancy among the Hsp70, Hsp40 and NEF family components of this machinery (), which provides a large number of combinations that could potentially tune the activity of the disaggregation reaction in many ways. Even transient or subtle alterations in the activity of many of these factors, including several of the J-protein and nucleotide exchange factor families, can significantly affect how efficiently prions propagate.Citation4,7,20–26 These effects could reflect alterations in such tuning. Moreover, compensatory alterations in activity of the same or different chaperones or co-chaperones can offset such effects.Citation21,27 Since small changes in efficiency of prion propagation can be reflected in differences of readily observable prion phenotypes, prions provide a sensitive way to monitor the status of chaperone activity. Furthermore, many chaperone alterations that inhibit prion propagation have imperceptible effects on cell physiology, so prions also offer a unique way to uncover variations in chaperone function that otherwise would be difficult to detect. Therefore, yeast prions provide not only a useful way to study how chaperones can influence propagation of amyloid in cells, but also a very sensitive way to identify and monitor various aspects of chaperone function and cooperation.
Table 1. Redundancy of cytosolic Hsp70 system components
Roles for Hsp104 and the Hsp70 System in Prion Propagation
Altering abundance or function of Hsp104 and Hsp70 can have counteracting effects on formation and propagation of prions, suggesting that a balance of these chaperones is important for maintaining both native and prion forms of prion proteins.Citation28–31 Yet, while many different point mutations in the Hsp70s Ssa1 and Ssa2 reduce the strength and mitotic stability of prions,Citation20,21,32 there is little evidence indicating a direct role of Hsp70 in prion propagation beyond the finding that Ssa1 binds preferentially to the prion form of Sup35 in cell lysates.Citation33 Moreover, data showing anti-prion effects of Hsp70 alterations can be interpreted as resulting from indirect effects on the protein disaggregation process.Citation9,10,25 Thus, altering Hsp70 could affect prion propagation by upsetting the balance between the Hsp104 and Hsp70 activities that are important for certain disaggregation machinery functions. Additionally, the noticeable effects that many Hsp70 co-chaperones have on prions are dependent on their interactions with Hsp70. These findings indicate that efficient prion propagation depends on proper regulation of Hsp70, and raise the possibility that the co-chaperones also influence prions indirectly by affecting the Hsp104-based disaggregation machinery.
Using purified components, however, Hsp104 alone was reported to promote both assembly and fragmentation of amyloid polymers composed of Sup35 or Ure2.Citation34,35 It also was reported that ClpB, the bacterial Hsp100 family homolog of Hsp104, was unable to fragment amyloid in similar reactions or to support propagation of [PSI+] prions in yeast.Citation34,36 These findings led to suggestions that Hsp104 could act unaided to promote prion replication in yeast and that this activity had evolved beyond the functions of its bacterial homolog. If Hsp104 acts alone to propagate prions in cells, then an important question would be how Hsp70 and its many co-chaperones are acting to influence prion propagation so strongly.
Hsp70 Regulates Hsp104 FUNCTION IN VIVO
Overexpressing Hsp104 is normally benign because Hsp104 activity is tightly regulated in vivo. Mutations in the middle (M) region of Hsp104 can disrupt this regulation and inappropriately activate Hsp104, making its overexpression toxic.Citation37 More recently, this M region, which is conserved among Hsp100 family proteins, was shown to mediate species-specific interactions with Hsp70 that regulate Hsp100 activity.Citation10,38–40 Replacing the M region of Hsp104 with that of ClpB allows Hsp104 to function in place of ClpB in E. coli, while ClpB with the M-region of Hsp104 can function in place of Hsp104 in yeast.Citation36,41 Similarly, purified Hsp100s with the swapped M regions cooperate with the cross-species Hsp70 system to reactivate aggregated proteins.Citation36,41,42 Thus, the in vivo results likely reflect an ability of the endogenous yeast and bacterial Hsp70 systems to cooperate with the hybrid Hsp100s through an interaction with the cognate M regions.
Yeast Prions Help Define Chaperone Machinery Interactions
We reasoned that inability of ClpB to interact with the yeast Hsp70 system due to this species-specificity for Hsp70-Hsp100 interaction explained the failure of ClpB to drive protein disaggregation in yeast. To test this idea we expressed components of the E. coli Hsp70 system in yeast cells with ClpB in place of Hsp104.Citation10 Adding the E. coli Hsp70 DnaK did not restore prion propagation, but adding both DnaK and its NEF GrpE enabled ClpB to support propagation of [PSI+] prions (). [PSI+] does not propagate as efficiently in these cells as it does in cells expressing Hsp104, which probably reflects other limitations of the bacterial disaggregation machinery in the more complex eukaryotic environment. Nevertheless, the fact that ClpB, DnaK and GrpE can propagate [PSI+] shows conservation of the ability of this machinery to act on amyloid substrates.
Because J-proteins are thought to be obligatory co-chaperones for Hsp70, we were surprised that the major bacterial J-protein, DnaJ, was not necessary for ClpB function in yeast. To test the possibility that the bacterial chaperones were working with a yeast J-protein, DnaK was engineered to contain a point mutation (R167H) that specifically disrupts interaction with the conserved histidine-proline-aspartate (HPD) motif in the J-domain of J-proteins.Citation43 The E. coli machinery with DnaKR167H in place of wild type DnaK failed to propagate yeast prions, implying a J-protein is indeed a necessary component in the disaggregation reaction for replicating yeast prions, and that a yeast J-protein must be involved ().
Figure 2. Prion propagation by the disaggregation machinery requires Hsp40 (J-protein). (A) To determine if E. coli DnaK (green circle) requires interaction with a yeast Hsp40 (triangles), DnaK with a mutation (red x, DnaK*) that specifically disrupts interaction with J-proteins, was expressed with ClpB and GrpE (BK*E) in cells lacking chromosomal Hsp104 (hsp104Δ). Loss of ability of BK*E to propagate prions shows an unknown Hsp40 (?) must be cooperating with BKE to promote prion replication. (B) In order to identify which yeast Hsp40 is involved, a compensatory mutation in J-proteins (red +) that restores interaction with DnaK* was made in yeast Hsp40s Sis1, Ydj1 and the E. coli Hsp40 DnaJ (indicated Sis1*, Ydj1* and DnaJ*, respectively). The compensatory mutations direct the interaction of these mutants with DnaK* even in the presence of wild type versions of the endogenous Hsp70s (circles) and Hsp40s (triangles). Each Hsp40 mutant was expressed separately with BK*E in hsp104Δ cells. Only Sis1* cooperates with BK*E to restore prion replication. Medium shown allows growth only if cells propagate prions.
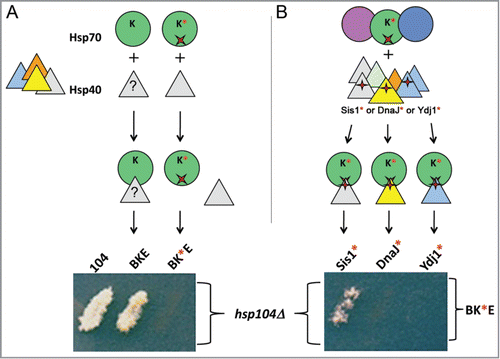
A useful feature of the R167H mutant is that a compensatory mutation in the HPD of E. coli DnaJ (D35N) restores the J-domain interaction producing a functional Hsp70/J-protein pair.Citation43 By engineering the major yeast Hsp40 J-proteins (Ydj1 and Sis1) to contain the analogous HPD substitution (D36N), we were able to direct DnaKR167H to interact specifically with the modified Hsp40s, even in the presence of their wild type counterparts (). We found that Sis1D36N restores ability of DnaKR167H to cooperate with ClpB and GrpE to propagate [PSI+]. Ydj1D36N does not function in this process, but it cooperates more effectively than Sis1D36N with the bacterial chaperones to protect yeast from exposure to lethal heat. These findings confirm that functions of the disaggregation machinery in vivo require Hsp40 and that this requirement is mediated by a physical interaction between the J-protein and Hsp70 components. They also show that ClpB, DnaK and GrpE cooperate specifically with Sis1 to propagate yeast prions, and with Ydj1 to provide thermotolerance.
This work shows that functions of the disaggregation machinery in different cellular processes minimally require cooperation of Hsp100 with Hsp70 and its Hsp40 (J-protein) and NEF co-chaperones. Since GrpE and ClpB bind DnaK on an overlapping surface, it is unlikely they interact with Hsp70 simultaneously,Citation40,44 which would limit ways that GrpE could participate in the disaggregation reaction. Evidence suggests GrpE is not required before DnaK engages with ClpB,Citation45 so it might act on DnaK after substrates are threaded through ClpB. Regardless of the actual mechanism, while interactions of the Hsp100, Hsp70 and NEF components are species-specific, J-proteins, even across life domains, are less discriminate. Yet, despite this promiscuity in their ability to regulate Hsp70s, different Hsp40s govern distinctions in the activity of the machines with which they interact. The findings are in line with the view that J-proteins confer such specificity by targeting the chaperone machinery to its substrates.Citation40,45,46
Yeast Prions Help Identify Interactions that Specify Activity of Chaperone Machines
Because yeast, like most organisms, encode several functionally overlapping Hsp70s, targeting of the disaggregation machinery to different substrates by cytosolic J-proteins could also occur by recruitment of specific Hsp70s. Without addressing this question directly, earlier studies assessing specificity of functions of Ydj1 and Sis1 structural domains provide some insight,Citation47–49 although they came to different conclusions depending on whether the domains were swapped or deleted. Swapping domains between Ydj1 and Sis1 showed that the Sis1 substrate-binding region is necessary for essential Sis1 function, even when attached to the Ydj1 J-domain.Citation49 Additionally, the C-terminal substrate-binding regions (CTDs) of Ydj1 and Sis1 prefer substrates with distinct amino acid compositions.Citation50 These findings suggest that Sis1-specific functions are determined by specificity of substrate binding. However, a truncated Sis1 lacking its substrate-binding domain supports growth in place of intact Sis1, suggesting Sis1-specific functions are determined more by Hsp70 regulation than by substrate binding.Citation47,48
Having established a yeast system for distinguishing J-protein functions using the bacterial disaggregation machinery, we swapped conserved domains of Ydj1 and Sis1 and used full-length hybrid proteins directed to interact with DnaKR167H to identify which regions of these Hsp40s conferred their functional distinctions. Their ability to cooperate with ClpB in prion propagation relied primarily on the CTD of Sis1, while thermotolerance required the CTD of Ydj1.Citation51 We also confirmed that Sis1 and Ydj1 also specify functions of the endogenous Hsp104 machine for prion propagation and thermotolerance, respectively, and that these differences were similarly mediated primarily by the Hsp40 CTDs. These findings were recapitulated when we tested the ability of purified chaperones to reactivate denatured protein in reactions that depend specifically on Sis1 or Ydj1, which strengthens the conclusion that substrate binding by J-proteins can direct activity of the same machinery in different processes. We further showed the CTDs support Hsp40-specific growth requirements and that the CTD of Ydj1 determines the essential role of Ydj1 for the function of the Hsp90 machinery in activation of galactose-induced transcription.Citation51,52 Thus, the targeting of different chaperone machines to particular substrates is specified more by interaction of J-proteins with the substrates than with a specific Hsp70.
While these findings clarify specific activities and interactions of various chaperones that are required for them to perform effectively in some of their cellular roles, the complexity arising from genomic amplification of chaperone families leaves much to decipher. For example, despite their near identity and overall functional interchangeability, the Ssa1 and Ssa2 Hsp70s have distinct functions that are revealed by noticeable differences in phenotypes of [PSI+] and [URE3] prions.Citation53,54 The basis of this distinction is only partially characterized, but it also underlies differences in their functions in an unrelated vesicle-mediated protein degradation pathway.Citation55 Uncovering the particular interactions underlying these functional distinctions will help us understand how exquisitely subtle changes in structure can provide functional diversity and fine tuning of the chaperone machinery. Prions also might help studies of certain role-specific Hsp70s and co-chaperones that are designated for specific cellular processes. For example, specific J-proteins and Hsp70s associated with ribosomes cooperate to assist folding of nascent polypeptides, and alterations of these chaperones affect prion appearance or propagation.Citation56–59 Further work with this system could provide new insight into the functions of these chaperones. Prions are also affected by TPR-containing co-chaperones that bind the C-terminus of Hsp70 to regulate its activity and recruit it to other chaperone machines.Citation21,27,60–63 Finally, many other factors involved in a variety of cellular processes such as the Btn2/Cur1 system, protein trafficking, sorting and compartmentalization, ubiquitin/proteasome system and actin dynamics also influence yeast prion phenotypes.Citation64–68 Yeast prions should also provide a unique and sensitive tool to study functions of these protein quality control systems.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank our NIH colleagues for critical review of the manuscript.
Funding
This work was supported by the Intramural Research Program of the National Institutes of Health (NIH), National Institute of Diabetes, Digestive and Kidney Diseases.
REFERENCES
- Wickner RB. Evidence for a prion analog in S. cerevisiae: the [URE3] non-Mendelian genetic element as an altered URE2 protein. Science 1994; 264:566–9; PMID:7909170; http://dx.doi.org/10.1126/science.7909170.
- Inoue Y, Kishimoto A, Hirao J, Yoshida M, Taguchi H. Strong growth polarity of yeast prion fiber revealed by single fiber imaging. J Biol Chem 2001; 276:35227–30; PMID:11473105; http://dx.doi.org/10.1074/jbc.C100304200.
- Collins SR, Douglass A, Vale RD, Weissman JS. Mechanism of prion propagation: amyloid growth occurs by monomer addition. PLoS Biol 2004; 2:e321; PMID:15383837; http://dx.doi.org/10.1371/journal.pbio.0020321.
- Chernoff YO, Lindquist SL, Ono B, Inge-Vechtomov SG, Liebman SW. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+]. Science 1995; 268:880–4.
- Paushkin SV, Kushnirov VV, Smirnov VN, Ter-Avanesyan MD. Propagation of the yeast prion-like [psi+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J 1996; 15:3127–34; PMID:8670813.
- Derkatch IL, Bradley ME, Zhou P, Chernoff YO, Liebman SW. Genetic and environmental factors affecting the de novo appearance of the [PSI+] prion in Saccharomyces cerevisiae. Genetics 1997; 147:507–19; PMID:9335589.
- Moriyama H, Edskes HK, Wickner RB. [URE3] prion propagation in Saccharomyces cerevisiae: requirement for chaperone Hsp104 and curing by overexpressed chaperone Ydj1p. Mol Cell Biol 2000; 20:8916–22; PMID:11073991; http://dx.doi.org/10.1128/MCB.20.23.8916-8922.2000.
- Inoue Y, Taguchi H, Kishimoto A, Yoshida M. Hsp104 binds to yeast Sup35 prion fiber but needs other factor(s) to sever it. J Biol Chem 2004; 279:52319–23; PMID:15448141; http://dx.doi.org/10.1074/jbc.M408159200.
- Hung GC, Masison DC. N-terminal domain of yeast Hsp104 chaperone is dispensable for thermotolerance and prion propagation but necessary for curing prions by Hsp104 overexpression. Genetics 2006; 173:611–20; PMID:16582428; http://dx.doi.org/10.1534/genetics.106.056820.
- Reidy M, Miot M, Masison DC. Prokaryotic Chaperones Support Yeast Prions and Thermotolerance and Define Disaggregation Machinery Interactions. Genetics 2012; 192:185–93; PMID:22732191; http://dx.doi.org/10.1534/genetics.112.142307.
- Parsell DA, Kowal AS, Singer MA, Lindquist S. Protein disaggregation mediated by heat-shock protein Hsp104. Nature 1994; 372:475–8; PMID:7984243; http://dx.doi.org/10.1038/372475a0.
- Glover JR, Lindquist S. Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell 1998; 94:73–82; PMID:9674429; http://dx.doi.org/10.1016/S0092-8674(00)81223-4.
- Lum R, Tkach JM, Vierling E, Glover JR. Evidence for an unfolding/threading mechanism for protein disaggregation by Saccharomyces cerevisiae Hsp104. J Biol Chem 2004; 279:29139–46; PMID:15128736; http://dx.doi.org/10.1074/jbc.M403777200.
- Schlieker C, Tews I, Bukau B, Mogk A. Solubilization of aggregated proteins by ClpB/DnaK relies on the continuous extraction of unfolded polypeptides. FEBS Lett 2004; 578:351–6; PMID:15589844; http://dx.doi.org/10.1016/j.febslet.2004.11.051.
- Weibezahn J, Tessarz P, Schlieker C, Zahn R, Maglica Z, Lee S, Zentgraf H, Weber-Ban EU, Dougan DA, Tsai FT, et al. Thermotolerance requires refolding of aggregated proteins by substrate translocation through the central pore of ClpB. Cell 2004; 119:653–65; PMID:15550247; http://dx.doi.org/10.1016/j.cell.2004.11.027.
- Tessarz P, Mogk A, Bukau B. Substrate threading through the central pore of the Hsp104 chaperone as a common mechanism for protein disaggregation and prion propagation. Mol Microbiol 2008; 68:87–97; PMID:18312264; http://dx.doi.org/10.1111/j.1365-2958.2008.06135.x.
- Mayer MP, Bukau B. Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol Life Sci 2005; 62:670–84; PMID:15770419; http://dx.doi.org/10.1007/s00018-004-4464-6.
- Jung G, Masison DC. Guanidine hydrochloride inhibits Hsp104 activity in vivo: a possible explanation for its effect in curing yeast prions. Curr Microbiol 2001; 43:7–10; PMID:11375656; http://dx.doi.org/10.1007/s002840010251.
- Ferreira PC, Ness F, Edwards SR, Cox BS, Tuite MF. The elimination of the yeast [PSI+] prion by guanidine hydrochloride is the result of Hsp104 inactivation. Mol Microbiol 2001; 40:1357–69; PMID:11442834; http://dx.doi.org/10.1046/j.1365-2958.2001.02478.x.
- Jung G, Jones G, Wegrzyn RD, Masison DC. A role for cytosolic hsp70 in yeast [PSI+] prion propagation and [PSI+] as a cellular stress. Genetics 2000; 156:559–70; PMID:11014806.
- Jones GW, Masison DC. Saccharomyces cerevisiae Hsp70 mutations affect [PSI+] prion propagation and cell growth differently and implicate Hsp40 and tetratricopeptide repeat cochaperones in impairment of [PSI+]. Genetics 2003; 163:495–506; PMID:12618389.
- Lopez N, Aron R, Craig EA. Specificity of class II Hsp40 Sis1 in maintenance of yeast prion [RNQ+]. Mol Biol Cell 2003; 14:1172–81; PMID:12631732; http://dx.doi.org/10.1091/mbc.E02-09-0593.
- Kryndushkin D, Wickner RB. Nucleotide exchange factors for Hsp70s are required for [URE3] prion propagation in Saccharomyces cerevisiae. Mol Biol Cell 2007; 18:2149–54; http://dx.doi.org/10.1091/mbc.E07-02-0128.
- Fan Q, Park KW, Du Z, Morano KA, Li L. The role of Sse1 in the de novo formation and variant determination of the [PSI+] prion. Genetics 2007; 177:1583–93; PMID:18039878; http://dx.doi.org/10.1534/genetics.107.077982.
- Kirkland PA, Reidy M, Masison DC. Functions of yeast Hsp40 chaperone Sis1p dispensable for prion propagation but important for prion curing and protection from prion toxicity. Genetics 2011; 188:565–77; PMID:21555396; http://dx.doi.org/10.1534/genetics.111.129460.
- Kumar N, Gaur D, Masison DC, Sharma D. The BAG homology domain of Snl1 cures yeast prion [URE3] through regulation of Hsp70 chaperones. G3 (Bethesda) 2014; 4:461–70; PMID:24408033.
- Jones G, Song Y, Chung S, Masison DC. Propagation of Saccharomyces cerevisiae [PSI+] prion is impaired by factors that regulate Hsp70 substrate binding. Mol Cell Biol 2004; 24:3928–37; PMID:15082786; http://dx.doi.org/10.1128/MCB.24.9.3928-3937.2004.
- Newnam GP, Wegrzyn RD, Lindquist SL, Chernoff YO. Antagonistic interactions between yeast chaperones Hsp104 and Hsp70 in prion curing. Mol Cell Biol 1999; 19:1325–33; PMID:9891066.
- Allen KD, Wegrzyn RD, Chernova TA, Muller S, Newnam GP, Winslett PA, Wittich KB, Wilkinson KD, Chernoff YO. Hsp70 chaperones as modulators of prion life cycle: novel effects of Ssa and Ssb on the Saccharomyces cerevisiae prion [PSI+]. Genetics 2005; 169:1227–42; PMID:15545639; http://dx.doi.org/10.1534/genetics.104.037168.
- Newnam GP, Birchmore JL, Chernoff YO. Destabilization and recovery of a yeast prion after mild heat shock. J Mol Biol 2011; 408:432–48; PMID:21392508; http://dx.doi.org/10.1016/j.jmb.2011.02.034.
- Winkler J, Tyedmers J, Bukau B, Mogk A. Hsp70 targets Hsp100 chaperones to substrates for protein disaggregation and prion fragmentation. J Cell Biol 2012; 198:387–404; PMID:22869599; http://dx.doi.org/10.1083/jcb.201201074.
- Loovers HM, Guinan E, Jones GW. Importance of the Hsp70 ATPase domain in yeast prion propagation. Genetics 2007; 175:621–30; PMID:17151238; http://dx.doi.org/10.1534/genetics.106.066019.
- Bagriantsev SN, Gracheva EO, Richmond JE, Liebman SW. Variant-specific [PSI+] infection is transmitted by Sup35 polymers within [PSI+] aggregates with heterogeneous protein composition. Mol Biol Cell 2008; 19:2433–43; PMID:18353968; http://dx.doi.org/10.1091/mbc.E08-01-0078.
- Shorter J, Lindquist S. Hsp104 catalyzes formation and elimination of self-replicating Sup35 prion conformers. Science 2004; 304:1793–7; PMID:15155912; http://dx.doi.org/10.1126/science.1098007.
- Shorter J, Lindquist S. Destruction or potentiation of different prions catalyzed by similar Hsp104 remodeling activities. Mol Cell 2006; 23:425–38; PMID:16885031; http://dx.doi.org/10.1016/j.molcel.2006.05.042.
- Tipton KA, Verges KJ, Weissman JS. In vivo monitoring of the prion replication cycle reveals a critical role for Sis1 in delivering substrates to Hsp104. Mol Cell 2008; 32:584–91; PMID:19026788; http://dx.doi.org/10.1016/j.molcel.2008.11.003.
- Schirmer EC, Homann OR, Kowal AS, Lindquist S. Dominant gain-of-function mutations in Hsp104p reveal crucial roles for the middle region. Mol Biol Cell 2004; 15:2061–72; PMID:14978213; http://dx.doi.org/10.1091/mbc.E02-08-0502.
- Sielaff B, Tsai FT. The M-domain controls Hsp104 protein remodeling activity in an Hsp70/Hsp40-dependent manner. J Mol Biol 2010; 402:30–7; PMID:20654624; http://dx.doi.org/10.1016/j.jmb.2010.07.030.
- Haslberger T, Weibezahn J, Zahn R, Lee S, Tsai FT, Bukau B, Mogk A. M domains couple the ClpB threading motor with the DnaK chaperone activity. Mol Cell 2007; 25:247–60; PMID:17244532; http://dx.doi.org/10.1016/j.molcel.2006.11.008.
- Rosenzweig R, Moradi S, Zarrine-Afsar A, Glover JR, Kay LE. Unraveling the mechanism of protein disaggregation through a ClpB-DnaK interaction. Science 2013; 339:1080–3; PMID:23393091; http://dx.doi.org/10.1126/science.1233066.
- Miot M, Reidy M, Doyle SM, Hoskins JR, Johnston DM, Genest O, Vitery MC, Masison DC, Wickner S. Species-specific collaboration of heat shock proteins (Hsp) 70 and 100 in thermotolerance and protein disaggregation. Proc Natl Acad Sci U S A 2011; 108:6915–20; PMID:21474779; http://dx.doi.org/10.1073/pnas.1102828108.
- Carroni M, Kummer E, Oguchi Y, Wendler P, Clare DK, Sinning I, Kopp J, Mogk A, Bukau B, Saibil HR. Head-to-tail interactions of the coiled-coil domains regulate ClpB activity and cooperation with Hsp70 in protein disaggregation. ELife 2014; 3:e02481; PMID:24843029; http://dx.doi.org/10.7554/eLife.02481.
- Suh WC, Lu CZ, Gross CA. Structural features required for the interaction of the Hsp70 molecular chaperone DnaK with its cochaperone DnaJ. J Biol Chem 1999; 274:30534–9; PMID:10521435; http://dx.doi.org/10.1074/jbc.274.43.30534.
- Doyle SM, Shastry S, Kravats AN, Shih YH, Miot M, Hoskins JR, Stan G, Wickner S. Interplay between E. coli DnaK, ClpB and GrpE during Protein Disaggregation. J Mol Biol 2015; 427:312–27; PMID:25451597; http://dx.doi.org/10.1016/j.jmb.2014.10.013.
- Zietkiewicz S, Krzewska J, Liberek K. Successive and synergistic action of the Hsp70 and Hsp100 chaperones in protein disaggregation. J Biol Chem 2004; 279:44376–83; PMID:15302880; http://dx.doi.org/10.1074/jbc.M402405200.
- Haslberger T, Zdanowicz A, Brand I, Kirstein J, Turgay K, Mogk A, Bukau B. Protein disaggregation by the AAA+ chaperone ClpB involves partial threading of looped polypeptide segments. Nat Struct Mol Biol 2008; 15:641–50; PMID:18488042; http://dx.doi.org/10.1038/nsmb.1425.
- Yan W, Craig EA. The glycine-phenylalanine-rich region determines the specificity of the yeast Hsp40 Sis1. Mol Cell Biol 1999; 19:7751–8; PMID:10523664.
- Johnson JL, Craig EA. An essential role for the substrate-binding region of Hsp40s in Saccharomyces cerevisiae. J Cell Biol 2001; 152:851–6; PMID:11266475; http://dx.doi.org/10.1083/jcb.152.4.851.
- Fan CY, Lee S, Ren HY, Cyr DM. Exchangeable chaperone modules contribute to specification of type I and type II Hsp40 cellular function. Mol Biol Cell 2004; 15:761–73; PMID:14657253; http://dx.doi.org/10.1091/mbc.E03-03-0146.
- Lee S, Fan CY, Younger JM, Ren H, Cyr DM. Identification of essential residues in the type II Hsp40 Sis1 that function in polypeptide binding. J Biol Chem 2002; 277:21675–82; PMID:11919183; http://dx.doi.org/10.1074/jbc.M111075200.
- Reidy M, Sharma R, Shastry S, Roberts BL, Albino-Flores I, Wickner S, Masison DC. Hsp40s specify functions of Hsp104 and Hsp90 protein chaperone machines. PLoS Genet 2014; 10:e1004720; PMID:25329162; http://dx.doi.org/10.1371/journal.pgen.1004720.
- Floer M, Bryant GO, Ptashne M. HSP90/70 chaperones are required for rapid nucleosome removal upon induction of the GAL genes of yeast. Proc Natl Acad Sci U S A 2008; 105:2975–80; PMID:18287040; http://dx.doi.org/10.1073/pnas.0800053105.
- Schwimmer C, Masison DC. Antagonistic interactions between yeast [PSI+] and [URE3] prions and curing of [URE3] by Hsp70 protein chaperone Ssa1p but not by Ssa2p. Mol Cell Biol 2002; 22:3590–8; PMID:11997496; http://dx.doi.org/10.1128/MCB.22.11.3590-3598.2002.
- Sharma D, Masison DC. Single methyl group determines prion propagation and protein degradation activities of yeast heat shock protein (Hsp)-70 chaperones Ssa1p and Ssa2p. Proc Natl Acad Sci U S A 2011; 108:13665–70; PMID:21808014; http://dx.doi.org/10.1073/pnas.1107421108.
- Brown CR, McCann JA, Chiang HL. The heat shock protein Ssa2p is required for import of fructose-1, 6-bisphosphatase into Vid vesicles. J Cell Biol 2000; 150:65–76; PMID:10893257; http://dx.doi.org/10.1083/jcb.150.1.65.
- Chernoff YO, Newnam GP, Kumar J, Allen K, Zink AD. Evidence for a protein mutator in yeast: role of the Hsp70-related chaperone ssb in formation, stability, and toxicity of the [PSI] prion. Mol Cell Biol 1999; 19:8103–12; PMID:10567536.
- Gautschi M, Mun A, Ross S, Rospert S. A functional chaperone triad on the yeast ribosome. Proc Natl Acad Sci U S A 2002; 99:4209–14; PMID:11929994; http://dx.doi.org/10.1073/pnas.062048599.
- Kaschner LA, Sharma R, Shrestha OK, Meyer AE, Craig EA. A conserved domain important for association of eukaryotic J-protein co-chaperones Jjj1 and Zuo1 with the ribosome. Biochim Biophys Acta 2015; 1853(5):1035–45; PMID:25639645.
- Kiktev DA, Melomed MM, Lu CD, Newnam GP, Chernoff YO. Feedback control of prion formation and propagation by the ribosome-associated chaperone complex. Mol Microbiol 2015; PMID:25649498.
- Song Y, Masison DC. Independent regulation of Hsp70 and Hsp90 chaperones by Hsp70/Hsp90-organizing protein Sti1 (Hop1). J Biol Chem 2005; 280:34178–85; PMID:16100115; http://dx.doi.org/10.1074/jbc.M505420200.
- Wegele H, Haslbeck M, Reinstein J, Buchner J. Sti1 is a novel activator of the Ssa proteins. J Biol Chem 2003; 278:25970–6; PMID:12716905; http://dx.doi.org/10.1074/jbc.M301548200.
- Hainzl O, Wegele H, Richter K, Buchner J. Cns1 is an activator of the Ssa1 ATPase activity. J Biol Chem 2004; 279:23267–73; PMID:15044454; http://dx.doi.org/10.1074/jbc.M402189200.
- Wegele H, Wandinger SK, Schmid AB, Reinstein J, Buchner J. Substrate transfer from the chaperone Hsp70 to Hsp90. J Mol Biol 2006; 356:802–11; PMID:16403523; http://dx.doi.org/10.1016/j.jmb.2005.12.008.
- Ganusova EE, Ozolins LN, Bhagat S, Newnam GP, Wegrzyn RD, Sherman MY, Chernoff YO. Modulation of prion formation, aggregation, and toxicity by the actin cytoskeleton in yeast. Mol Cell Biol 2006; 26:617–29; PMID:16382152; http://dx.doi.org/10.1128/MCB.26.2.617-629.2006.
- Allen KD, Chernova TA, Tennant EP, Wilkinson KD, Chernoff YO. Effects of ubiquitin system alterations on the formation and loss of a yeast prion. J Biol Chem 2007; 282:3004–13; PMID:17142456; http://dx.doi.org/10.1074/jbc.M609597200.
- Kryndushkin DS, Shewmaker F, Wickner RB. Curing of the [URE3] prion by Btn2p, a Batten disease-related protein. EMBO J 2008; 27:2725–35; PMID:18833194; http://dx.doi.org/10.1038/emboj.2008.198.
- Kiktev DA, Patterson JC, Muller S, Bariar B, Pan T, Chernoff YO. Regulation of chaperone effects on a yeast prion by cochaperone Sgt2. Mol Cell Biol 2012; 32:4960–70; PMID:23045389; http://dx.doi.org/10.1128/MCB.00875-12.
- Wolfe KJ, Ren HY, Trepte P, Cyr DM. The Hsp70/90 cochaperone, Sti1, suppresses proteotoxicity by regulating spatial quality control of amyloid-like proteins. Mol Biol Cell 2013; 24:3588–602; PMID:24109600; http://dx.doi.org/10.1091/mbc.E13-06-0315.
