Abstract
ABSTRACT. The molecular basis by which fungal and mammalian prions arise spontaneously is poorly understood. A number of different environmental stress conditions are known to increase the frequency of yeast [PSI+] prion formation in agreement with the idea that conditions which cause protein misfolding may promote the conversion of normally soluble proteins to their amyloid forms. A recent study from our laboratory has shown that the de novo formation of the [PSI+] prion is significantly increased in yeast mutants lacking key antioxidants suggesting that endogenous reactive oxygen species are sufficient to promote prion formation. Our findings strongly implicate oxidative damage of Sup35 as an important trigger for the formation of the heritable [PSI+] prion in yeast. This review discusses the mechanisms by which the direct oxidation of Sup35 might lead to structural transitions favoring conversion to the transmissible amyloid-like form. This is analogous to various environmental factors which have been proposed to trigger misfolding of the mammalian prion protein (PrPC) into the aggregated scrapie form (PrPSc).
De novo Formation of Yeast and Mammalian Prions
Most cases of prion diseases are sporadic, that is they form spontaneously without any underlying genetic change. The emergence of disease correlates with a novel conformational form (PrPSc) of the cellular PrPC protein.Citation1 This prion form replicates through a cycle of seeded polymerisation and fragmentation and it is assumed that genetic or environmental factors trigger the initial conformational change in the absence of any pre-existing PrPSc ‘seeds’. The mechanism(s) underlying this switch from a normally soluble protein to a misfolded, infectious form are poorly understood. It is important that these mechanism(s) are established if we are to develop effective preventative measures for human and animal amyloidoses.
The initial alternative conformational state may be instigated by spontaneous misfolding events, triggered by mutation, mistranslation, environmental stresses and/or by disruption of the chaperone network.Citation2,3 This is a relatively rare event and for example the de novo appearance of yeast [PSI+] prion formation has been estimated to occur at frequencies ranging between ∼10−5–10−7.Citation3–5 This frequency can be elevated 1000-fold by overexpression of Sup35, presumably due to the excess Sup35 increasing the possibility for prion seed formation.Citation6–8 Additionally, alterations in the protein homeostasis network including mutations in chaperones and the ubiquitin proteasome system can increase the spontaneous rate of [PSI+] prion formation.Citation9 This suggests that spontaneous prion formation might arise as a consequence of random protein misfolding events which are normally dealt with by the cellular protein homeostasis systems. Conditions which cause the formation of misfolded or partially folded proteins may therefore promote the conversion of soluble proteins to their amyloid forms and several different environmental stress conditions, including oxidative stress, have been shown to increase the frequency of [PSI+] prion formation.Citation10 Further support for oxidative stress conditions playing a role in de novo prion formation has come from the finding that [PSI+] prion formation is significantly increased in yeast mutants lacking the peroxiredoxins, Tsa1 and Tsa2.Citation11,12 Peroxiredoxins are major cellular antioxidants and we have recently extended this analysis to show that the frequency of [PSI+] prion formation is commonly elevated in a range of antioxidant mutants, as well as in response to oxidant exposure.Citation13 These findings strongly implicate oxidative damage of Sup35 as an important trigger for the formation of the heritable [PSI+] prion in yeast.
Oxidative Stress and [PSI+] Prion Formation
Oxidative stress is a ubiquitous stress which occurs during the course of normal aerobic metabolism or following exposure to radical-generating compounds.Citation14 Reactive oxygen species (ROS) cause wide-ranging damage to macromolecules and an oxidative stress is said to occur when ROS overwhelm the cellular antioxidant defense systems. Oxidative stress is frequently implicated in disease and pathological conditions, although it is not always easy to separate cause from effect. ROS are also a well-established trigger of protein misfolding and damage proteins via oxidative modification of several different amino acid residues.Citation15
Increasing evidence suggests that protein oxidative damage can trigger misfolding events resulting in prion formation. Oxidative stress induced by exposure to hydrogen peroxide was found to significantly increase [PSI+] prion induction in a screen which used a Sup35 variant with increased [PSI+] formation rates relative to wild-type Sup35.Citation10 This study found that exposure to higher; more lethal concentrations of hydrogen peroxide (10 mM), were required for maximal [PSI+] induction. We have found that growth in the presence of lower concentrations of hydrogen peroxide (100 μM), which only modestly affect wild-type yeast growth, induce de novo formation of [PSI+] from wild-type Sup35.Citation13 Stronger induction of [PSI+] prion formation is also observed in response to oxidative stress caused by the superoxide anion.Citation13 However, the superoxide anion can be dismutated to hydrogen peroxide by the activity of superoxide dismutases and both hydrogen peroxide and the superoxide anion are thought to oxidize amino acid residues through the generation of the highly reactive hydroxyl radical.Citation16 More work will be required to determine the sources and types of ROS which promote protein damage leading to prion formation, which is important since no single oxidant is typical of ‘oxidative stress’.Citation17
An unbiased genome-wide screen for factors which increase [PSI+] induction did not identify any antioxidants.Citation10 However, this screen differentiated [PSI+] formation from nuclear gene mutations by their irreversible elimination in guanidine hydrochloride (GdnHCl). Reversible curability is a well-established genetic criterion for yeast prionsCitation6 and is commonly tested using GdnHCl which blocks the propagation of prions by inhibiting the key ATPase activity of Hsp104, a molecular chaperone that is absolutely required for yeast prion propagation.Citation18,19 The gene deletions identified in this screen were all curable at levels above 95% suggesting that they did not cause any increase in nuclear mutations. All of the antioxidant mutants which we have shown increase the frequency of spontaneous [PSI+] formation also exhibit significantly increased rates of nuclear mutations.Citation13 ROS are a well-known cause of DNA damage resulting in increased rates of mutagenesis and genomic instability. For example, loss of TSA1 results in an increased rate of spontaneous mutations which arise as a result of endogenous ROS production.Citation20 However, there does not appear to be any correlation between the frequency of nuclear mutations and the frequency of [PSI+] prion formation in antioxidant mutants.Citation13
Methionine Oxidation Triggers [PSI+] Prion Formation
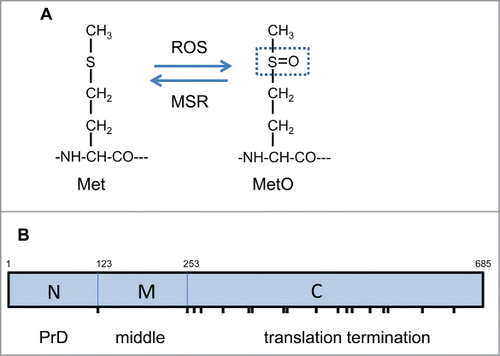
All amino acids are potential targets for oxidation, but methionine residues are particularly sensitive forming a racemic mixture of methionine-S-sulphoxide and methionine-R-sulphoxide in cells.Citation21 Oxidation converts the moderately hydrophobic thioester side chain of methionine into the hydrophilic sulphoxide form (MetO) which can significantly influence protein folding and structure (). Methionine oxidation can be reversed by the activity of methionine sulfoxide reductases (MSR) and hence methionine oxidation has been proposed to play an antioxidant role in scavenging ROS.Citation22 A number of proteins have been identified which form MetO in response to oxidative stress, including many examples where oxidation is thought to play a regulatory role modulating protein activity.Citation23 Thus, MetO formation is increasingly thought of as a reversible post translational modification, as well as a marker of protein oxidative damage.
Figure 1. Methionine oxidation and Sup35 functional domains. (A) Oxidation converts the moderately hydrophobic thioester side chain of methionine into the hydrophilic sulphoxide form (MetO). Methionine oxidation can be reversed by the activity of MSR enzymes which catalyze the thiol-dependent reduction of MetO. (B) The Sup35 protein can be divided into 3 distinct regions: an N-terminal prion forming domain (PrD), a highly charged middle region (M) and a C-terminal domain which functions in translation termination (C). The N-terminal domain lies between Met1 and Met123, the M-domain between Met123 and Met253 and the C-terminal domain from Met253 until the C-terminus of the protein. Sup35 contains a total of 19 Met residues (including its N-terminal Met residue) which are marked as vertical lines: the N and M regions contain no Met residues, the M region is flanked by Met residues and the C-terminal domain contains 16 Met residues.
We have used an anti-MetO antibody to detect methionine oxidation of Sup35. Oxidative stress conditions induced by exposure to hydrogen peroxide or the superoxide anion significantly increased MetO levels and increased the frequency of [PSI+] prion formation in a [PIN+][psi−] strain.Citation13 Similar increases in MetO levels were detected in antioxidant mutants, including mutants deficient in superoxide dismutases, catalases and peroxiredoxins, which all display increased [PSI+] prion formation. Increased MetO levels were detected in Sup35 from antioxidant mutants grown under normal non-stress conditions indicating that endogenous ROS levels are sufficient to promote Sup35 oxidation. Increased methionine oxidation is commonly detected in a number of disease states as well as during the aging process. For example, MetO antibodies have been used to detect increased methionine oxidation in aged mouse samples and samples from patients with Alzheimer's disease.Citation24 The MetO content of proteins is also known to increase with age in various model systems.Citation25 Thus, increased MetO content appears to be a common feature of oxidative stress conditions during various pathological conditions.
It is not know which methionine residues in Sup35 are important for [PSI+] prion formation. Sup35 has 3 functional regions: an N-terminal region (residues 1–123) which is essential for prion formation, a highly charged middle (M) region (residues 124–253) and a C-terminal region (residues 254–685) which is required for translation termination activity.Citation26 Sup35 contains a total of 19 Met residues including its N-terminal Met residue (). The N and M regions contain no Met residues, but the M region is flanked by Met residues. Although the C domain of Sup35 does not form the [PSI+] prion in the absence of the NM region, oxidation of Met residues in the C domain might be expected to affect the overall protein structure/folding of Sup35 which in turn might promote the formation of misfolded structures that have a higher propensity to switch to the prion conformation. Alternatively Met mutations may affect specific Sup35-chaperone interactions. For example, the M region interacts with Hsp104 and is required for prion curing but not propagation.Citation27 Several chaperones are known to modulate de novo [PSI+] prion formation, although the molecular details are not well defined.Citation9 Hence, methionine oxidation and any resulting alterations in Sup35 secondary structure which alter protein-protein interactions may affect prion formation.
How does Oxidative Protein Damage Promote [PSI+] Formation?
The elevated frequency of [PSI+] formation observed in response to Sup35 overexpression requires the presence of another prion, the best studied example of which is the [PIN+] prion form of Rnq1.Citation28 [PIN+] is still required for the elevated frequency of [PSI+] prion formation observed in tsa1 tsa2 mutants, indicating that oxidative stress does not overcome the requirement of cells being [PIN+] to form the [PSI+] prion.Citation12 [PIN+] prion aggregates are thought to provide imperfect templates which seed the polymerization of Sup35, facilitating the formation of its transmissible prion form.Citation29 Recent studies have provided a better understanding of the pathway that leads to prion formation. Misfolded Sup35 molecules are targeted to the IPOD (Insoluble Protein Deposit), a component of the protein quality control machinery in eukaryotic cells that is located adjacent to the vacuole. The IPOD is a site of accumulation of both prion proteins, as well as oxidatively damaged proteins.Citation30,31 This is believed to facilitate the nucleation of prion protein polymerization in a [PIN+]-dependent manner.Citation32 The transmissible form of the prion is then formed via a 2 stage process which initially involves the formation of non-transmissible extended polymers of the prion protein followed by their fragmentation into shorter transmissible forms that are thought to define the propagon.Citation30
Our data indicate that while methionine oxidation can promote prion formation, it can be reversed by the activity of MSR enzymes preventing the switch to the heritable prion form. Yeast contains 3 MSR enzymes (fRMsr/MsrA/MsrB).Citation33 Overexpression of MsrA protects Sup35 against methionine oxidation and also prevents [PSI+] prion formation, confirming that Sup35 oxidation can cause the switch from a soluble to an aggregated prion-form of Sup35 in antioxidant mutants.Citation12,13 Taking these observations together we propose a model for ROS-induced [PSI+] prion formation as shown in . Oxidation of Sup35 promotes misfolding and the resulting misfolded/damaged forms of Sup35 are targeted to the IPOD where [PIN+] aggregates promote de novo formation of the [PSI+] prion. Sup35 oxidation may cause misfolding by altering the conformation of its polypeptide backbone and decreasing thermal stability, or alternatively, it may disrupt normal Sup35-chaperone interactions. It is unknown whether Sup35 oxidation occurs on the nascent polypeptide chain or in pre-existing Sup35 molecules (). Interestingly however, Tsa1 and Tsa2 are both ribosome-associated antioxidants where they may localize to protect nascent proteins as well as the protein biosynthetic machinery against oxidation.Citation11,34
Figure 2. Model depicting the role of antioxidants in protecting Sup35 against protein oxidation and de novo [PSI+] prion formation. Oxidative stress occurs when ROS overwhelm the cellular antioxidant defense systems. Such stress can damage all macromolecules in cells including amino acid residues in proteins. Methionine residues are particularly sensitive forming MetO. It remains to be established whether methionine oxidation occurs on the nascent Sup35 polypeptide chain or in pre-existing Sup35 proteins. Sup35 is shown as green dots on translating ribosomes or free in cells. Antioxidants therefore provide the first line of defense against Sup35 oxidation and misfolding. When MetO formation does occur on Sup35, it causes misfolding and aggregation which may underlie the switch from a soluble to an aggregated form of Sup35. Methionine sulphoxide reductases (MSR) provide a second line of defense by converting MetO back to methionine. Once oxidized, Sup35 misfolds and the resulting aggregates can be targeted to the IPOD where [PIN+] aggregates cross-seed de novo formation of the [PSI+] prion. Soluble Sup35 is converted into the [PSI+] state which interacts with additional monomeric forms of Sup35. The growing amyloid fibrils are fragmented and propagated by the disaggregase activity of the Hsp104 chaperone. The resulting propagons are inherited in daughter cells via cytoplasmic transfer which ensures continued prion propagation.
![Figure 2. Model depicting the role of antioxidants in protecting Sup35 against protein oxidation and de novo [PSI+] prion formation. Oxidative stress occurs when ROS overwhelm the cellular antioxidant defense systems. Such stress can damage all macromolecules in cells including amino acid residues in proteins. Methionine residues are particularly sensitive forming MetO. It remains to be established whether methionine oxidation occurs on the nascent Sup35 polypeptide chain or in pre-existing Sup35 proteins. Sup35 is shown as green dots on translating ribosomes or free in cells. Antioxidants therefore provide the first line of defense against Sup35 oxidation and misfolding. When MetO formation does occur on Sup35, it causes misfolding and aggregation which may underlie the switch from a soluble to an aggregated form of Sup35. Methionine sulphoxide reductases (MSR) provide a second line of defense by converting MetO back to methionine. Once oxidized, Sup35 misfolds and the resulting aggregates can be targeted to the IPOD where [PIN+] aggregates cross-seed de novo formation of the [PSI+] prion. Soluble Sup35 is converted into the [PSI+] state which interacts with additional monomeric forms of Sup35. The growing amyloid fibrils are fragmented and propagated by the disaggregase activity of the Hsp104 chaperone. The resulting propagons are inherited in daughter cells via cytoplasmic transfer which ensures continued prion propagation.](/cms/asset/d59c3479-3f70-46ba-80af-a924c9e97bbd/kprn_a_1065372_f0002_oc.gif)
Under normal non-stress growth conditions, MetO formation is not detected in Sup35 suggesting that cellular antioxidants are sufficient to protect against endogenous ROS exposure and MSR enzymes function to reduce any MetO that might be formed. The addition of an exogenous oxidant, which can overwhelm the cellular antioxidant defense systems, results in Sup35 oxidation and misfolding, prior to targeting to the IPOD where it is converted to the heritable [PSI+] prion form. Methionine oxidation does not appear to account for the formation of all yeast prions since we were unable to detect methionine oxidation of Rnq1 in a tsa1 tsa2 mutant.Citation12 However, the frequency of [PIN+] formation is elevated in tsa1 tsa2 and sod1 mutants suggesting that prion formation may be a common phenomenon in antioxidant mutants.Citation12,13 The spontaneous frequency of [PIN+] prion formation is significantly higher than [PSI+] prion formationCitation12 and hence it may be difficult to detect methionine oxidation once it has formed the [PIN+] prion form. Alternatively, other forms of protein oxidation may underlie the switch from soluble Rnq1 to its amyloid form.
Conflicting Evidence Linking Methionine Oxidation with Mammalian Prion Formation
Conflicting data has been reported regarding the potential role of methionine oxidation in the aggregation and pathogenicity of mammalian prions. Mass spectrometry was originally used to detect MetO in PrPSc purified from brain samples; although it was unclear whether oxidation actually occurred in the brain or during the protein purification and analysis protocol.Citation35 Exposing purified PrPC to an oxidant has been shown to result in methionine oxidation,Citation36 although other in vitro studies suggested that methionine oxidation of recombinant hamster and mouse PrPC interferes with its conversion into amyloid fibrils.Citation37 Further in vivo studies showed that MetO (particularly Met213 in humans) could be detected in PrPSc isolated from brain samples, but not in PrPC, leading to the suggestion that MetO is a specific marker for PrPSc.Citation38 Conversely, sensitive mass spectrometry based comparison of Met213 oxidation in PrPC and PrPSC isolated from a hamster-adapted scrapie model found comparable low levels of methionine oxidation suggesting that MetO213 should not be regarded as a prion-specific covalent signature.Citation39 Assuming that methionine oxidation does occur on the PrP protein in vivo, it raise the question as to whether methionine oxidation underlies the structural switch that converts PrPC to PrPSc, or is simply a biomarker for PrPSC formation. Specificity for PrPSc might arise if methionine oxidation requires partial denaturation of PrPC before methionine residues can become oxidized, or else they may be oxidized only after PrPSC forms.
None of the existing in vivo studies are able address the importance of methionine oxidation in sporadic PrPSC formation. In vitro studies have attempted to examine the importance of methionine oxidation as a destabilizing event that triggers PrPC misfolding leading to spontaneous prion formation. For example, methionine analogs have been used to correlate the degree of PrPC methionine oxidation with the structural transitions that underlie its conversion to the infectious PrPSC form.Citation40 More recently, methionine oxidation within the core of PrPC has been shown to reduce its thermal stability, promoting generation of a molten globule fold which is thought to be a key step in the misfolding pathway.Citation41 Similarly, molecular dynamic simulations have suggested that oxidation of Met213 triggers destabilization of PrPC which can trigger its conversion to the pathogenic PrPSC form.Citation42,43 Pseudosulfoxidation mutants have been used to mimic methionine oxidation, confirming that oxidation of surface exposed methionine residues perturbs the PrPC structure resulting in destabilization and misfolding.Citation44 Interestingly, the D178N variant of PrPC was recently found to be more susceptible to methionine oxidation.Citation45 This PrP mutant has been linked with the inherited human prion disease fatal famililial insomnia (FFI) and methionine oxidation of D178N PrPC decreased its structural stability, enhanced its aggregation and increased neurotoxicity. This suggests that methionine oxidation can have synergistic effects with other pathogenic mutations in PrPC which cause prion diseases.
Further work will be required to establish a causal role for methionine oxidation in PrP misfolding and the formation of toxic oligomers and amyloid plaques. This may occur via PrPC destabilization or PrPSC stabilization such as inhibiting clearance of misfolded forms and favoring misfolding pathways which result in amyloid formation. This will however, be experimentally difficult without the tractability of the yeast model system.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
FUNDING
This work was funded by a Biotechnology and Biological Sciences Research Council grant (BB/J000183/1).
REFERENCES
- Prusiner SB. Biology and genetics of prions causing neurodegeneration. Annu Rev Genet 2013; 47:601-23; PMID:24274755; http://dx.doi.org/10.1146/annurev-genet-110711-155524
- DeMarco ML, Daggett V. Local environmental effects on the structure of the prion protein. C R Biol 2005; 328:847-62; PMID:16286076; http://dx.doi.org/10.1016/j.crvi.2005.05.001
- Allen KD, Chernova TA, Tennant EP, Wilkinson KD, Chernoff YO. Effects of ubiquitin system alterations on the formation and loss of a yeast prion. J Biol Chem 2007; 282:3004-13; PMID:17142456; http://dx.doi.org/10.1074/jbc.M609597200
- Lancaster AK, Bardill JP, True HL, Masel J. The spontaneous appearance rate of the yeast prion [PSI+] and its implications for the evolution of the evolvability properties of the [PSI+] system. Genetics 2010; 184:393-400; PMID:19917766; http://dx.doi.org/10.1534/genetics.109.110213
- Lund PM, Cox BS. Reversion analysis of [psi-] mutations in Saccharomyces cerevisiae. Genet Res 1981; 37:173-82; PMID:7021322; http://dx.doi.org/10.1017/S0016672300020140
- Wickner RB. [URE3] as an altered URE2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science 1994; 264:566-5699; PMID:7909170; http://dx.doi.org/10.1126/science.7909170
- Chernoff YO, Derkach IL, Inge-Vechtomov SG. Multicopy SUP35 gene induces de-novo appearance of psi-like factors in the yeast Saccharomyces cerevisiae. Curr Genet 1993; 24:268-70; PMID:8221937; http://dx.doi.org/10.1007/BF00351802
- Derkatch IL, Chernoff YO, Kushnirov VV, Inge-Vechtomov SG, Liebman SW. Genesis and variability of [PSI] prion factors in Saccharomyces cerevisiae. Genetics 1996; 144:1375-86; PMID:8978027
- Chernova TA, Wilkinson KD, Chernoff YO. Physiological and environmental control of yeast prions. FEMS Microbiol Rev 2014; 38:326-44; PMID:24236638; http://dx.doi.org/10.1111/1574-6976.12053
- Tyedmers J, Madariaga ML, Lindquist S. Prion switching in response to environmental stress. PLoS Biol 2008; 6:e294; PMID:19067491; http://dx.doi.org/10.1371/journal.pbio.0060294
- Sideri TC, Stojanovski K, Tuite MF, Grant CM. Ribosome-associated peroxiredoxins suppress oxidative stress-induced de novo formation of the [PSI+] prion in yeast. Proc Natl Acad Sci U S A 2010; 107:6394-9; PMID:20308573; http://dx.doi.org/10.1073/pnas.1000347107
- Sideri TC, Koloteva-Levine N, Tuite MF, Grant CM. Methionine oxidation of Sup35 protein induces formation of the [PSI+] prion in a yeast peroxiredoxin mutant. J Biol Chem 2011; 286:38924-31; PMID:21832086; http://dx.doi.org/10.1074/jbc.M111.272419
- Doronina VA, Staniforth GL, Speldewinde SH, Tuite MF, Grant CM. Oxidative stress conditions increase the frequency of de novo formation of the yeast [PSI(+) ] prion. Mol Microbiol 2015; 96:163-74; PMID:25601439; http://dx.doi.org/10.1111/mmi.12930
- Morano KA, Grant CM, Moye-Rowley WS. The Response to Heat Shock and Oxidative Stress in Saccharomyces cerevisiae. Genetics 2011; 190:1157-95; PMID:22209905; http://dx.doi.org/10.1534/genetics.111.128033
- Dalle-Donne I, Giustarini D, Colombo R, Rossi R, Milzani A. Protein carbonylation in human diseases. Trends Mol Med 2003; 9:169-76; PMID:12727143; http://dx.doi.org/10.1016/S1471-4914(03)00031-5
- Berlett BS, Stadtman ER. Protein oxidation in aging, disease, and oxidative stress. J Biol Chem 1997; 272:20313-6; PMID:9252331; http://dx.doi.org/10.1074/jbc.272.33.20313
- Temple MD, Perrone GG, Dawes IW. Complex cellular responses to reactive oxygen species. Trends Cell Biol 2005; 15:319-26; PMID:15953550; http://dx.doi.org/10.1016/j.tcb.2005.04.003
- Jung G, Masison DC. Guanidine hydrochloride inhibits Hsp104 activity in vivo: a possible explanation for its effect in curing yeast prions. Curr Microbiol 2001; 43:7-10; PMID:11375656; http://dx.doi.org/10.1007/s002840010251
- Ferreira PC, Ness F, Edwards SR, Cox BS, Tuite MF. The elimination of the yeast [PSI+] prion by guanidine hydrochloride is the result of Hsp104 inactivation. Mol Microbiol 2001; 40:1357-69; PMID:11442834; http://dx.doi.org/10.1046/j.1365-2958.2001.02478.x
- Iraqui I, Kienda G, Soeur J, Faye G, Baldacci G, Kolodner RD, Huang ME. Peroxiredoxin Tsa1 is the key peroxidase suppressing genome instability and protecting against cell death in Saccharomyces cerevisiae. PLoS Genet 2009; 5:e1000524; PMID:19543365; http://dx.doi.org/10.1371/journal.pgen.1000524
- Stadtman ER, Levine RL. Free radical-mediated oxidation of free amino acids and amino acid residues in proteins. Amino Acids 2003; 25:207-18; PMID:14661084; http://dx.doi.org/10.1007/s00726-003-0011-2
- Stadtman ER J. M, Levine RL. Oxidation of methionine residues of proteins: biological consequences. Antioxid Redox Signal 2003; 5:577-82; PMID:14580313; http://dx.doi.org/10.1089/152308603770310239
- Oien DB, Moskovitz J. Substrates of the methionine sulfoxide reductase system and their physiological relevance. Curr Top Dev Biol 2008; 80:93-133; PMID:17950373; http://dx.doi.org/10.1016/S0070-2153(07)80003-2
- Oien DB, Canello T, Gabizon R, Gasset M, Lundquist BL, Burns JM, Moskovitz J. Detection of oxidized methionine in selected proteins, cellular extracts and blood serums by novel anti-methionine sulfoxide antibodies. Arch Biochem Biophys 2009; 485:35-40; PMID:19388147; http://dx.doi.org/10.1016/j.abb.2009.01.020
- Stadtman ER, Van Remmen H, Richardson A, Wehr NB, Levine RL. Methionine oxidation and aging. Biochim Biophys Acta 2005; 1703:135-40; PMID:15680221; http://dx.doi.org/10.1016/j.bbapap.2004.08.010
- Tessier PM, Lindquist S. Unraveling infectious structures, strain variants and species barriers for the yeast prion [PSI+]. Nat Struct Mol Biol 2009; 16:598-605; PMID:19491937; http://dx.doi.org/10.1038/nsmb.1617
- Helsen CW, Glover JR. Insight into molecular basis of curing of [PSI+] prion by overexpression of 104-kDa heat shock protein (Hsp104). J Biol Chem 2012; 287:542-56; PMID:22081611; http://dx.doi.org/10.1074/jbc.M111.302869
- Derkatch IL, Bradley ME, Masse SV, Zadorsky SP, Polozkov GV, Inge-Vechtomov SG, Liebman SW. Dependence and independence of [PSI(+)] and [PIN(+)]: a two-prion system in yeast? EMBO J 2002; 19:1942-52; http://dx.doi.org/10.1093/emboj/19.9.1942
- Derkatch IL, Uptain SM, Outeiro TF, Krishnan R, Lindquist SL, Liebman SW. Effects of Q/N-rich, polyQ, and non-polyQ amyloids on the de novo formation of the [PSI+] prion in yeast and aggregation of Sup35 in vitro. Proc Natl Acad Sci U S A 2004; 101:12934-9; PMID:15326312; http://dx.doi.org/10.1073/pnas.0404968101
- Tyedmers J, Treusch S, Dong J, McCaffery JM, Bevis B, Lindquist S. Prion induction involves an ancient system for the sequestration of aggregated proteins and heritable changes in prion fragmentation. Proc Natl Acad Sci U S A 2010; 107:8633-8; PMID:20421488; http://dx.doi.org/10.1073/pnas.1003895107
- Kaganovich D, Kopito R, Frydman J. Misfolded proteins partition between two distinct quality control compartments. Nature 2008; 454:1088-95; PMID:18756251; http://dx.doi.org/10.1038/nature07195
- Arslan F, Hong JY, Kanneganti V, Park SK, Liebman SW. Heterologous aggregates promote de novo prion appearance via more than one mechanism. PLoS Genet 2015; 11:e1004814; PMID:25568955; http://dx.doi.org/10.1371/journal.pgen.1004814
- Le DT, Lee BC, Marino SM, Zhang Y, Fomenko DE, Kaya A E. H, Kwak GH, Koc A, Kim HY, et al. Functional analysis of free methionine-R-sulfoxide reductase from Saccharomyces cerevisiae. J Biol Chem 2009; 284:4354-64; PMID:19049972; http://dx.doi.org/10.1074/jbc.M805891200
- Trotter EW, Rand JD, Vickerstaff J, Grant CM. The yeast Tsa1 peroxiredoxin is a ribosome-associated antioxidant. Biochem J 2008; 412:73-80; PMID:18271751; http://dx.doi.org/10.1042/BJ20071634
- Stahl N, Baldwin MA, Teplow DB, Hood L, Gibson BW, Burlingame AL, Prusiner SB. Structural studies of the scrapie prion protein using mass spectrometry and amino acid sequencing. Biochemistry 1993; 32:1991-2002; PMID:8448158; http://dx.doi.org/10.1021/bi00059a016
- Requena JR, Dimitrova MN, Legname G, Teijeira S, Prusiner SB, Levine RL. Oxidation of methionine residues in the prion protein by hydrogen peroxide. Arch Biochem Biophys 2004; 432:188-95; PMID:15542057; http://dx.doi.org/10.1016/j.abb.2004.09.012
- Breydo L, Bocharova OV, Makarava N, Salnikov VV, Anderson M, Baskakov IV. Methionine oxidation interferes with conversion of the prion protein into the fibrillar proteinase K-resistant conformation. Biochemistry 2005; 44:15534-43; PMID:16300402; http://dx.doi.org/10.1021/bi051369+
- Canello T, Engelstein R, Moshel O, Xanthopoulos K, Juanes ME, Langeveld J, Sklaviadis T, Gasset M, Gabizon R. Methionine sulfoxides on PrPSc: a prion-specific covalent signature. Biochemistry 2008; 47:8866-73; PMID:18680312; http://dx.doi.org/10.1021/bi800801f
- Silva CJ, Onisko BC, Dynin I, Erickson ML, Vensel WH, Requena JR, Antaki EM, Carter JM. Assessing the role of oxidized methionine at position 213 in the formation of prions in hamsters. Biochemistry 2010; 49:1854-61; PMID:20121218; http://dx.doi.org/10.1021/bi901850n
- Wolschner C, Giese A, Kretzschmar HA, Huber R, Moroder L, Budisa N. Design of anti- and pro-aggregation variants to assess the effects of methionine oxidation in human prion protein. Proc Natl Acad Sci U S A 2009; 106:7756-61; PMID:19416900; http://dx.doi.org/10.1073/pnas.0902688106
- Younan ND, Nadal RC, Davies P, Brown DR, Viles JH. Methionine oxidation perturbs the structural core of the prion protein and suggests a generic misfolding pathway. J Biol Chem 2012; 287:28263-75; PMID:22654104; http://dx.doi.org/10.1074/jbc.M112.354779
- Colombo G, Meli M, Morra G, Gabizon R, Gasset M. Methionine sulfoxides on prion protein Helix-3 switch on the alpha-fold destabilization required for conversion. PLoS One 2009; 4:e4296; PMID:19172188; http://dx.doi.org/10.1371/journal.pone.0004296
- Lisa S, Meli M, Cabello G, Gabizon R, Colombo G, Gasset M. The structural intolerance of the PrP alpha-fold for polar substitution of the helix-3 methionines. Cell Mol Life Sci 2010; 67:2825-38; PMID:20454997; http://dx.doi.org/10.1007/s00018-010-0363-1
- Elmallah MI, Borgmeyer U, Betzel C, Redecke L. Impact of methionine oxidation as an initial event on the pathway of human prion protein conversion. Prion 2013; 7:404-11; PMID:24121542; http://dx.doi.org/10.4161/pri.26745
- Feng B, Wang Z, Liu T, Jin R, Wang S, Wang W, Xiao G, Zhou Z. Methionine oxidation accelerates the aggregation and enhances the neurotoxicity of the D178N variant of the human prion protein. Biochim Biophys Acta 2014; 1842:2345-56; PMID:25281825; http://dx.doi.org/10.1016/j.bbadis.2014.09.012
