ABSTRACT
Disease-related prion protein (PrPSc), which is a structural isoform of the host-encoded cellular prion protein, is thought to be a causative agent of transmissible spongiform encephalopathies. However, the specific role of PrPSc in prion pathogenesis and its relationship to infectivity remain controversial. A time-course study of prion-affected mice was conducted, which showed that the prion infectivity was not simply proportional to the amount of PrPSc in the brain. Centrifugation (20,000 ×g) of the brain homogenate showed that most of the PrPSc was precipitated into the pellet, and the supernatant contained only a slight amount of PrPSc. Interestingly, mice inoculated with the obtained supernatant showed incubation periods that were approximately 15 d longer than those of mice inoculated with the crude homogenate even though both inocula contained almost the same infectivity. Our results suggest that a small population of fine PrPSc may be responsible for prion infectivity and that large, aggregated PrPSc may contribute to determining prion disease duration.
ABBREVIATIONS
| PrPSc | = | disease-related prion protein |
| PrPC | = | cellular prion protein |
| TSE | = | transmissible spongiform encephalopathy |
| mBSE | = | mouse-adapted bovine spongiform encephalopathy |
| PK | = | proteinase K |
| LD50 | = | 50% lethal dose |
| I.U | = | infectious unit |
INTRODUCTION
Transmissible spongiform encephalopathies (TSEs), or prion diseases, are fatal neurodegenerative disorders, including scrapie in sheep and goats, bovine spongiform encephalopathy (BSE) in cattle, chronic wasting disease in cervids, and Creutzfeldt-Jakob disease in humans. Although the entity of the infectious agent, referred to as the prion, is still obscure, disease-related prion protein (PrPSc) is thought to be a main component of the causative agent of TSEs.Citation1,2 PrPSc is generated by post-transcriptional modification of host cellular prion protein (PrPC) and was originally defined by its proteinase K (PK) resistance and detergent insolubility.Citation3-5 PK resistance is a widely used property for discriminating between PrPSc and PrPC, and this characteristic is critical for the diagnosis of prion diseases. Many studies have focused on the relationship between prion infectivity and PrPSc, but the results have been conflicting. Some studies have demonstrated a reasonably close relationship between the amount of PrPSc and prion infectivity,Citation6-8 whereas others have demonstrated that tissues or cells harboring high prion infectivity showed little or no detectable PrPSc.Citation9-11 PrPSc is currently believed to consist of multiple misfolded prion proteins, including protease-sensitive isoforms.Citation12-17 PrPSc also displays various aggregate sizesCitation18,19 with the most infectious PrPSc particles reported to be small aggregates approximately 17–27 nm in diameter.Citation20 More recently, 2 distinct PrPSc populations were reported: one was associated with prion infectivity, and the other was associated with its neurotoxicity.Citation21 Thus, the role of PrPSc in prion pathogenesis remains controversial.
In this study, the dynamics of PrPSc and prion infectivity in mouse-adapted BSE (mBSE) were examined by immunohistochemistry, western blot analysis, enzyme-linked immunosorbent assay (ELISA), and endpoint titration assay to reveal the role of PrPSc populations during prion disease progression.
RESULTS
No Involvement of PK-Sensitive PrPSc in mBSE Prion Infectivity
To investigate the possibility that PK-sensitive PrPSc contributes to prion infectivity, the mBSE-brain homogenate was first digested with 5, 40, and 100 μg/ml of PK and then the PrPSc was detected by western blot. The samples were diluted to 1:100 with phosphate-buffered saline (PBS) and subjected to a bioassay to estimate their infectivity. As shown in , the N-terminal truncated PK-resistant core of PrPSc (PrP 27–30) was clearly detected, and no intact PrPSc signal was detected from the samples digested with 5 and 40 μg/ml of PK. The incubation period of the mice inoculated with these samples was approximately 183 days, which was similar to that of mice inoculated with the undigested brain homogenate (). In the sample digested with 100 μg/ml of PK, faint PrPSc signals were detectable and the incubation period of the inoculated mice was extended to 254 ± 3.3 d (). These results indicated that the prion infectivity of mBSE is not associated with PK-sensitive PrPSc.
FIGURE 1. Comparison of PrPSc in brains digested with various PK concentrations. PrPSc was detected with anti-PrP mAb T2. The PK concentrations are shown on the top of each lane. The % PrPSc values are indicated at the bottom of each lane. Molecular marker sizes are indicated on the left.
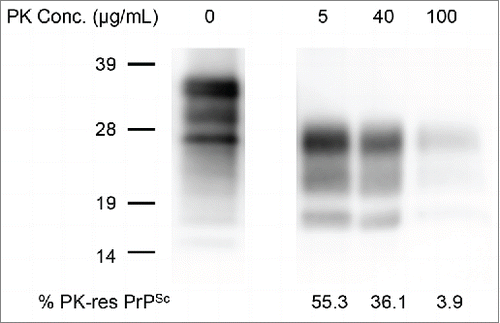
Table 1. Comparison of survival days between mice inoculated with crude homogenate and the PK-digested mBSE homogenates
PrPSc Accumulation in The Brain
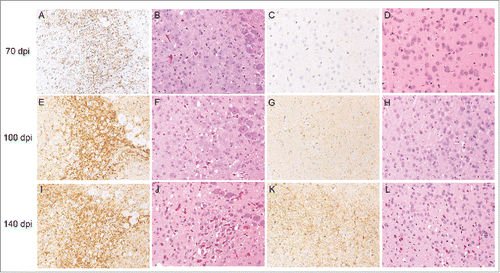
The accumulation of PrPSc was confirmed by immunohistochemistry in the vestibular nuclei at 40 d post-inoculation (dpi) in all of the mice examined (data not shown). There were clear PrPSc deposits observed in the dorsal medulla at 70 dpi, but subtle or no spongiform changes were observed in the brain at this time point (). The number of PrPSc deposits increased, and moderate spongiform changes were observed in the dorsal medulla at 100 dpi (). In addition, weak PrPSc immunoreactivities were detected in the cerebral cortex, but there were few spongiform changes observed in the same region at 100 dpi (). A large number of PrPSc deposits and severe vacuolation in whole brain regions were observed at 140 dpi (). These results indicate that neuropathological changes occurred after the accumulation of PrPSc in the brain.
FIGURE 2. Pathological changes in the brain occurring during mBSE disease progression. Groups of mice were sacrificed at the defined time points indicated on the left side of the panels. Fixed brain sections from each culling time were subjected to hematoxylin and eosin staining (B, D, F, H, J, and L) together with immunohistochemistry for PrPSc (A, C, E, G, I, and K). PrPSc immunoreactivities were detected with the anti-PrP monoclonal antibody SAF84. Micrographs show the dorsal medulla (A, B, E, F, I, and J) and cerebral cortex (C, D, G, H, K, and L).
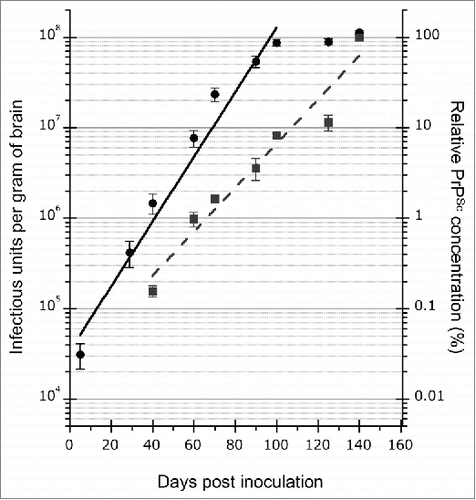
In the western blot, PrPSc was also detected at 40 dpi in all of the examined mice, and the amount of PrPSc increased in accordance with the disease time course (). ELISA was used for quantification of the dynamics of PrPSc. This assay revealed that PrPSc was amplified exponentially until the end of the disease with a doubling time of 12.4 d (, blue dotted line), supporting the results of the western blot. The prion infectivity exponentially increased with a doubling time of 8.4 d (, black solid line) until 100 dpi. The doubling time of infectivity was faster than that of PrPSc at this disease stage. However, the prion infectivity reached a plateau at 100 dpi and was stable until the end of the disease (). These results indicate a discrepancy between the replication rate of prion infectivity and PrPSc accumulation. Therefore, to further examine the prion infectivity in small PrPSc aggregates, the inoculum was centrifuged to remove the large aggregated PrPSc.
FIGURE 3. Detection of PrPSc in the brains of mice by western blot. PrPSc was detected with anti-PrP mAb T2. The defined time points (days post-inoculation, dpi) are indicated on the top of the blot. Mo and Nl indicate mouse scrapie ObihiroCitation39 and the uninfected mouse brain, respectively. The relative protein load in each lane is indicated on the bottom; 1x indicates 0.25 mg brain equivalent. Molecular markers are indicated on the left side of the blot.
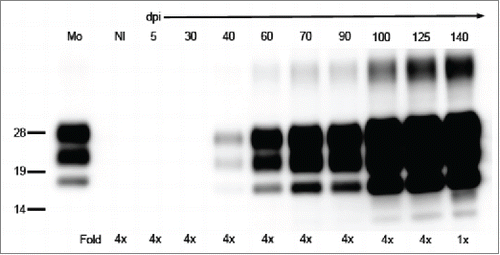
FIGURE 4. Kinetics of mBSE prion replication and PrPSc accumulation in the mouse brain during disease progression. Black circles and blue squares show the infectious units and PrPSc accumulation in the mouse brains. The infectious dose of mBSE prion at each time point was estimated by the incubation period bioassay method. ELISA was used for measuring the amount of PrPSc at the same time points, and the relative values (%) are indicated against the amount of PrPSc in the brain at the terminal stage. The kinetics of mBSE prion replication and PrPSc accumulation are shown as a semi-logarithmic plot. An exponential correlation was observed between the incubation time and mBSE prion infectivity from 5 to 100 dpi (y = 33557* e^(0.082279x) R2 = 0.94593, where y is the infectious unit and x is the survival days). A similar correlation was observed between the incubation time and PrPSc accumulation from 40 to 140 dpi (y = 0.024371* e^(0.056019x) R2 = 0.90149, where y is the relative value (%) compared to the PrPSc values of 140-dpi brains and x is the survival days).
Prion Titers and Incubation Periods of Brain Homogenates After Centrifugation
The brain homogenates of mBSE-affected mice were centrifuged at 20,000 ×g for 10 min at 4°C, and the supernatant (20K-sup) was subjected to western blot and an end-point bioassay. Most of the PrPSc was precipitated into the pellet, and the 20K-sup contained only 0.4–2.3% of PrPSc (). On the other hand, the endpoint titration assays revealed that the infectivity of the crude homogenate and 20K-sup was 108.6 and 108.4 LD50, respectively (). In spite of the decline of PrPSc due to centrifugation, similar infectivity was restored in the 20K-sup. Interestingly, the incubation period of the 20K-sup was extended by approximately 15 d compared to that of the crude homogenate, even though both samples showed the same level of prion infectivity (). These results indicated that a small population of PrPSc may contribute to the infectivity, whereas large, aggregated PrPSc may play a role in accelerating the disease progression.
FIGURE 5. Comparison of PrPSc and prion infectivity between the crude homogenate and 20K-sup. (A) Comparison of PrPSc amounts between the crude homogenate and 20K-sup. The relative protein load in each lane is indicated on the top. Nl indicates the uninfected mouse brain. PrP was detected with anti-PrP mAb SAF84; 1x indicates 0.25 mg brain equivalent. Molecular markers are indicated on the left side of the blot. (B) The relationship between incubation time (mean days ± standard error) and infectious units (log LD50/gram of the brain) in mice inoculated with serial dilutions of the crude homogenate (black circles and solid line; y = 16.642 – 0.061312x, where y is log LD50 and x is the incubation period) and the 20K-sup (blue squares and dotted line y = 17.474 – 0.060916x).
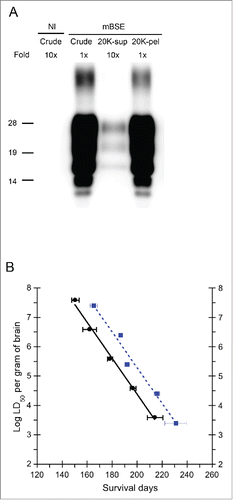
Table 2. Comparison of mBSE prion infectious units between the crude homogenate and 20K-sup by an endpoint titration assay
DISCUSSION
There are several reports suggesting that PK-sensitive PrPSc (PK-sen PrPSc) is responsible for prion infectivity.Citation12,16,Citation17,22 However, in the present study mBSE prion infectivity was not altered following digestion with 40 µg/mL of PK (). Even with 5 μg/ml of PK digestion, the PrPSc signal completely shifted to that of PrP27–30, which could indicate elimination of PK-sen PrPSc. Although our results do not rule out the existence of PK-sen PrPSc in mBSE, they do suggest that it has little or no contribution to mBSE prion infectivity. The involvement of PK-sensitive PrPSc in prion infectivity may differ among prion strains.
We examined the PrPSc dynamics in the mouse brain by immunohistochemistry, western blot, and ELISA, using basic criteria such as deposition or PK resistance.Citation23 PrPSc amplified continuously until the terminal stage with a doubling time of 12.4 d. On the other hand, prion infectivity replicated with a doubling time of 8.4 d. These results demonstrated that prion infectivity is not simply proportional to the number of PrPSc molecules. The formation of a quaternary structure of PrPSc molecules is important for prion infectivity. Prion infectivity and toxicity could be associated with multiple aggregate states of PrPSc molecules.Citation19,24,Citation25 One study reported that the most infectious PrPSc particles consisted of 14–28 PrP molecules.Citation20 In our study, we quantified the total amount of monomeric PrPSc, which was denatured with sodium dodecyl sulfide boiling or guanidine treatment, by western blot and ELISA. Based on the protein-only hypothesis, this might have caused the observed difference in the doubling time between prion infectivity and PrPSc accumulation.
Previous experiments have found that the infectivity reached a plateau even when PrPSc continuously increased until the end of illness.Citation21,26-28 In contrast, other studies reported the continuous increase of both PrPSc and infectivity to the end of diseases.Citation6-8 Our results demonstrated that the mBSE prion infectivity reached a plateau at 100 dpi, which supports the idea that prion infectivity is independent of the total amount of PrPSc. We also examined the PrPSc accumulation and spongiform changes in situ. As shown in , few spongiform changes were observed at 100 dpi. However, the number of spongiform changes increased in the whole brain area after 100 dpi, and this period matched with the plateau infectivity stage. We propose that a portion of de novo synthesis of fine PrPSc in brain cells may be associated with prion infectivity. In addition, polymerized and/or elongated PrPSc aggregation in neuropils at the late stage may be associated with neurotoxicity rather than prion infectivity. Acceleration of the production of toxic, large PrPSc aggregates may be responsible for the plateau of prion infectivity. Indeed, it has been reported that 2 distinct types of PrPSc isoforms may be involved in prion infectivity and neurotoxicity, respectively.Citation28 On the other hand, it is possible that the size of the infectious PrPSc aggregates may differ among prion strains.Citation19 Variations in host species, prion strains, and the route of inoculation may also affect the amplification patterns of total PrPSc and prion infectivity in the brain.
These results highlight the importance of determining the size of aggregated PrPSc to clarify prion pathogenesis. Therefore, the inoculum was simply centrifuged at 20,000 ×g for 10 min at 4°C. With this process, most of the PrPSc was precipitated into the pellet (97.7–99.6%), and a small population of PrPSc remained in the supernatant (0.4–2.3%). We consider that the large PrPSc aggregates and relatively fine PrPSc aggregates could be separated with this simple centrifugation process. The end-point titration assay revealed that the 20K-sup and original homogenate harbored similar infectivity (108.4 vs. 108.6 LD50). Several studies have supported the idea that infectious prions are comprised of a minority of PrPSc,Citation9,10,Citation28 and our data support this notion. Of particular note, the mice inoculated with the 20K-sup showed a delayed incubation period of approximately 15 d as compared to mice inoculated with the original homogenate with a similar titer (). The same phenomenon was observed in a previous study using an inoculum prepared from the spleens of prion-affected mice, which showed a longer incubation time than that prepared from brain samples containing the same infectious titer.Citation29 No large PrPSc aggregates were observed in the spleen by immunohistochemistry, suggesting that the inoculum prepared from the spleen consisted of relatively fine PrPSc aggregates, similar to the 20K-sup. Although the precise mechanism is obscure, our study suggests that large PrPSc aggregates may act as a progression factor of prion disease. In addition, it has been reported that the detergent treatment of scrapie materials can lengthen the incubation period without reducing prion infectivity.Citation30,31 Moreover, PMCA-generated PrPSc, with an infectious dose comparable to that of the infected brain, has been reported to extend the disease incubation period.Citation32 Although we need to consider the involvement of cofactors, such as RNA moleculesCitation33 and lipids,Citation34 in the infectivity and toxicity of PMCA-generated PrPSc, these previous reports and our results suggest that the change of PrPSc aggregate states has a big impact on prion pathogenicity, including infectivity and disease duration. Analyses of the associations of infectivity with the molecular size of PrPSc aggregates are required to clarify the relationship between PrPSc and its infectivity and toxicity. Furthermore, attention should be paid to the optimal sample conditions for comparing infectivity using an incubation time assay. Two distinct populations of PrPSc isoforms were recently reported to be involved in prion infectivity and neurotoxicity, respectively.Citation28 Our results also imply the existence of 2 types of PrPSc: one that is responsible for prion infectivity and the other that is associated with disease duration. However, it remains to be determined whether the 2 PrPSc populations identified in this study are identical to those reported previously.
In conclusion, we demonstrated that the 20K-sup contained highly infectious units in spite of only a small amount of PrPSc. The incubation period of the 20K-sup was delayed by 15 d compared to that of the crude brain homogenate with the same prion titer. Further study is required to clarify the different types of PrPSc and their role in prion pathogenesis.
MATERIALS AND METHODS
Animals and Prions
Mouse-adapted classical BSE (mBSE) prions, which had been passaged in ICR mice (Japan SLC, Hamamatsu, Japan) more than 10 times, were prepared from infected brains homogenized in PBS and intracerebrally inoculated into 3-week-old specific pathogen-free female ICR mice. The mice were examined daily and their survival periods were determined as the time from inoculation to the clinical endpoint or death. At appropriate times, the brain was collected and the left hemispheres were immediately stored at −80°C for the bioassay and biochemical analysis. The other hemispheres were fixed with 10% neutral-buffered formalin (pH 7.4) containing 10% methanol for histopathology.
All animal experiments were approved by the Animal Ethical Committee and the Animal Care and Use Committee of the National Institute of Animal Health (authorization 11–008 and 11–012). All experiments were conducted in biosafety level 3 facilities with strict adherence to safety protocols.
Prion Infectivity Assay
To determine the prion infectivity, an endpoint titration assay was conducted. The mouse brain homogenate was serially diluted with PBS, and then each dilution was intracerebrally inoculated into 5 or 6 ICR mice. Infectious units (LD50 per gram of the brain) were determined according to Behrens-Karber's formula.Citation35 In some experiments, the brain homogenate was solubilized with 1% N-lauroylsarcosine (Sarkosyl, Sigma-Aldrich, Japan) and 1% n-dodecyl-β-d-maltoside (Dojindo Laboratories, Kumamoto, Japan) for 15 min at room temperature (R/T) and then digested with PK (Roche Diagnosis Japan, Tokyo, Japan) under the following conditions: 5 or 40 µg/ml at 37°C for 30 min, or 100 µg/ml at 37°C overnight. The samples were subjected to western blot for the detection of PrPSc as described below. The aliquots of these samples were also diluted to 1:100 in PBS and then subjected to the incubation time assayCitation36 to compare the infectivity.
Time-Course Analysis of mBSE-Affected Mice
Twenty microliters of mBSE brain homogenates, which contained 105.9 LD50 of mBSE, was intracerebrally inoculated into ICR mice. Four to 6 recipient mice were sacrificed at designated time points (dpi), and the brains were collected and examined.
ELISA
The ELISA method using the seprion ligand system (Microsens Biotechnologies, London, UK) was performed for the relative quantification of PrPSc according to previously described protocols.Citation37,38 ELISA plates were coated with a solution containing the seprion ligand (Microsens Biotechnologies, London, UK, 150 μl/well) at R/T for 1 h and then blocked with 5% bovine serum albumin (BSA, 200 μl/well; Nacalai Tesque, Kyoto, Japan) at R/T for 30 min. Aliquots of the brain homogenate were lysed in 100 μl of lysis buffer (50 mM Tris-HCl [pH 8.3], 1% Sarkosyl, 1% Triton X-100, 1% BSA, 0.5 mg/ml trypsin) and then transferred to the seprion ligand-coated plates. After incubation at R/T for 2 h to allow PrPSc isoforms to be captured on the seprion ligand, the plates were incubated with denaturation buffer (4 M guanidine hydrochloride, 20% polyethylene glycol 8000) at R/T for 10 min and then further incubated with horseradish peroxidase (HRP)-conjugated anti-PrP mAb T2Citation38 at 4°C for 2 h. Development was performed with a chemiluminescent substrate (Super Signal ELISA, Thermo Fisher Scientific, Rockland, IL) at R/T. The chemiluminescent signals were measured with a microplate reader (ARVO SV; PerkinElmer, Boston, MA). Two-fold serial dilutions of the mBSE-brain homogenate were subjected to ELISA to construct a standard curve for semi-quantitative analysis of PrPSc (Fig. S1).
Western Blot
Following PK digestion, the samples were mixed with an equal volume of a 2-butanol:methanol mixture (5:1), and PrPSc was precipitated by 20,000 ×g centrifugation. The pellets were resuspended in Laemmli sample buffer and boiled for 5 min. Samples were electrophoresed on NuPAGE Novex 12% Bis-Tris gels and NuPAGE MOPS-SDS running buffer in accordance with the manufacturer's instructions (Life Techonologies, Carlsbad, CA, USA). The proteins were transferred onto an Immobilon-P membrane (Millipore, Billerica, MA, USA). The blotted membrane was incubated with anti-PrP monoclonal antibodies T2, 6H4 (Prionics, Zurich, Switzerland), and SAF84 (Bertin Pharma, Montigny le Bretonneux, France) at 4°C overnight. After washing with PBS containing Tween 20 twice, the membrane was incubated with HRP-conjugated anti-mouse IgG (Jackson ImmunoResearch, West Grove, PA, USA) for 60 min at R/T. Signals were developed with a chemiluminescent substrate (SuperSignal; Thermo Fisher Scientific). For semi-quantitation, blots were imaged using a Fluorchem system (Alpha Innotech, San Leandro, CA, USA) and analyzed using image reader software (AlphaEaseFC; Alpha Innotech) according to the manufacturer's instructions.
Histopathology and Immunohistochemistry
Formalin-fixed left hemispheres were immersed in 98% formic acid to reduce infectivity and then embedded in paraffin wax. Serial sections were stained with hematoxylin and eosin for evaluation of neuropathological changes. After epitope retrieval, PrPSc immunocytochemistry was performed using the monoclonal antibody SAF84. Immunoreactions were developed using the anti-mouse universal immunoperoxidase polymer (Nichirei Histofine Simple Stain MAX-PO (M); Nichirei, Tokyo, Japan) as the secondary antibody and were visualized with 3,3′-diaminobenzedine tetrachloride as the chromogen.
DISCLOSURE OF POTENTIAL CONFLICTS OF INTEREST
The authors indicated no potential conflicts of interest.
SUPPLEMENTAL MATERIAL
Supplemental data for this article can be accessed on the publisher's website.
Supplemental_Material.zip
Download Zip (103.8 KB)ACKNOWLEDGMENTS
We are grateful to Naoko Tabeta, Ritsuko Miwa, Naomi Furuya, Junko Yamada, and the animal laboratory staff of NIAH for their technical and animal care assistance.
Funding
This work was supported by a Grant-in-Aid from the BSE and other Prion Disease Control Project of the Ministry of Agriculture, Forestry, and Fisheries; in part by Grants-in-Aid from the Ministry of Health, Labor, and Welfare of Japan; and in part by a Grant-in-Aid for Young Scientists (Category B) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
REFERENCES
- Prusiner SB. Prions. Proc Natl Acad Sci U S A 1998; 95:13363-83; PMID:9811807; http://dx.doi.org/10.1073/pnas.95.23.13363.
- Safar JG, Kellings K, Serban A, Groth D, Cleaver JE, Prusiner SB, Riesner D. Search for a prion-specific nucleic acid. J Virol 2005; 79:10796-806; PMID:16051871; http://dx.doi.org/10.1128/JVI.79.16.10796-10806.2005.
- Bolton DC, McKinley MP, Prusiner SB. Identification of a protein that purifies with the scrapie prion. Science 1982; 218:1309-11; PMID:6815801; http://dx.doi.org/10.1126/science.6815801.
- Meyer RK, McKinley MP, Bowman KA, Braunfeld MB, Barry RA, Prusiner SB. Separation and properties of cellular and scrapie prion proteins. Proc Natl Acad Sci U S A 1986; 83:2310-4; PMID:3085093; http://dx.doi.org/10.1073/pnas.83.8.2310.
- Pan KM, Baldwin M, Nguyen J, Gasset M, Serban A, Groth D, Mehlhorn I, Huang Z, Fletterick RJ, Cohen FE, et al. Conversion of α-helices into β-sheets features in the formation of the scrapie prion proteins. Proc Natl Acad Sci U S A 1993; 90:10962-6; PMID:7902575; http://dx.doi.org/10.1073/pnas.90.23.10962.
- Beekes M, Baldauf E, Diringer H. Sequential appearance and accumulation of pathognomonic markers in the central nervous system of hamsters orally infected with scrapie. J Gen Virol 1996; 77 ( Pt 8):1925-34; PMID:8760444; http://dx.doi.org/10.1099/0022-1317-77-8-1925.
- Jendroska K, Heinzel FP, Torchia M, Stowring L, Kretzschmar HA, Kon A, Stern A, Prusiner SB, DeArmond SJ. Proteinase-resistant prion protein accumulation in Syrian hamster brain correlates with regional pathology and scrapie infectivity. Neurology 1991; 41:1482-90; PMID:1679911; http://dx.doi.org/10.1212/WNL.41.9.1482.
- Bolton DC, Rudelli RD, Currie JR, Bendheim PE. Copurification of Sp33-37 and scrapie agent from hamster brain prior to detectable histopathology and clinical disease. J Gen Virol 1991; 72 ( Pt 12):2905-13; PMID:1684986; http://dx.doi.org/10.1099/0022-1317-72-12-2905.
- Barron RM, Campbell SL, King D, Bellon A, Chapman KE, Williamson RA, Manson JC. High titers of transmissible spongiform encephalopathy infectivity associated with extremely low levels of PrPSc in vivo. J Biol Chem 2007; 282:35878-86; PMID:17923484; http://dx.doi.org/10.1074/jbc.M704329200.
- Lasmezas CI, Deslys JP, Robain O, Jaegly A, Beringue V, Peyrin JM, Fournier JG, Hauw JJ, Rossier J, Dormont D. Transmission of the BSE agent to mice in the absence of detectable abnormal prion protein. Science 1997; 275:402-5; PMID:8994041; http://dx.doi.org/10.1126/science.275.5298.402.
- Miyazawa K, Emmerling K, Manuelidis L. High CJD infectivity remains after prion protein is destroyed. J Cell Biochem 2011; 112:3630-7; PMID:21793041; http://dx.doi.org/10.1002/jcb.23286.
- D'Castro L, Wenborn A, Gros N, Joiner S, Cronier S, Collinge J, Wadsworth JD. Isolation of proteinase K-sensitive prions using pronase E and phosphotungstic acid. PloS One 2010; 5:e15679; PMID:21187933; http://dx.doi.org/10.1371/journal.pone.0015679.
- Sajnani G, Silva CJ, Ramos A, Pastrana MA, Onisko BC, Erickson ML, Antaki EM, Dynin I, Vázquez-Fernández E, Sigurdson CJ, et al. PK-sensitive PrP is infectious and shares basic structural features with PK-resistant PrP. PLoS Pathog 2012; 8:e1002547; PMID:22396643; http://dx.doi.org/10.1371/journal.ppat.1002547.
- Owen JP, Rees HC, Maddison BC, Terry LA, Thorne L, Jackman R, Whitelam GC, Gough KC. Molecular profiling of ovine prion diseases by using thermolysin-resistant PrPSc and endogenous C2 PrP fragments. J Virol 2007; 81:10532-9; PMID:17652380; http://dx.doi.org/10.1128/JVI.00640-07.
- Tremblay P, Ball HL, Kaneko K, Groth D, Hegde RS, Cohen FE, DeArmond SJ, Prusiner SB, Safar JG. Mutant PrPSc conformers induced by a synthetic peptide and several prion strains. J Virol 2004; 78:2088-99; PMID:14747574; http://dx.doi.org/10.1128/JVI.78.4.2088-2099.2004.
- Tzaban S, Friedlander G, Schonberger O, Horonchik L, Yedidia Y, Shaked G, Gabizon R, Taraboulos A. Protease-sensitive scrapie prion protein in aggregates of heterogeneous sizes. Biochemistry 2002; 41:12868-75; PMID:12379130; http://dx.doi.org/10.1021/bi025958g.
- Krasemann S, Neumann M, Szalay B, Stocking C, Glatzel M. Protease-sensitive prion species in neoplastic spleens of prion-infected mice with uncoupling of PrP(Sc) and prion infectivity. J Gen Virol 2013; 94:453-63; PMID:23136363; http://dx.doi.org/10.1099/vir.0.045922-0.
- Kasai K, Iwamaru Y, Masujin K, Imamura M, Mohri S, Yokoyama T. Heterogeneity of the Abnormal Prion Protein (PrPSc) of the Chandler Scrapie Strain. Pathogens 2013; 2:92- 104; PMID:25436883; http://dx.doi.org/10.3390/pathogens2010092.
- Tixador P, Herzog L, Reine F, Jaumain E, Chapuis J, Le Dur A, Laude H, Béringue V. The physical relationship between infectivity and prion protein aggregates is strain-dependent. PLoS Pathog 2010; 6:e1000859; PMID:20419156; http://dx.doi.org/10.1371/journal.ppat.1000859.
- Silveira JR, Raymond GJ, Hughson AG, Race RE, Sim VL, Hayes SF, Caughey B. The most infectious prion protein particles. Nature 2005; 437:257-61; PMID:16148934; http://dx.doi.org/10.1038/nature03989.
- Sandberg MK, Al-Doujaily H, Sharps B, Clarke AR, Collinge J. Prion propagation and toxicity in vivo occur in two distinct mechanistic phases. Nature 2011; 470:540-2; PMID:21350487; http://dx.doi.org/10.1038/nature09768.
- Sajnani G, Requena JR. Prions, proteinase K and infectivity. Prion 2012; 6:430-2; PMID:23044510; http://dx.doi.org/10.4161/pri.22309.
- Gavier-Widen D, Stack MJ, Baron T, Balachandran A, Simmons M. Diagnosis of transmissible spongiform encephalopathies in animals: a review. J Vet Diagn Invest 2005; 17:509-27; PMID:16475509; http://dx.doi.org/10.1177/104063870501700601.
- Simoneau S, Rezaei H, Sales N, Kaiser-Schulz G, Lefebvre-Roque M, Vidal C, Fournier JG, Comte J, Wopfner F, Grosclaude J, et al. In vitro and in vivo neurotoxicity of prion protein oligomers. PLoS Pathog 2007; 3:e125; PMID:17784787; http://dx.doi.org/10.1371/journal.ppat.0030125.
- Caughey B, Lansbury PT. Protofibrils, pores, fibrils, and neurodegeneration: separating the responsible protein aggregates from the innocent bystanders. Annu Rev Neurosci 2003; 26:267-98; PMID:12704221; http://dx.doi.org/10.1146/annurev.neuro.26.010302.081142.
- Czub M, Braig HR, Diringer H. Pathogenesis of scrapie: study of the temporal development of clinical symptoms, of infectivity titres and scrapie-associated fibrils in brains of hamsters infected intraperitoneally. J Gen Virol 1986; 67 ( Pt 9):2005-9; PMID:2875123; http://dx.doi.org/10.1099/0022-1317-67-9-2005.
- Manuelidis L, Fritch W. Infectivity and host responses in Creutzfeldt-Jakob disease. Virology 1996; 216:46-59; PMID:8615006; http://dx.doi.org/10.1006/viro.1996.0033.
- Sandberg MK, Al-Doujaily H, Sharps B, De Oliveira MW, Schmidt C, Richard-Londt A, Lyall S, Linehan JM, Brandner S, Wadsworth JD, et al. Prion neuropathology follows the accumulation of alternate prion protein isoforms after infective titre has peaked. Nat Commun 2014; 5:4347; PMID:25005024; http://dx.doi.org/10.1038/ncomms5347.
- Robinson MM, Cheevers WP, Burger D, Gorham JR. Organ-specific modification of the dose-response relationship of scrapie infectivity. J Infect Dis 1990; 161:783-6; PMID:2138656; http://dx.doi.org/10.1093/infdis/161.4.783.
- Lax AJ, Millson GC, Manning EJ. Can scrapie titres be calculated accurately from incubation periods? J Gen Virol 1983; 64 (Pt 4):971-3; PMID:6403663; http://dx.doi.org/10.1099/0022-1317-64-4-971.
- Somerville RA, Carp RI. Altered scrapie infectivity estimates by titration and incubation period in the presence of detergents. J Gen Virol 1983; 64 (Pt 9):2045-50; PMID:6411862; http://dx.doi.org/10.1099/0022-1317-64-9-2045.
- Shikiya RA, Bartz JC. In vitro generation of high-titer prions. J Virol 2011; 85:13439-42; PMID:21957291; http://dx.doi.org/10.1128/JVI.06134-11.
- Deleault NR, Lucassen RW, Supattapone S. RNA molecules stimulate prion protein conversion. Nature 2003; 425:717-20; PMID:14562104; http://dx.doi.org/10.1038/nature01979.
- Deleault NR, Piro JR, Walsh DJ, Wang F, Ma J, Geoghegan JC, Supattapone S. Isolation of phosphatidylethanolamine as a solitary cofactor for prion formation in the absence of nucleic acids. Proc Natl Acad Sci U S A 2012; 109:8546-51; PMID:22586108; http://dx.doi.org/10.1073/pnas.1204498109.
- Gilles HJ. [Calculation of the index of acute toxicity by the method of linear regression. Comparison with the method of "Karber and Behrens"]. Eur J Toxicol Environ Hyg 1974; 7:77-84; PMID:4408718.
- Prusiner SB, Cochran SP, Downey DE, Groth DF. Determination of scrapie agent titer from incubation period measurements in hamsters. Adv Exp Med Biol 1981; 134:385-99; PMID:7194570; http://dx.doi.org/10.1007/978-1-4757-0495-2_35.
- Lane AC, Stanley S, Dealler S, Wilson SM. Polymeric ligands with specificity for aggregated prion proteins. Clin Chem 2003; 49:1774-75; http://dx.doi.org/10.1373/49.10.1774.
- Shimizu Y, Kaku-Ushiki Y, Iwamaru Y, Muramoto T, Kitamoto T, Yokoyama T, Mohri S, Tagawa Y. A novel anti-prion protein monoclonal antibody and its single-chain fragment variable derivative with ability to inhibit abnormal prion protein accumulation in cultured cells. Microbiol Immunol 2010; 54:112-21; PMID:20377745; http://dx.doi.org/10.1111/j.1348-0421.2009.00190.x.
- Shinagawa M, Matsuda A, Sato G, Takeuchi M, Ichijo S, Ono T. Occurrence of ovine scrapie in Japan: clinical and histological findings in mice inoculated with brain homogenates of an affected sheep. Nihon Juigaku Zasshi 1984; 46:913-6; PMID:6441054; http://dx.doi.org/10.1292/jvms1939.46.913.
